Key Points
In vitro CRISPR screens can be used to identify potential biomarkers for adoptive T-cell therapies.
CD64 and components of the SAGA complex were identified and functionally validated as important regulators of DNT-AML interactions.
Abstract
Acute myeloid leukemia (AML) remains a devastating disease in need of new therapies to improve patient survival. Targeted adoptive T-cell therapies have achieved impressive clinical outcomes in some B-cell leukemias and lymphomas but not in AML. Double-negative T cells (DNTs) effectively kill blast cells from the majority of AML patients and are now being tested in clinical trials. However, AML blasts obtained from ∼30% of patients show resistance to DNT-mediated cytotoxicity; the markers or mechanisms underlying this resistance have not been elucidated. Here, we used a targeted clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) screen to identify genes that cause susceptibility of AML cells to DNT therapy. Inactivation of the Spt-Ada-Gcn5-acetyltransferase (SAGA) deubiquitinating complex components sensitized AML cells to DNT-mediated cytotoxicity. In contrast, CD64 inactivation resulted in resistance to DNT-mediated cytotoxicity. Importantly, the level of CD64 expression correlated strongly with the sensitivity of AML cells to DNT treatment. Furthermore, the ectopic expression of CD64 overcame AML resistance to DNTs in vitro and in vivo. Altogether, our data demonstrate the utility of CRISPR/Cas9 screens to uncover mechanisms underlying the sensitivity to DNT therapy and suggest CD64 as a predictive marker for response in AML patients.
Introduction
Acute myeloid leukemia (AML) is the most common form of adult acute leukemia with increasing incidence and poor overall long-term survival as the result of a high rate of disease relapse after standard chemotherapy.1-4 The advent of next-generation sequencing (NGS) has allowed for greater understanding of the genetic landscape present in AML.5,6 Based on genetic changes and gene/protein expression, it is now possible to more accurately predict a patient’s prognosis in the context of conventional treatments.7,8 Despite these improvements, there have been few changes in treatment strategy and little improvement in outcome for many years.9-11 As there is a shift to individualization of therapy, it is necessary to understand the ways in which AML gene expression can influence the success of emerging therapies.
Double-negative T cells (DNTs) are peripheral mature T cells that express CD3, but not CD4, CD8, or invariant natural killer T-cell markers. We have demonstrated that human DNTs expanded ex vivo from AML patients or healthy donors are able to selectively target AML cells in vitro12-15 and reduce leukemia load in patient-derived xenograft models without observable toxicities.13-15 Furthermore, we have demonstrated that allogeneic DNTs have the potential to be used as an “off-the-shelf” cellular therapy, because they do not induce graft-versus-host or host-versus-graft responses in preclinical studies.14 As such, first-in-human phase 1 clinical trials using allogeneic DNTs expanded from healthy donors to treat patients with high-risk AML have been initiated (NCT03027102 and ChiCTR-IPR-1900022795).
However, although AML cells from the majority of patients were killed effectively, ∼30% of AML samples were resistant to DNT-mediated cytotoxicity.15 Hence, biomarkers that identify AML patients who are more likely to respond to DNT therapy can increase its therapeutic efficacy by guiding treatment to the appropriate population. With recent advances in genetic engineering, advanced screening methods can provide vital information that might identify new biomarkers for patient stratification, ultimately improving the therapeutic benefits of DNTs.
Clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) is an RNA-guided platform that can efficiently knock out genes one at a time or in a high throughput screen to identify positive and negative regulators of a phenotype.16,17 Here, we used a pooled single-guide RNA (sgRNA) library and the CRISPR/Cas9 system to identify and functionally validate CD64 and components of the Spt-Ada-Gcn5-acetyltransferase (SAGA) complex as important regulators of DNT-AML interactions.
Methods
Ex vivo expansion of human DNTs
Peripheral blood was obtained from healthy volunteers, and DNTs were enriched by depleting CD4+ and CD8+ cells with RosetteSep Depletion Kits (STEMCELL Technologies). The enriched DNTs were expanded ex vivo as described previously.12
Cytotoxicity assays
sgRNA pooled library design and synthesis
The CRISPR screen used in our pooled epidrug library consisted of ∼12 500 sgRNAs targeting 317 epigenetic regulators, 657 US Food and Drug Administration (FDA)-approved drug targets based on Drugbank v4.3,18 and control genes, with an average of 10 sgRNAs per gene (supplemental Table 1, available on the Blood Web site). The sgRNAs were designed using the CRISPR-DO tool that accounted for sgRNA specificity and cutting efficiency.19 sgRNAs were synthesized as 73-mer oligonucleotides (CustomArray), GAAAGGACGAAACACCGNNNNNNNNNNNNNNNNNNNNGTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGC (Ns denote sgRNA 20-nucleotide target sequence; sense orientation) with a total of 12 472 sequences and amplified by polymerase chain reaction (PCR) as a pool using the following primers: TAACTTGAAAGTATTTCGATTTCTTGGCTTTATATATCTT GTGGAAAGGACGAAACACCG (forward) and ACTTTTTCAAGTTGATAACG GACTAGCCTTATTTTAACTTGCTATTTCTAGCTCTAAAAC (reverse). The PCR product was purified and cloned in the lentiGuide-Puro vector (gift from Feng Zhang; Addgene Plasmid #52963) using BsmBI (New England Biolabs). Ligation was performed using a NEBuilder HiFi DNA Assembly Cloning Kit and plasmids were transformed into an electrocompetent strain (Stbl4; Thermo Fisher Scientific) to achieve ∼300× coverage. Colonies were scraped off agar plates using LB medium. Plasmid DNA was extracted using an NA0310 Sigma GenElute HP Plasmid Maxiprep Kit, and adequate library representation of each sgRNA was confirmed by NGS.
CRISPR pooled screening, sequencing, and analysis
Stable Cas9-expressing cell lines (OCI-AML2 [AML2] and OCI-AML3 [AML3]) were generated using the pCDH-EF1-Cas9(NLS)-T2A-copGFP plasmid (Cellecta), which was kindly provided by Steven Chan’s laboratory (University Health Network). In brief, Cas9-copGFP lentiviral particles were generated in HEK293FT cells (Invitrogen) using the pMDG.2 and psPAX2 packaging plasmids (Addgene #12259 and #12260; gifts from Didier Trono). Cell lines were transduced for 24 to 48 hours, and GFP+ cells were sorted by fluorescence-activated cell sorting (FACS) to generate a purified Cas9-copGFP population. Similarly, library viruses were produced in HEK293FT cells and multiplicity of infection (MOI) was determined for each cell line screened, as previously described.20 The pRSI9-RFP plasmid (Cellecta) expressing sgRNAs against copGFP (sgGFP) or expressing sgRNAs against a control (sgControl) was used to assess Cas9 activity in stably expressing AML cell lines (sgRNA sequences are listed in Table 1).
List of sgRNA/shRNA target sequences for validation of CRISPR screen hits
| Target name . | sgRNA/shRNA target sequences . |
|---|---|
| sgGFP | GAGATCGAGTGCCGCATCAC |
| sgControl | GCCCGTTTCGTCATTCCCAC |
| sgAAVS | ATTCCCAGGGCCGGTTAATG |
| sgATXN7L3-1 | ACAGCACACAGATGAGCAG |
| sgATXN7L3-2 | TTGTTGGAACCTAGGCCGG |
| sgENY2-1 | GTGGCTGAAATCACTCCAAA |
| sgENY2-2 | ACACGTTACTGTTGATGACT |
| sgUSP22-1 | AGACATGGAAATAATCGCCA |
| sgUSP22-2 | AAGGCGAAGCGGCACAACCT |
| sgFCGR1A-1 | GACATCCCCACTCCTGGAG |
| sgFCGR1A-2 | TGGGAGCAGCTCTACACAG |
| shControl | CGAGGGCGACTTAACCTTAGG |
| shUSP22-1 | CCTACCTGCTGTAAGATTATG |
| shUSP22-2 | AGCTACCAGGAGTCCACAAAG |
| shENY2-1 | CCAGCCTTTAAGATTGAATTA |
| shENY2-2 | GCACACTGTAAAGAGGTAATT |
| shATXN7L3-1 | AGGCGAACCGTACGGATTTAT |
| shATXN7L3-2 | GCAGGCAGAGGATCCGATTAT |
| Target name . | sgRNA/shRNA target sequences . |
|---|---|
| sgGFP | GAGATCGAGTGCCGCATCAC |
| sgControl | GCCCGTTTCGTCATTCCCAC |
| sgAAVS | ATTCCCAGGGCCGGTTAATG |
| sgATXN7L3-1 | ACAGCACACAGATGAGCAG |
| sgATXN7L3-2 | TTGTTGGAACCTAGGCCGG |
| sgENY2-1 | GTGGCTGAAATCACTCCAAA |
| sgENY2-2 | ACACGTTACTGTTGATGACT |
| sgUSP22-1 | AGACATGGAAATAATCGCCA |
| sgUSP22-2 | AAGGCGAAGCGGCACAACCT |
| sgFCGR1A-1 | GACATCCCCACTCCTGGAG |
| sgFCGR1A-2 | TGGGAGCAGCTCTACACAG |
| shControl | CGAGGGCGACTTAACCTTAGG |
| shUSP22-1 | CCTACCTGCTGTAAGATTATG |
| shUSP22-2 | AGCTACCAGGAGTCCACAAAG |
| shENY2-1 | CCAGCCTTTAAGATTGAATTA |
| shENY2-2 | GCACACTGTAAAGAGGTAATT |
| shATXN7L3-1 | AGGCGAACCGTACGGATTTAT |
| shATXN7L3-2 | GCAGGCAGAGGATCCGATTAT |
Cas9-expressing AML cell lines were infected with the library at an MOI ∼ 0.3 and coverage of 300×. At 24 to 48 hours postinfection, cells were selected with puromycin for 72 hours (1 or 3 μg/mL – AML2 and AML3) and then cultured for ∼7 days, maintaining 300× coverage prior to our screening assay.
For the live-vs-dead screening assay, the sgRNA library–transduced Cas9+ AML3 cells were incubated with DNTs at a 1:1 E:T ratio to induce apoptosis in 40% to 50% of the targets. DNTs were depleted after a 2-hour coculture by anti-CD3–coated magnetic beads (EasySep Human CD3 Positive Selection Kit; STEMCELL Technologies), and Annexin V+ (live) and Annexin V− (dead or dying) AML cells were separated by FACS. For the DNT-vs-no DNT screening assay, ∼100 million of the transduced AML2 or AML3 cells were cultured alone or with DNTs at a 0.5:1 E:T ratio for 18 hours to induce apoptosis in ∼80% of AML cells. Subsequently, DNTs were removed by magnetic separation, and then necrotic cells and cell debris were removed by density gradient centrifugation. Genomic DNA was extracted and sgRNA inserts were amplified by PCR, as previously described.21
The input amount of genomic DNA was calculated to achieve 250× coverage of the library, and resulting libraries were sequenced on an Illumina HiSeq 2500 System. The screens were performed in duplicates for the live-vs-dead screens and in triplicates for the DNT-vs-no DNT screens.
The NGS data from CRISPR screens were first aligned to the library sgRNAs using bowtie (version 1.2.2).22 The read count for each sgRNA was computed using a custom python script. The resulting count matrix was the input to the tool MAGeCK,23 which estimates the enrichment/depletion of individual sgRNAs using a negative binomial model and estimates the enrichment of genes in live or dead cells using a robust rank aggregation model. Normalized read counts for all screens are listed in supplemental Table 2. The correlation analysis of sgRNA count between replicates was determined and visualized using custom R scripts. The results were plotted using ggplot224 in R (version 3.2.2). To perform gene set enrichment among the genes enriched in dead cells, Gene Ontology (GO) gene sets (version 6.2) were obtained from the MSigDB (http://software.broadinstitute.org/gsea/downloads.jsp). Library genes were used as background for enrichment analysis. The gene set enrichment analysis and permutation-based P value of the enrichment were estimated using the R package clusterProfiler25 in R (version 3.2.2) by comparing genes dropped out (false-discovery rate [FDR] ≤0.1) in dead cells with all genes in the library.
Xenograft model
NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice (The Jackson Laboratory) were maintained at the University Health Network animal facility. On day 0, 6- to 8-week-old female NSG mice were irradiated (225 cGy) and then injected IV with 1 × 106 KG1a cells. On days 1, 4, and 7, mice were treated with 20 × 106 DNTs. Human recombinant interleukin-2 (Proleukin; 104 IU per mouse) was given to all mice IV on days 1, 4, and 7 and intraperitoneally weekly from day 14 until euthanization, which occurred when bone marrow engraftment reached ∼70%.
Statistical analyses
Statistical analyses for in vitro and in vivo assays were performed using GraphPad Prism version 6 (GraphPad Software). Data were expressed as means ± standard deviation. Two-tailed, unpaired Student t tests and 1-way analysis of variance with the Dunnett test for multiple comparisons were performed, where appropriate, to identify significant differences between groups in our experiments.
Data and software availability
Cell line RNA-Seq and CRISPR screen data were deposited in the Gene Expression Omnibus (GEO) under the accession number GSE157618 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE157618).
Additional methods for RNA sequencing, western blot, flow cytometry, and CRISPR validation, as well as details about study approval, are included in supplemental Methods.
Results
Targeted CRISPR screen coupled with cell sorting identifies essential genes for DNT therapy in AML
To understand how AML cells are being targeted and killed by DNTs, we used a CRISPR/Cas9-based screen to knock out a library of genes in AML cells before subjecting them to DNT-mediated cytotoxicity. AML3 and AML2 cells were transduced with Cas9-GFP lentiviral particles and sorted by FACS to obtain a GFP+ population (supplemental Figure 1A-B). To assess the activity of Cas9, we transduced both AML Cas9-GFP lines with a viral vector containing an RFP gene along with sgGFP or sgControl. As expected, we observed a significant loss of GFP expression in RFP+ cells transduced with sgGFP compared with sgControl (AML3: 21.3% vs 86.4% GFP+ and AML2: 9.44% vs 68.3% GFP+; supplemental Figure 1C-D), confirming the activity of Cas9 in both AML cell lines.
Mutations in epigenetic modifiers have been documented in AML patients,26 and there has been an emergence of epigenetic therapeutics in immuno-oncology.27 However, the effect of epigenetic modifications on AML susceptibility to immune cell–mediated killing has not been studied. Therefore, we designed a focused sgRNA library (epidrug) against 317 epigenetic regulators (10 sgRNAs per gene) and also included 657 genes that are targets of FDA-approved drugs (Figure 1A). To identify genes involved in sensitizing or developing resistance to DNTs, Cas9-expressing AML3 cells were transduced with the epidrug sgRNA library and cocultured with DNTs for 2 hours, which induced apoptosis in 40% to 50% of the AML cells (Figure 1B). FACS was used to separate DNT-targeted (Annexin V+/dead-dying cells) and untargeted (Annexin V−/live cells) AML populations after coculture. NGS was then performed on genomic DNA isolated from the DNT-targeted and untargeted AML cells to identify sgRNAs that were enriched/depleted in each cell population. A strong correlation was observed between biological replicates across dead/dying and live AML cells (supplemental Figure 2A-C).
CRISPR-Cas9–mediated DNT cytotoxicity screen reveals gene targets for DNT therapy in AML. (A) Pie chart illustrates the composition of the epidrug sgRNA library that contains ∼1000 genes. “Other” refers to m6A-related genes (eg, METTL3, METTL14, ALKBH5, YTHDF1). (B) Experimental design of the in vitro CRISPR screen with AML3 cells. An epigenetic drug library with ∼12 500 sgRNAs was cloned into lentiviral constructs and Cas9-expressing AML3 cells were transduced and selected with the library at an MOI ∼ 0.3. Ten days postinfection, transduced AML3 cells were cocultured with DNTs for 2 hours, which resulted in 40% to 50% Annexin V staining of target cells. Annexin V+ and Annexin V− AML cells were subsequently sorted using FACS. Two independent experiments were performed, and samples were collected for PCR amplification followed by NGS. The data were analyzed by MAGeCK.
CRISPR-Cas9–mediated DNT cytotoxicity screen reveals gene targets for DNT therapy in AML. (A) Pie chart illustrates the composition of the epidrug sgRNA library that contains ∼1000 genes. “Other” refers to m6A-related genes (eg, METTL3, METTL14, ALKBH5, YTHDF1). (B) Experimental design of the in vitro CRISPR screen with AML3 cells. An epigenetic drug library with ∼12 500 sgRNAs was cloned into lentiviral constructs and Cas9-expressing AML3 cells were transduced and selected with the library at an MOI ∼ 0.3. Ten days postinfection, transduced AML3 cells were cocultured with DNTs for 2 hours, which resulted in 40% to 50% Annexin V staining of target cells. Annexin V+ and Annexin V− AML cells were subsequently sorted using FACS. Two independent experiments were performed, and samples were collected for PCR amplification followed by NGS. The data were analyzed by MAGeCK.
SAGA deubiquitinating complex confers resistance to DNTs in AML
sgRNAs against genes conferring resistance to DNT-mediated cytotoxicity would be enriched in the Annexin V+/dead cell population. Among the most significantly enriched sgRNAs in this dead/dying cell population (17 genes; FDR ≤0.1), we identified 3 closely related genes, ATXN7L3 (ranked #2), ENY2 (ranked #11), and USP22 (ranked #16), which are all components of the deubiquitinating module (DUBm) of the SAGA complex28 (Figure 2A; supplemental Table 3). The involvement of SAGA-related pathways was further supported by GO pathway analyses performed using genes significantly enriched in dead AML3 cells (Figure 2B). Importantly, negative-control sgRNAs (eg, LacZ, Luciferase) were not significantly enriched in live or dead populations, which confirmed the quality of our screening approach (supplemental Figure 2D-E).
Members of the SAGA complex regulate susceptibility of AML cells to DNT-mediated cytotoxicity. (A) Volcano plot of all genes enriched in dead (Annexin V+) AML3 cells after DNT treatment. X-axis represents the log2-transformed fold changes in normalized sgRNA tag counts in dead vs live cells. Red circles denote genes that are significantly enriched (FDR ≤0.10) in dead cells; genes associated with the SAGA complex are labeled. (B) The top 6 GO gene sets represented by the 17 enriched genes in AML3 dead cells (left panel). Enriched genes in these gene sets are shown (right panel). (C) Western blot analysis of AML2 Cas9-expressing cells transduced with pairs of sgRNAs against ATXN7L3, ENY2, USP22, or AAVS (a targeting genome-wide control). (D-E) Percentage of specific killing of SAGA-deficient AML3 (D) and AML2 (E) Cas9-expressing cells following coculture with DNTs at various E:T ratios for 2 hours. These experiments were conducted independently 3 times, with similar results. (F) Immunoblot analysis of H2B ubiquitination (H2Bubq) levels in ATXN7L3, ENY2, and USP22 in AML2 and AML3-Cas9 cells. Data are the mean ± standard deviation of replicates. *P < .05, **P < .01, ***P < .001, ****P < .0001. n.s., nonsignificant.
Members of the SAGA complex regulate susceptibility of AML cells to DNT-mediated cytotoxicity. (A) Volcano plot of all genes enriched in dead (Annexin V+) AML3 cells after DNT treatment. X-axis represents the log2-transformed fold changes in normalized sgRNA tag counts in dead vs live cells. Red circles denote genes that are significantly enriched (FDR ≤0.10) in dead cells; genes associated with the SAGA complex are labeled. (B) The top 6 GO gene sets represented by the 17 enriched genes in AML3 dead cells (left panel). Enriched genes in these gene sets are shown (right panel). (C) Western blot analysis of AML2 Cas9-expressing cells transduced with pairs of sgRNAs against ATXN7L3, ENY2, USP22, or AAVS (a targeting genome-wide control). (D-E) Percentage of specific killing of SAGA-deficient AML3 (D) and AML2 (E) Cas9-expressing cells following coculture with DNTs at various E:T ratios for 2 hours. These experiments were conducted independently 3 times, with similar results. (F) Immunoblot analysis of H2B ubiquitination (H2Bubq) levels in ATXN7L3, ENY2, and USP22 in AML2 and AML3-Cas9 cells. Data are the mean ± standard deviation of replicates. *P < .05, **P < .01, ***P < .001, ****P < .0001. n.s., nonsignificant.
To delineate AML-specific vs DNT-AML–specific essential gene targets, we compared the sgRNA libraries in AML cells on the day of transduction (day 0; baseline) vs 7 days posttransduction, without DNT coincubation (supplemental Figure 3A; supplemental Table 4). Fourteen of the 17 DNT-AML hits were identified as AML essential genes (supplemental Figure 3B), and ATXN7L3, ENY2, and USP22 were ranked at the bottom of the 144 essential gene list (supplemental Figure 3C).
To further determine the role of SAGA DUBm components in conferring AML resistance to DNTs, we generated AML2 and AML3 cells devoid of ATXN7L3, ENY2, and USP22 using 2 independent sgRNAs along with a genome targeting control (sgAAVS). Immunoblot analysis demonstrated efficient reduction of ATXN7L3 and USP22 in both cell lines and ENY2 in AML2 cells (Figure 2C; supplemental Figure 4A). Although the level of ENY2 expression was inconclusive in AML3 cells (supplemental Figure 4A), TIDE analysis confirmed the on-target activity of both ENY2 sgRNAs (supplemental Figure 4B). A modest and comparable degree of spontaneous cell death in the absence of DNT was observed across ENY2, ATXN7L3, and USP22–deficient AML cells (3-6%) and sgAAVS control cells (4%) (supplemental Figure 5). To account for such basal level spontaneous apoptosis changes, the percentage of specific killing was normalized by the number of Annexin V− cells in non-DNT–treated conditions (see Methods). In AML3 cells, depletion of ATXN7L3, ENY2, and USP22 significantly increased their susceptibility to DNT-mediated cytotoxicity at varying DNT/AML ratios compared with the AAVS control (Figure 2D). A similar trend was observed for ATXN7L3 and ENY2 in AML2 cells (Figure 2E).
Because the DUBm of the SAGA complex is known to respond to double-stranded breaks that occur during CRISPR/Cas9 cleavage of target proteins,29 we used orthogonal approaches to validate our hits. By depleting ENY2, USP22, and ATXN7L3 using short hairpin RNA (shRNA; supplemental Figure 6A-C) in AML2 cells, we observed increased susceptibility to DNT-mediated cytotoxicity at varying DNT/AML ratios compared with a nontargeting shControl (supplemental Figure 6D-F). A similar trend was observed in AML3 cells depleted of ENY2 and USP22 (supplemental Figure 6G-I), providing supporting evidence that the SAGA complex could be a general regulator of AML-DNT interactions.
The SAGA complex exerts multiple functions, including acetylation and deubiquitination, to modify chromatin and, hence, gene expression.30 Specifically, DUBm is the functional subunit of SAGA that catalyzes the reaction to cleave monoubiquitin from histone H2A and H2B (supplemental Figure 7). Importantly, increased global levels of H2B ubiquitination were observed in AML2 cells and AML3 cells deficient for all 3 SAGA DUBm components (Figure 2F), suggesting that the SAGA complex may enhance the resistance of AML cells to DNT-mediated cytotoxicity through the deubiquitination of histones across the genome.
To further investigate the broader immunological effect of DUBm subunits, genes differentially expressed on Cas9+ AML3 cells transduced with sgRNAs against DUBm subunits (USP22, ENY2, or ATXN7L3) vs an AAVS control were identified through RNA sequencing analysis (supplemental Table 5). Following standard RNA sequencing quality control analysis, we observed comparable mapped reads and count distribution (supplemental Figure 8A-C) with differential genes showing good consistency in fold changes and overlap among the 3 knockout samples (supplemental Figure 8D-I). GO analysis of genes upregulated (P < .05 and fold increase >1.5) in AML3 cells upon genetic inhibition of DUBm subunits identified 2 immune response–related pathways among the top 10 most significant terms, including “defense response to other organism” and “defense response to bacterium” (supplemental Figure 8J). Genes involved in immune cell recognition and T-cell costimulation, such as HLA-DRA and CD40, were among the upregulated genes. In contrast, none of the pathways identified from GO analysis of the downregulated genes in DUBm knockouts was directly immune related (supplemental Figure 8K). This suggests that the DUBm complex renders AML cells immune quiescent by suppressing immune response–related genes rather than upregulating inhibitory receptors.
CD64 sensitizes DNT-mediated cytotoxicity in AML cells
Having uncovered genes that confer AML resistance to DNTs, we next examined targets that promote DNT-mediated cell death. sgRNAs against genes involved in sensitizing AML would be enriched in the cells that are not killed by DNTs. However, it is possible that some AML cells survived after 2 hours of coincubation with DNTs because of a lack of interaction. To mitigate this possibility and identify cells that were highly resistant to DNT-mediated killing, Cas9+ AML2 cells and Cas9+ AML3 cells transduced with the epidrug sgRNA library viruses were cultured or not with DNTs overnight (supplemental Figure 9). Although 40% to 50% AML cell death was observed after 2 hours, ∼80% death occurred in AML cells after coculture with DNTs overnight. sgRNAs targeting FCGR1A were the most enriched in Annexin V− AML3 cells sorted after a 2-hour coculture with DNTs, as described in Figure 1B (Figure 3A). Similarly, FCGR1A sgRNAs were top ranked in AML2 and AML3 cells surviving prolonged DNT exposure (Figure 3B-3C; supplemental Tables 6 and 7). FCGR1A encodes a high-affinity activating Fc receptor, also known as CD64. To determine whether CD64 expression correlates with AML susceptibility to DNT killing, we performed cytotoxicity assays with 6 AML cell lines. We observed a significant positive correlation between the level of CD64 expression on AML cells and their susceptibility to DNT-mediated cytotoxicity (Figure 3D). To further validate the role of CD64 in conferring AML susceptibility, we silenced CD64 expression in AML cells by targeting FCGR1A using sgRNAs. Although >50% of AML2 and AML3 cells with sgRNAs targeting AAVS expressed CD64, <8% of cells transduced with FCGR1A sgRNAs expressed CD64 (Figure 3E). Importantly, we observed a significant reduction in DNT-mediated killing of CD64-silenced cells for AML2 cells (Figure 3F) and AML3 cells (Figure 3G) at various DNT/AML ratios compared with the respective sgAAVS controls.
CD64 expression on AML correlates with their susceptibility to DNT-mediated cytotoxicity in vitro. (A) Volcano plot of all genes enriched in live (Annexin V−) AML3 cells following DNT treatment. X-axis represents the log2-transformed fold changes in normalized sgRNA tag counts in dead vs live cells. The gene targets with significantly enriched sgRNAs (FDR ≤0.10) in live cells are represented by red circles. (B-C) CRISPR screen of essential genes in the epidrug CRISPR library in remaining live AML cells vs untreated AML cells for AML2 and AML3 cells. Genes were rank-ordered by robust rank aggregation (RRA) scores calculated by MAGeCK24 ; a higher RRA score indicates genes that are enriched in LIVE cells following DNT coculture (eg, genes conferring AML susceptibility to DNTs). FCGR1A is the top-ranked gene in AML2 and AML3 cell screens. (D) CD64 expression on 6 AML cell lines was analyzed by flow cytometry. Each cell line was cocultured with DNTs for 2 hours at an E:T ratio of 2:1 in triplicates. Scatterplot depicts Pearson’s correlation coefficient for the mean killing of AML against surface CD64 expression. (E) Surface expression of CD64 by flow cytometry in AML2 and AML3 Cas9-expressing cells transduced with sgRNAs against FCGR1A and a control (sgAAVS). (F-G) Cytotoxicity assays with DNTs at various E:T ratios (2 hours) in FCGR1A-deficient AML2 cells (F) and AML3 cells (G). Experiments were repeated 3 times with similar results, and statistical significance was determined by 1-way analysis of variance with the Dunnett test for multiple comparisons. ****P < .0001. MFI, mean fluorescence intensity.
CD64 expression on AML correlates with their susceptibility to DNT-mediated cytotoxicity in vitro. (A) Volcano plot of all genes enriched in live (Annexin V−) AML3 cells following DNT treatment. X-axis represents the log2-transformed fold changes in normalized sgRNA tag counts in dead vs live cells. The gene targets with significantly enriched sgRNAs (FDR ≤0.10) in live cells are represented by red circles. (B-C) CRISPR screen of essential genes in the epidrug CRISPR library in remaining live AML cells vs untreated AML cells for AML2 and AML3 cells. Genes were rank-ordered by robust rank aggregation (RRA) scores calculated by MAGeCK24 ; a higher RRA score indicates genes that are enriched in LIVE cells following DNT coculture (eg, genes conferring AML susceptibility to DNTs). FCGR1A is the top-ranked gene in AML2 and AML3 cell screens. (D) CD64 expression on 6 AML cell lines was analyzed by flow cytometry. Each cell line was cocultured with DNTs for 2 hours at an E:T ratio of 2:1 in triplicates. Scatterplot depicts Pearson’s correlation coefficient for the mean killing of AML against surface CD64 expression. (E) Surface expression of CD64 by flow cytometry in AML2 and AML3 Cas9-expressing cells transduced with sgRNAs against FCGR1A and a control (sgAAVS). (F-G) Cytotoxicity assays with DNTs at various E:T ratios (2 hours) in FCGR1A-deficient AML2 cells (F) and AML3 cells (G). Experiments were repeated 3 times with similar results, and statistical significance was determined by 1-way analysis of variance with the Dunnett test for multiple comparisons. ****P < .0001. MFI, mean fluorescence intensity.
CD64 expression facilitates DNT-mediated cytotoxicity of AML cells in vitro and in vivo
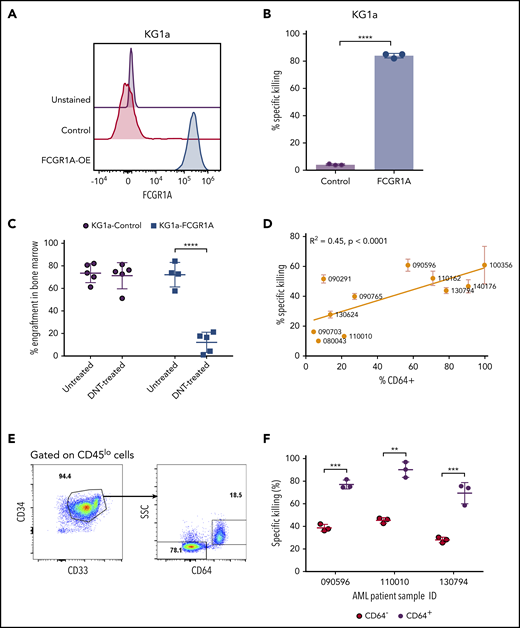
To examine CD64 as a bona fide AML susceptibility marker to DNT-mediated cytotoxicity, we next determined whether CD64 expression was sufficient to confer upon AML cells susceptibility to DNT-mediated killing. Because KG1a is a stem-like AML line that is resistant to DNT-mediated cytotoxicity,13 and it does not express CD64 (Figure 4A), we transduced KG1a cells with a vector expressing FCGR1A. Overexpression of CD64 in KG1a cells, confirmed by flow cytometry (Figure 4A), converted KG1a cells from being DNT resistant to being highly sensitive to DNT-mediated killing, as determined by in vitro cytotoxicity assays (Figure 4B). To further validate the importance of AML CD64 expression in DNT therapy in vivo, NSG mice were engrafted with wild-type or CD64-expressing KG1a cells, followed by DNT infusion. Interestingly, DNT treatment significantly reduced the leukemia burden by 83.2% ± 12.4% in NSG mice engrafted with CD64-expressing KG1a cells compared with only 3.2% ± 15.8% in mice engrafted with wild-type KG1a cells (Figure 4C).
FCGR1A (CD64) expression on AML increases its sensitivity to DNTs. (A) CD64 expression in KG1a cells transduced with FCGR1A or a control vector. (B) Killing of the control or FCGR1A-overexpressing KG1a cells by DNTs at a 1:1 E:T ratio. This experiment was repeated 3 times with similar results, and a 2-tailed Student t test was used to examine differences. (C) NSG mice (4-5 per group) were engrafted with control or FCGR1A- overexpressing KG1a cells, followed by treatment with 3 doses of DNTs or phosphate-buffered saline (with interleukin-2) on days 1, 4, and 7 postinoculation of leukemic cells. Mice were euthanized when bone marrow engraftment of KG1a cells reached ∼70%. A 2-tailed Student t test was performed to determine statistical differences. (D) Eleven primary AML samples were cocultured with DNTs for 2 hours at an E:T ratio of 4:1 in triplicates. Scatterplot depicts the Pearson’s correlation coefficient for the mean killing of primary AML against the proportion of CD64+ cells. (E-F) In vitro cytotoxicity assay conducted against 3 primary AML samples (090596, 110010, and 130794) with CD64+ and CD64− leukemic blast populations at a 4:1 E:T ratio. (E) Representative flow plot showing gating strategy for leukemic subpopulations with or without CD64 expression. (F) Representative percentage of specific killing induced by DNTs from 1 donor. Two-tailed Student t tests were performed to detect statistical differences between the CD64− and CD64+ populations within each patient sample. This experiment was performed using DNTs expanded from 3 donors. **P < .01, ***P < .001, ****P < .0001.
FCGR1A (CD64) expression on AML increases its sensitivity to DNTs. (A) CD64 expression in KG1a cells transduced with FCGR1A or a control vector. (B) Killing of the control or FCGR1A-overexpressing KG1a cells by DNTs at a 1:1 E:T ratio. This experiment was repeated 3 times with similar results, and a 2-tailed Student t test was used to examine differences. (C) NSG mice (4-5 per group) were engrafted with control or FCGR1A- overexpressing KG1a cells, followed by treatment with 3 doses of DNTs or phosphate-buffered saline (with interleukin-2) on days 1, 4, and 7 postinoculation of leukemic cells. Mice were euthanized when bone marrow engraftment of KG1a cells reached ∼70%. A 2-tailed Student t test was performed to determine statistical differences. (D) Eleven primary AML samples were cocultured with DNTs for 2 hours at an E:T ratio of 4:1 in triplicates. Scatterplot depicts the Pearson’s correlation coefficient for the mean killing of primary AML against the proportion of CD64+ cells. (E-F) In vitro cytotoxicity assay conducted against 3 primary AML samples (090596, 110010, and 130794) with CD64+ and CD64− leukemic blast populations at a 4:1 E:T ratio. (E) Representative flow plot showing gating strategy for leukemic subpopulations with or without CD64 expression. (F) Representative percentage of specific killing induced by DNTs from 1 donor. Two-tailed Student t tests were performed to detect statistical differences between the CD64− and CD64+ populations within each patient sample. This experiment was performed using DNTs expanded from 3 donors. **P < .01, ***P < .001, ****P < .0001.
Having confirmed that CD64 expression levels correlate with AML cell line susceptibility to DNTs in in vitro and xenograft models, we further validated the potential role of CD64 in AML patient-derived primary samples. We first observed variable expression of CD64 among different primary AML blasts. Furthermore, we found that the higher expression of CD64 on leukemic blasts correlated with greater cytotoxicity induced by DNTs (Figure 4D). Some individual patient samples contained CD64+ and CD64− leukemic blast populations (Figure 4E), which allowed us to determine the effect of CD64 expression on AML cell sensitivity to DNTs in the same patient, with minimal influence from other factors. Interestingly, consistent among all 3 AML patient samples tested, significantly higher killing of CD64+ leukemic blasts compared with CD64− leukemic blasts was observed after 2 hours of coculture with DNTs (Figure 4F). These data demonstrate the crucial role of CD64 in sensitizing AML blasts to DNT-mediated antileukemic effects. Taken together, our findings support the potential of using CD64 as a susceptibility marker for selecting AML patients for DNT therapy.
Discussion
In this study, we used an in vitro CRISPR screen to identify genes that regulate AML cell susceptibility to DNT-mediated cytotoxicity. Several recent studies have used CRISPR/Cas9 screens to identify genes involved in immunotherapy31,32 and AML pathobiology.33-36 Herein, we used FACS to differentiate between AML cells that were susceptible or resistant to DNT exposure. The highest frequencies of sgRNAs in live or apoptotic AML cells after coculture with DNTs were studied as potential susceptibility or resistance markers. Identification of such susceptibility markers will help us to understand how DNTs target AML cells and may act as potential positive biomarkers for patient selection. On the other hand, resistance markers can be negative biomarkers and offer an avenue to use drug inhibitors to target these molecules as potential combination therapies that can be used in conjunction with DNTs.
Our screen revealed a number of genes potentially involved in susceptibility and resistance mechanisms of AML to DNT-mediated cytotoxicity. Subsequent validation experiments underlined the functional involvement of the identified genes, thereby demonstrating the accuracy and potential utility of this screening method. Patel et al also used a CRISPR screen involving cell cocultures to investigate the mechanisms behind cancer immunotherapy.32 Although their study used a whole-genome screen, we used a targeted CRISPR library that was chosen for greater reliability and clinical relevance, such that our findings may be promptly applied to clinical trials. Furthermore, by performing a shorter 2-hour cytotoxicity assay, we were able to sort cells undergoing early-stage apoptosis from live cells, allowing us to investigate mechanisms involved in susceptibility and resistance. This represents one of the first studies to use a CRISPR screen that incorporates cytotoxicity assays and FACS sorting to study distinct molecular mechanisms conferring susceptibility and resistance of leukemic cells to T-cell therapy simultaneously. This approach could also be applicable to other chemotherapy and adoptive cellular therapies in which there is rapid cell killing.
We identified multiple members of the SAGA DUBm as potential resistance markers: ATXN7L3, ENY2, and USP22. This module is involved in the deubiquitination of histone H2A and H2B, which has implications in oncogenesis,37-41 and there is evidence that USP22 supports the proto-oncogenic function of MYC.28 Ubiquitination can have pleiotropic effects on AML cells. A recent study by Cartel et al showed that inhibition of USP7, the largest subfamily of deubiquitinating enzymes, reduces the proliferation of AML cells and sensitizes them to chemotherapy.42 Here, we found that silencing of ENY2 or ATXN7L3 increased susceptibility of AML2 and AML3 cells to DNTs. The slight differences between CRISPR knockout and shRNA knockdown may be due to different levels of target silencing or the confounding effect caused by the DNA-repair activity of the SAGA DUBm responding to double-stranded breaks that occur during CRISPR/Cas9 cleavage of target proteins. Because ENY2 and ATXN7L3 are essential genes, inhibiting them may have dual benefits of reducing cell proliferation and enhancing DNT killing. Although no specific inhibitors exist for the SAGA DUBm, there are a number of FDA-approved drugs that modulate ubiquitination43 and could be tested in combination with DNT therapy in future studies. Additionally, deubiquitinating enzyme complexes play important roles in DNA-repair mechanisms.44 Hence, sgRNA knockout of SAGA DUBm components may induce DNA damage response and cellular stress pathways, which can ultimately render AML cells more immunogenic.
Furthermore, reports have shown that some of the SAGA DUBm components, such as USP22, can suppress FOXP3 expression and enhance antitumor immunity.45,46 In addition to GO analysis supporting the immunomodulatory role of SAGA DUBm, genetic inhibition of SAGA DUBm subunits on AML cells induced expression of genes involved in immune cell recognition (HLA-DRA) and T-cell costimulation (CD40), suggesting that DUBm may regulate these immune-related genes to inhibit AML-DNT interaction, DNT activation, and/or cytotoxicity to AML cells. Further studies will elucidate the pathways by which SAGA DUBm regulates AML-DNT interactions in vitro and in vivo.
Our screens in AML2 and AML3 cells suggested that CD64 plays a significant role in conferring susceptibility to DNTs. This receptor preferentially binds to immunoglobulin to initiate and modulate an immunological reaction.47 CD64 expression has been shown to correlate with improved overall survival in AML patients,48 although the underlying mechanism remains unclear. Previously, CD64 was shown to be involved in targeting of AML by lymphokine-activated killer cells in vitro.49 Here, we demonstrated that CD64 expression sensitizes AML to DNT-mediated cytotoxicity. There are ≥2 possible explanations for how CD64 confers AML susceptibility to DNT-mediated cytotoxicity. One possibility is that CD64 signaling leads to cellular changes in AML cells that confer susceptibility to DNTs. Another possibility is that CD64 can cross-link antibodies that are present on effector cells, as described by Notter et al.49 In that study, they observed that anti-CD3 antibody–coated lymphokine-activated killer cells selectively targeted AML cells that expressed CD64. They proposed a mechanism whereby CD64 on target cells would bind and cross-link anti-CD3 antibody bound on T cells, resulting in increased lytic activity. These findings may have implications for many T-cell–based immunotherapies because anti-CD3 antibodies are commonly used for T-cell expansion. Therefore, additional studies are warranted to clarify the function of CD64 in increasing DNT cytotoxic function.
CD64 is often used as part of immunophenotyping panels to characterize AML cases.50,51 The presence of CD64 on AML blasts is associated with a more mature phenotype and can help to distinguish M3, M4, and M5 forms of the disease from other subtypes.50 We previously showed that AML-M5 or monocytic AML, which is known to more commonly express CD64, is more susceptible to DNT-mediated cytotoxicity than are other AML subtypes.15 More importantly, we found that the susceptibility of multiple AML cell lines and primary AML samples to DNT-mediated cytotoxicity directly correlated with surface CD64 expression. Collectively, these findings indicate that the level of CD64 expression on AML cells is important for DNT-mediated cytotoxicity. In our phase 1 clinical trial (NCT03027102), we will further confirm that CD64 can be used as a biomarker for patient stratification by correlating its expression by AML blasts with clinical response to DNT therapy.
Overall, our findings emphasize the utility of CRISPR/Cas9 screens to investigate mechanisms underlying immunotherapy, which may help to improve the efficacy of DNT therapy for AML patients. Further, a similar screening strategy may be applicable for identifying genes involved in interactions between other cytotoxic cells and various cancer types.
Data sharing requests should be sent to Li Zhang (lzhang@uhnresearch.ca) or Housheng Hansen He (hansenhe@uhnresearch.ca).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Aaron Schimmer for helpful comments on project design and critical reading of the manuscript. Figures were created with BioRender.com.
This work was supported by the Canadian Cancer Society (grant 704121) (L.Z.) and the Princess Margaret Cancer Foundation (grant 886012001223) (H.H.H.). L.Z. is the Maria H. Bacardi Chair of Transplantation. H.H.H. holds an Office of Management, Information, and Research Early Researcher Award, a Canadian Institutes of Health Research (CIHR) New Investigator Salary Award, and 4 Canadian Institutes of Health Research project grants, and is the Joey and Toby Tanenbaum Brazilian Ball Chair in Prostate Cancer.
Authorship
Contribution: F.S., B.C., H.H.H., and L.Z. conceived and designed the study; F.S., B.C., J.B.L., H.K., E.T., N.A., and D.L. performed experiments; F.S., B.C., J.B.L., Y.Z., and M.A. analyzed and interpreted data; M.D.M. provided AML patient samples for the study; B.C. and F.S. wrote the manuscript; M.D.M., H.H.H., J.B.L., D.L., and L.Z. provided feedback and edited the manuscript; H.H.H. and L.Z. provided funding support; and all authors read and approved the final version of the manuscript.
Conflict-of-interest disclosure: L.Z. is one of the inventors on 7 DNT technology-related patents and serves as a scientific consultant for WYZE Biotech Co. Ltd. B.C., J.B.L., D.L., and H.K. are inventors on patent applications using DNTs to treat cancers. The remaining authors declare no competing financial interests.
Correspondence: Li Zhang, University Health Network, Princess Margaret Cancer Research Tower, 101 College St, Room 2-807, Toronto, ON M5G 1L7, Canada; e-mail: lzhang@uhnresearch.ca; and Housheng Hansen He, University Health Network, Princess Margaret Cancer Research Tower, 101 College St, Room 11-305, Toronto, ON M5G 1L7, Canada; e-mail: hansenhe@uhnresearch.ca.
REFERENCES
Author notes
F.S., B.C., J.B.L., and M.A. contributed equally to this work.