Key Points
HSC fitness response to stress depends on Yap1/Taz.
Scribble links Cdc42 and the cytosolic functions of the Hippo signaling cascade in HSC fate determination.
Abstract
Yap1 and its paralogue Taz largely control epithelial tissue growth. We have identified that hematopoietic stem cell (HSC) fitness response to stress depends on Yap1 and Taz. Deletion of Yap1 and Taz induces a loss of HSC quiescence, symmetric self-renewal ability, and renders HSC more vulnerable to serial myeloablative 5-fluorouracil treatment. This effect depends on the predominant cytosolic polarization of Yap1 through a PDZ domain-mediated interaction with the scaffold Scribble. Scribble and Yap1 coordinate to control cytoplasmic Cdc42 activity and HSC fate determination in vivo. Deletion of Scribble disrupts Yap1 copolarization with Cdc42 and decreases Cdc42 activity, resulting in increased self-renewing HSC with competitive reconstitution advantages. These data suggest that Scribble/Yap1 copolarization is indispensable for Cdc42-dependent activity on HSC asymmetric division and fate. The combined loss of Scribble, Yap1, and Taz results in transcriptional upregulation of Rac-specific guanine nucleotide exchange factors, Rac activation, and HSC fitness restoration. Scribble links Cdc42 and the cytosolic functions of the Hippo signaling cascade in HSC fate determination.
Introduction
The search of molecular targets to expand functional hematopoietic stem cells (HSC) for translational application remains paramount. HSC will respond to microenvironmental cues and self-renew or differentiate according to changes in cellular polarity. HSC frequently display an asymmetric distribution of the proliferation and fate determinant Cdc42.1,2 Cdc42 activity must be tightly regulated for proper HSC function.3,4 Very few polarity proteins, outside of Cdc42, have such a convincing functional role in HSC.5-8 The observation that HSC isolated from mice lacking Numb and Numb-like,9 upstream Notch1,10 or aPKC11 behave normally have proven that Numb asymmetries are unlikely to control HSC divisional fate. Searching for functionally relevant HSC fate determinants, we analyzed the role of the Scribble polarity complex. The Scribble complex, consisting of Lethal Giant Larvae (Lgl), Discs Large, and Scribble, is 1 of 3 major evolutionarily conserved polarity complexes that coordinates the spatial organization of intracellular proteins. Conditional deletion of Lgl1 in HSC leads to an expansion in the HSC population with an acquired increase in competitive repopulation capacity.12 On the contrary, deficiency of Scribble has been reported to result in decreased competitive reconstitution ability.13 The mechanisms underlying the seemingly antagonistic roles of Lgl1 and Scribble in HSC activity is unknown. Both Scribble and Lgl1 complex partners lack any known intrinsic enzymatic activity; however, they contain a number of well-characterized protein–protein interacting domains that enable them to bind signaling node proteins.14,15 An accumulating body of evidence has shown that apical–basal polarity proteins may regulate the Hippo signaling pathway16-18 and consequently the downstream transcriptional coactivators Yes-associated protein 1 (YAP1) and transcriptional coactivator with a PDZ-binding domain (TAZ).19-22
Yap1 is believed to play a role in mammalian hematopoietic specification23 and in cord blood-derived human HSC, Yap1, and its obligatory nuclear partner TEAD1 regulate differentiation of B-cell progenitors.24 Additionally, Yap1 signaling has recently been shown to instruct primitive HSC specification, production, and maturation in vivo.25 However, formal demonstration of the role of Yap1 and Taz in adult HSC activity is lacking.26,27 Thus, we wanted to explore the concept of Yap1/Taz interacting with and coordinating polarity fate determinants in HSC. Indeed, polarized Scribble coordinates the organization and activation of the Hippo signaling pathway resulting with cytosolic Yap1. We provide the first functional evidence that Scribble and Yap1 coordinate to control Cdc42 location while positively regulating its activity, driving HSC quiescence and fate decisions in vivo. Our data indicate that the Scribble/Yap1 copolarization is indispensable for Cdc42-dependent activity on HSC asymmetric division and fate.
Materials and methods
Animals
Exons 2 through 8 of the wild type (Wt) Scribble locus were conditionally targeted to create Scribbleflox/flox alleles. Details on the creation of these animals and subsequent crossings can be found in supplemental Materials and methods, available on the Blood Web site.
Immunofluorescence studies of intracellular localization, cell isolation, and cell-cycle analysis
Performed by combination of flow cytometry, sorting, confocal microscopy, and proximity ligation assays. Experimental details can be found in supplemental Materials and methods.
Functional assays
Colony-forming unit, liquid cultures, cobblestone-area-forming cell, serial competitive repopulation, 2-bromodeoxyuridine uptake, 5-fluorouracil administration, and paired-daughter cell assays were performed. Experimental details can be found in supplemental Materials and Methods.
Biochemical assays and generation of reconstitute mutant Scribble expressing HSC
GST-PAK pulldown was performed using p21-activated kinase effector containing beads and analyzed in immunoblots. Expression of HSC with full-length and structure-function mutants of Scribble was performed by lentiviral transduction. Experimental details can be found in supplemental Materials and Methods.
Transcriptome and bioinformatic analyses
Purified complementary DNA was captured on an Illumina flow cell for cluster generation and libraries were sequenced on the Illumina HiSeq2500 following the manufacturer's protocol. Bioinformatic analysis and details on transcriptome generation can be found in supplemental Materials and Methods.
Results
Combined activity of the paralogues Yap1 and Taz is necessary for HSC function
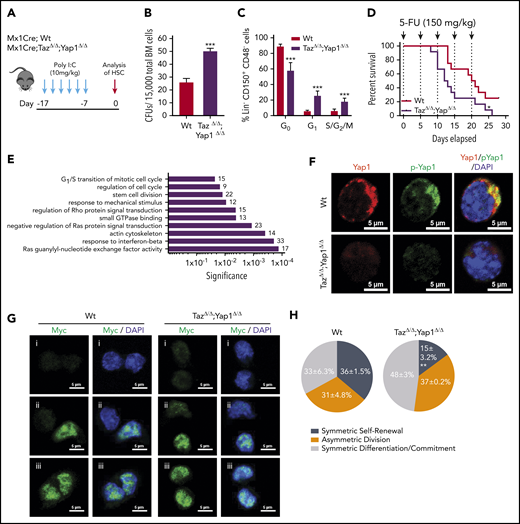
In determining whether Yap1 and Taz control hematopoiesis, we first observed that the complete loss of Yap1/Taz results in nonviable pups, whereas the incomplete loss of 1 to 3 alleles did not impair the expected Mendelian inheritance ratios from hematopoietic-specific Vav1CreTg/-;Yap1f/f;Tazf/f mice (supplemental Figure 1A). These data suggested that the complete loss of Yap1/Taz severely impairs hematopoietic development resulting in fetal death. Because recombination in Vav-Cre transgenic mice can also affect endothelial cells,28 we generated a hematopoietic, inducible (Mx1-CreTg/−) Yap1f/f;Tazf/f mouse model. Deficiency of Yap1/Taz was induced by administration of polyinositide:polycytidine (poly I:C, Figure 1A; supplemental Figure 1B). No significant changes were observed in peripheral blood (PB) (white blood cell, neutrophil, platelet, reticulocyte counts, and hemoglobin level) 1 week after induced deletion of Yap1/Taz (supplemental Figure 1C). Yap1/TazΔ/Δ hematopoiesis displayed no difference in bone marrow (BM) cellularity (supplemental Figure 1C); however, Yap1/TazΔ/Δ BM contains a 50% to 100% increase in the frequency of colony-forming units from total BM or BM Lin− Sca-1+ c-kit+ (LSK) cells (Figure 1B; supplemental Figure 1D). Yap1/TazΔ/Δ HSC exhibit a loss of quiescence (∼50% decrease in the frequency of G0 HSC and concomitant increase in the G1 and S/G2/M phases of the cell cycle) (Figure 1C). Although no relevant alteration in the competitive repopulation ability of Yap1/TazΔ/Δ hematopoiesis in primary recipient mice was observed (supplemental Figure 1E-F), Yap1/TazΔ/Δ HSC displayed reduced reconstitution abilities during serial competitive repopulation, evidenced by decreased PB chimerism throughout secondary reconstitution with a 50% decrease in the BM content of HSC (supplemental Figure 1E-F). Consistent with increased cellular proliferation and cell-cycle progression of BM HSC/progenitor (P) (Figure 1C), Yap1/TazΔ/Δ HSC/P were more susceptible to exhaustion upon serial myeloablation resulting from 5-fluorouracil (5-FU). Yap1/TazΔ/Δ mice succumb to BM failure significantly earlier than their Wt counterparts (12.5 vs 20.5 days) with significantly lower absolute neutrophil counts in PB (Figure 1D; supplemental Figure 1G). To rule out the effects of potential nonhematopoietic Yap1 and Taz deficiency, induced with poly I:C-mediated deletion, we confirmed these results using fully hematopoietic chimeric (>90%) mice that had been reconstituted with Mx1Cre;Wt and Mx1Cre;Tazfl/fl;Yap1fl/fl BM before deletion induction and 5-FU administration (supplemental Figure 1H-I). Taken together, the genetic deletion of Yap1 and its paralogue Taz in HSC resulted in loss of HSC quiescence and increased HSC/P proliferation leading toward exhaustion.
Yap1/Taz are necessary for HSC function. (A) Schematic representing the inducible deletion of Yap1/Taz in the hematopoietic system followed by 1 week of recovery before subjecting mice to experimental testing. (B) Number of colony-forming units (CFUs) from Mx1Cre;Wt and Mx1Cre;TazΔ/Δ;Yap1Δ/Δ total BM cells. (C) Cell-cycle analysis of Lin− CD48− CD150+ HSC by fluorescence-activated cell sorting (FACS). (D) Kaplan-Meier survival analysis after serial myeloablation with 5-fluorouracil (150 mg/kg) 5 days apart. (E) Gene ontology (GO) pathway analysis of differentially regulated genes (P < .05) between Mx1Cre;Wt and Mx1Cre;TazΔ/Δ;Yap1Δ/Δ HSC (cutoff, 1.5-fold). Numbers represent the percentage of genes within each GO pathway that are differentially regulated. (F) Immunofluorescence depicting Yap1 protein localization in Wt HSC (immunophenotypically defined as LSK CD150+ CD48−) (red pseudo-color) and Yap1 phosphorylation status at Serine 112 (green pseudo-color). Cells are counterstained with 4′,6-diamidino-2-phenylindole (DAPI) and merged images are shown in the right micrographs. Scale bar is 5 µm. (G) Immunofluorescence depicting fate determinant allocation of Myc and the corresponding HSC division mode in Wt and Yap1/TazΔ/Δ paired daughter HSC (nocodazole, 10 nM for 24 hours). Low Myc expression in paired daughter cells represents (i) symmetric self-renewal, (ii) high and low Myc expression between the 2 daughters represents an asymmetric division, whereas (iii) high Myc expression in both cells is indicative of symmetric commitment. (H) Quantification of fate determinant allocation and division mode among Wt and Yap1/TazΔ/Δ paired daughter HSC. *P < .05; **P < .01; ***P < .001.
Yap1/Taz are necessary for HSC function. (A) Schematic representing the inducible deletion of Yap1/Taz in the hematopoietic system followed by 1 week of recovery before subjecting mice to experimental testing. (B) Number of colony-forming units (CFUs) from Mx1Cre;Wt and Mx1Cre;TazΔ/Δ;Yap1Δ/Δ total BM cells. (C) Cell-cycle analysis of Lin− CD48− CD150+ HSC by fluorescence-activated cell sorting (FACS). (D) Kaplan-Meier survival analysis after serial myeloablation with 5-fluorouracil (150 mg/kg) 5 days apart. (E) Gene ontology (GO) pathway analysis of differentially regulated genes (P < .05) between Mx1Cre;Wt and Mx1Cre;TazΔ/Δ;Yap1Δ/Δ HSC (cutoff, 1.5-fold). Numbers represent the percentage of genes within each GO pathway that are differentially regulated. (F) Immunofluorescence depicting Yap1 protein localization in Wt HSC (immunophenotypically defined as LSK CD150+ CD48−) (red pseudo-color) and Yap1 phosphorylation status at Serine 112 (green pseudo-color). Cells are counterstained with 4′,6-diamidino-2-phenylindole (DAPI) and merged images are shown in the right micrographs. Scale bar is 5 µm. (G) Immunofluorescence depicting fate determinant allocation of Myc and the corresponding HSC division mode in Wt and Yap1/TazΔ/Δ paired daughter HSC (nocodazole, 10 nM for 24 hours). Low Myc expression in paired daughter cells represents (i) symmetric self-renewal, (ii) high and low Myc expression between the 2 daughters represents an asymmetric division, whereas (iii) high Myc expression in both cells is indicative of symmetric commitment. (H) Quantification of fate determinant allocation and division mode among Wt and Yap1/TazΔ/Δ paired daughter HSC. *P < .05; **P < .01; ***P < .001.
Unbiased transcriptomic analysis of Yap1/TazΔ/Δ HSC failed to identify differential regulation genes classically associated with Yap1 and its cofactors (TEAD, p73, ERBB4, Runx, β-catenin/Tbx5, or Egr-1), including Cyr61, Ctgf, Ankrd1, Myc, Gli2, Vimentin, Axl, Bax, and Birc5 marker genes (supplemental Figure 1J-K). Instead, RNA sequencing analysis revealed significant differential clustering among genes involved with regulation of the actin cytoskeleton and small GTPases (supplemental Figure 1J). Noteworthy, Gene Ontology (GO) pathway analysis of Yap1/TazΔ/Δ HSC pinpointed differentially regulated genes involved in several pathways that coordinate small GTPase activity, responses to mechanical stimulus and cell cycle (Figure 1E). GO pathways such as these validate the in vivo phenotype of Yap1/TazΔ/Δ HSC and suggests that the loss of quiescence might be linked with small GTPase activity. Consistent with unchanged Yap1 transcriptional targets in Yap1/TazΔ/Δ HSC, we observed that Yap1 protein and its phosphorylated form (S112) are abundantly expressed and exclusively polarized within the cytosol of Wt HSC compared with their absence in Yap1/TazΔ/Δ HSC (Figure 1F). Cytosolic location and function of murine Yap1 requires phosphorylation at serine 11229 and indeed, Yap1 colocalized with its phosphorylated form (Figure 1F), indicating that cytosolic Yap1 is the predominantly featured in polarized BM HSC. Forced expression of the exclusively cytosolic human Yap1 S127D, in which a Lats-mediated phosphorylation site (Ser127) was replaced by aspartic acid30 resulting in a phospho-mimetic mutant that will not translocate to the nucleus, restores the expression of polarized Yap1 in the cytosol of Yap1/Taz deficient LSK cells (supplemental Figure 1L) and the colony formation ability of Yap/Taz-deficient Lin− HSC/P (supplemental Figure 1M) to Wt levels.
In concordance with these functional and molecular data, the deletion of Yap1 and Taz influences HSC fate at the division level. Myc activation and translocation is a hallmark of HSC fate after division.31 Self-renewing HSC divisions are characterized by reduced activation and translocation of Myc (low Myc), whereas asymmetrically dividing daughter cells have dramatically distinct levels of Myc activation and committed symmetrically dividing cells will both have high levels of Myc activation and translocation (high Myc). Analysis of molecular fate determinant allocation within paired daughter cells provides evidence that Yap1/TazΔ/Δ HSC have a ∼50% increased frequency of high Myc commitment divisions with concomitant reductions in low Myc symmetric self-renewal divisions (Figure 1G-H; supplemental Figure 1N).
Scribble scaffolds cytosolic Yap1 with upstream inhibitory components of the Hippo signaling pathway in HSC
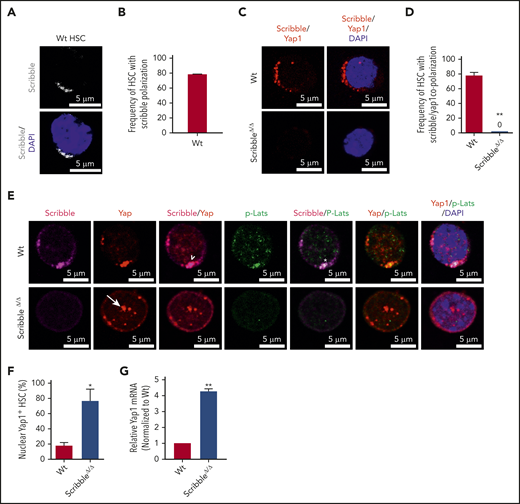
Accumulating evidence has shown that apical–basal polarity proteins from the Par complex (Par3/Par6/αPKC) and Scribble complex (Scribble/Discs Large/Lgl) have been implicated in the regulation of the Hippo signaling pathway, thereby controlling Yap1/Taz localization and function in epithelial cells.16-18,32-36 Our previous data have identified that the deficiency of the Par polarity complex member, atypical protein kinase C (aPKC), is dispensable for HSC activity under basal, stressed or regenerative hematopoiesis.11,37 Thus, we turned our attention toward the basal polarity complex member, Scribble, and provide the first evidence that Scribble is asymmetrically distributed and polarized in Wt HSC (Figures 2A-B). Both Yap1/Taz contain a class I PDZ-binding motif (-LTWL-COO−) at their extreme carboxyl-terminus that have been predicted to bind with Scribble.38 Using single-cell proximity ligation analysis (PLA), we demonstrated that Scribble interacts with Yap1 in ∼80% of HSC (Figure 2C-D; supplemental Figure 2A).
Scribble scaffolds components of the Hippo pathway in HSC and controls Yap1 cytoplasmic localization. (A) Immunofluorescence depicting Scribble protein localization in Wt HSC (immunophenotypically defined as LSK CD150+ CD48−) (white areas). Cells are counterstained with DAPI and merged images are shown in the bottom micrographs. Scale bar is 5 µm. (B) Quantification for the frequency of HSC with Scribble polarization. (C) Immunofluorescence depicting a proximity ligation assay (PLA) on Wt and ScribbleΔ/Δ HSC using anti-Scribble and anti-Yap1 primary antibodies subsequently targeted with corresponding probes for oligomerization. The detected dimers are pseudo-colored in red. Nuclei are counterstained with DAPI and merged images are shown in the right micrographs. Scale bar is 5 µm. (D) Quantification of PLA signal. (E) Immunofluorescence showing Scribble polarization and Yap1 colocalization (white arrowheads) in HSC isolated from Wt mice. White asterisks indicate areas of colocalization between Scribble and the activated upstream inhibitory kinase of Yap1, phosphorylated Lats1/2. White arrows denote Yap1 nuclear translocation in ScribbleΔ/Δ HSC. Nuclei are counterstained with DAPI; merged images are shown in the right micrographs. Scale bar is 5 µm. (F) Quantification for the frequency of HSC with Yap1 nuclear foci. (G) Quantitative reverse transcriptase polymerase chain reaction of Yap1 messenger RNA expression from HSC cultured for 40 hours; *P < .05; **P < .01.
Scribble scaffolds components of the Hippo pathway in HSC and controls Yap1 cytoplasmic localization. (A) Immunofluorescence depicting Scribble protein localization in Wt HSC (immunophenotypically defined as LSK CD150+ CD48−) (white areas). Cells are counterstained with DAPI and merged images are shown in the bottom micrographs. Scale bar is 5 µm. (B) Quantification for the frequency of HSC with Scribble polarization. (C) Immunofluorescence depicting a proximity ligation assay (PLA) on Wt and ScribbleΔ/Δ HSC using anti-Scribble and anti-Yap1 primary antibodies subsequently targeted with corresponding probes for oligomerization. The detected dimers are pseudo-colored in red. Nuclei are counterstained with DAPI and merged images are shown in the right micrographs. Scale bar is 5 µm. (D) Quantification of PLA signal. (E) Immunofluorescence showing Scribble polarization and Yap1 colocalization (white arrowheads) in HSC isolated from Wt mice. White asterisks indicate areas of colocalization between Scribble and the activated upstream inhibitory kinase of Yap1, phosphorylated Lats1/2. White arrows denote Yap1 nuclear translocation in ScribbleΔ/Δ HSC. Nuclei are counterstained with DAPI; merged images are shown in the right micrographs. Scale bar is 5 µm. (F) Quantification for the frequency of HSC with Yap1 nuclear foci. (G) Quantitative reverse transcriptase polymerase chain reaction of Yap1 messenger RNA expression from HSC cultured for 40 hours; *P < .05; **P < .01.
To understand the role of the aforementioned Yap1/Scribble complex in HSC, we complemented our epistasis studies by the development of a novel Scribble floxed animal model (supplemental Figure 2B-D). Given that components of the upstream Hippo signaling pathway, Lats1/2 and Mst1/2, have been shown to interact with Scribble in nonhematopoietic tissues,33,39,40 we confirmed that Scribble copolarizes with the activated (phosphorylated) upstream inhibitory kinase Lats1 in ∼60% of Wt HSC (Figure 2E; supplemental Figure 2E-G). The deletion of Scribble in HSC disrupted the phospho-Lats1/Yap1 complex, permitting Yap1 to translocate into the nucleus (Figures 2E-F). Yap1 messenger RNA expression, which can be dependent on its own transcriptional activity, is fourfold upregulated in ScribbleΔ/Δ HSC (Figure 2G). Importantly, Scribble remains polarized in Yap1/TazΔ/Δ HSC, suggesting that Scribble acts upstream of Yap1 polarization (supplemental Figures 2H-I), to control its cytosolic localization.
Scribble PDZ domains are necessary for Yap1 cytoplasmic polarization while active Lats recruitment is mediated through its LRR domain
We then hypothesized that Scribble acts as a molecular scaffold to facilitate upstream Hippo signaling and the maintenance of Yap1 cytosolic localization and function. To test this hypothesis, we generated structure/function mutants of Scribble41 in bicistronic (EF1α-IRES-RFP) lentiviral-expressing vectors (Figure 3A). We transduced LSK BM cells with an empty lentiviral vector, vectors expressing the full-length Scribble protein or structure function mutants of Scribble. The overexpression of full-length Scribble in Wt LSK BM cells maintains Scribble/Yap1 cytoplasmic polarization (Figure 3B,D), whereas ScribbleΔ/Δ LSK BM cells display predominantly translocated nuclear Yap1 (Figure 3C-D). The expression of full-length Scribble in ScribbleΔ/Δ LSK BM cells restored Scribble/Yap1 copolarization in the cytosol, effectively preventing Yap1 translocation to the nucleus (Figure 3C-D). Forced expression of the PDZ containing mutant of Scribble within ScribbleΔ/Δ LSK cells reverted the nuclear accumulation of Yap1 back to the Wt-like cytoplasmic polarized state (Figure 3C-D). The N-terminal LRR domain of Scribble successfully recruited p-Lats in the cytosol, however, was unable to revert the nuclear Yap1 translocation in ScribbleΔ/Δ LSK BM cells (Figure 3C-F). These data suggest that both Scribble PDZ domains and LRR domains are independently required for scaffolding Yap1 in proximity to its inhibitory kinase, Lats1. As expected, the expression of the extreme carboxyl-terminus section of Scribble (lacking any functional domains) cannot restore the copolarization of Scribble with Yap1. Taken together, Scribble is indispensable for Hippo-mediated cytosolic Yap1 activity in HSC.
Cytoplasmic polarization of Yap1 is restored in ScribbleΔ/Δ HSC/P with expression of full-length Scribble or PDZ-containing mutants. (A) Graphical representation of the functional domains in human full-length scribble protein and the truncation mutations incorporated into an Ef1α-IRES-RFP lentivirus. (B) Immunofluorescence showing Scribble polarization and Yap1 colocalization in Lin− Sca-1+ c-kit+ (LSK) BM cells isolated from Wt mice and transduced with Ef1α-IRES-RFP (EMPTY) lentivirus or human full-length Scribble as indicated. Nuclei are counterstained with DAPI and merged images are shown in the right micrographs. Scale bar is 5 µm. (C) Immunofluorescence showing Scribble expression and Yap1 localization in LSK BM cells isolated from ScribbleΔ/Δ mice and transduced with EMPTY lentivirus, human full-length Scribble, or structure-function mutants as indicated. Nuclei are counterstained with DAPI and merged images are shown in the right micrographs. Scale bar is 5 µm. (D) Quantifications for the frequency of transduced LSK cells with nuclear Yap1 accumulation. (E) Immunofluorescence showing Scribble and pLats1 expression in LSK BM cells isolated from ScribbleΔ/Δ mice and transduced with EMPTY lentivirus or the LRR mutant. Nuclei are counterstained with DAPI and merged images are shown in the right micrographs. Scale bar is 5 µm. (F) Quantification of pLats1 colocalization with Scribble when the N-terminal portion of Scribble (LRR) is reintroduced into Scribble null LSK cells. (G) Cartoon depicting the ternary complex between Scribble, activated Last1/2, and Yap1 in the cytosol of Wt HSC. **P < .01; ***P < .001.
Cytoplasmic polarization of Yap1 is restored in ScribbleΔ/Δ HSC/P with expression of full-length Scribble or PDZ-containing mutants. (A) Graphical representation of the functional domains in human full-length scribble protein and the truncation mutations incorporated into an Ef1α-IRES-RFP lentivirus. (B) Immunofluorescence showing Scribble polarization and Yap1 colocalization in Lin− Sca-1+ c-kit+ (LSK) BM cells isolated from Wt mice and transduced with Ef1α-IRES-RFP (EMPTY) lentivirus or human full-length Scribble as indicated. Nuclei are counterstained with DAPI and merged images are shown in the right micrographs. Scale bar is 5 µm. (C) Immunofluorescence showing Scribble expression and Yap1 localization in LSK BM cells isolated from ScribbleΔ/Δ mice and transduced with EMPTY lentivirus, human full-length Scribble, or structure-function mutants as indicated. Nuclei are counterstained with DAPI and merged images are shown in the right micrographs. Scale bar is 5 µm. (D) Quantifications for the frequency of transduced LSK cells with nuclear Yap1 accumulation. (E) Immunofluorescence showing Scribble and pLats1 expression in LSK BM cells isolated from ScribbleΔ/Δ mice and transduced with EMPTY lentivirus or the LRR mutant. Nuclei are counterstained with DAPI and merged images are shown in the right micrographs. Scale bar is 5 µm. (F) Quantification of pLats1 colocalization with Scribble when the N-terminal portion of Scribble (LRR) is reintroduced into Scribble null LSK cells. (G) Cartoon depicting the ternary complex between Scribble, activated Last1/2, and Yap1 in the cytosol of Wt HSC. **P < .01; ***P < .001.
Scribble scaffolds polarized Yap1 with activated Cdc42
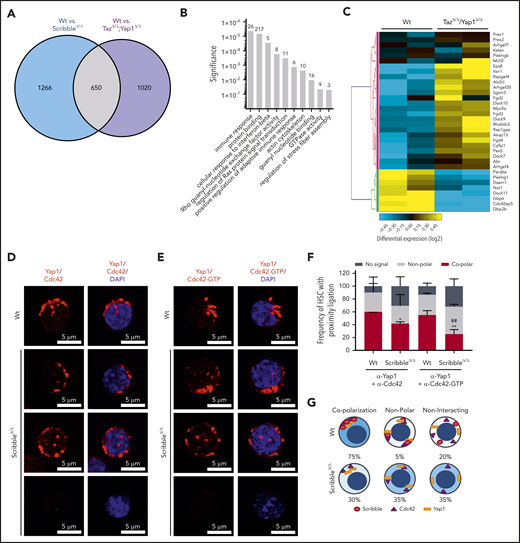
To understand further the requirement of Yap1 in HSC, we performed transcriptomic analysis on Wt and ScribbleΔ/Δ HSC (supplemental Figure 3A). Yap1 and Taz transcripts are expressed at very low frequency in Wt HSC (∼2000-fold less than the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase [GAPDH]) but are ∼25% to 50% upregulated upon deficiency of Scribble, whereas other bona fide Yap1 transcriptional targets remained unchanged (supplemental Figure 3B). However, RNA-sequencing and GO pathway analysis revealed significant differential clustering among genes pertaining to protein binding, actomyosin formation, and regulation of Rho GTPase activity, particularly Cdc42 (supplemental Figure 3A,C-D). This observation parallels recent mass spectroscopy data on putative Scribble interacting partners as regulators of the cytoskeleton network and GTPase activity from hematopoietic cell lines.13 To further funnel our analyses, we identified 650 overlapping genes (∼1/3 of all differentially expressed genes) when comparing the differentially regulated transcripts from ScribbleΔ/Δ HSC with those of Yap1/TazΔ/Δ HSC (Figure 4A). Pathway analysis run on the overlapping gene set highlighted pathways regulating small GTPase activity (Figures 4B-C). The GTPase and Rho guanyl-nucleotide exchange factor (GEF) activity genes include many upregulated GEFs that act specifically on RhoA, like Arhgef4, Arhgef28, Plekhg1, and Plekhg6,42 along with other GEFs that favor Cdc42 like Fgd2, Fgd3, Fgd4, Mcf2l, and Arhgef4 (Figure 4C; supplemental Figure 3D).42 Noticeably, Rac1-specific GEFs (Prex1, Prex2, Plekhg6)42 cluster together and display a modest upregulation in comparison with the upregulation of Cdc42 specific GEFs observed in theYap1/TazΔ/Δ HSC (Figure 4C). Given that both Scribble and Yap1 have independently been reported to regulate Cdc42 activation43,44 with the aforementioned data, we hypothesized that the scaffolding ability of Scribble on Yap1 coordinates Cdc42 activity and contributes to HSC function.
Scribble scaffolds Yap1 and Cdc42 in the cytoplasm of HSC. (A) Venn diagram highlighting the number of differentially regulated genes in common when comparing Mx1Cre;Wt and Mx1Cre;ScribbleΔ/Δ and comparing Mx1Cre;Wt and Mx1Cre;TazΔ/Δ;Yap1Δ/Δ HSC. (B) Gene ontology (GO) pathway analysis of the common differentially regulated genes between 2 independent analysis of Mx1Cre;Wt to Mx1Cre;ScribbleΔ/Δ HSC and of Mx1Cre;Wt to Mx1Cre;TazΔ/Δ;Yap1Δ/Δ HSC. Numbers represent the number of genes within each GO pathway that are differentially regulated. (C) Heat map depicting the differential regulation of common genes between Mx1Cre; ScribbleΔ/Δ and Mx1Cre;TazΔ/Δ;Yap1Δ/Δ HSC that pertain to the Rho guanyl nucleotide exchange factor activity and small GTPase activity gene ontology pathways. Proximity ligation assay (PLA) detection of endogenous (D) Yap1/Cdc42 and (E) Yap1/Cdc42-GTP interactions in HSC. The detected dimers are pseudo-colored in red. Nuclei are counterstained with DAPI and merged images are shown in the right micrographs. Scale bar is 5 µm. (F) Frequency of HSC in which PLA signal depicted in panels D and E was found in relation or not with polarization. *P < .05 and **P < .01 between Vav1Cre;Wt and Vav1Cre;ScribbleΔ/Δ; ##P < .01 between Cdc42 and Cdc42-GTP frequencies. (G) Cartoon representing the overall polarization status between Scribble, Yap1, and Cdc42 in Wt cells emphasizing the loss of copolarization and asymmetry in ScribbleΔ/Δ HSC.
Scribble scaffolds Yap1 and Cdc42 in the cytoplasm of HSC. (A) Venn diagram highlighting the number of differentially regulated genes in common when comparing Mx1Cre;Wt and Mx1Cre;ScribbleΔ/Δ and comparing Mx1Cre;Wt and Mx1Cre;TazΔ/Δ;Yap1Δ/Δ HSC. (B) Gene ontology (GO) pathway analysis of the common differentially regulated genes between 2 independent analysis of Mx1Cre;Wt to Mx1Cre;ScribbleΔ/Δ HSC and of Mx1Cre;Wt to Mx1Cre;TazΔ/Δ;Yap1Δ/Δ HSC. Numbers represent the number of genes within each GO pathway that are differentially regulated. (C) Heat map depicting the differential regulation of common genes between Mx1Cre; ScribbleΔ/Δ and Mx1Cre;TazΔ/Δ;Yap1Δ/Δ HSC that pertain to the Rho guanyl nucleotide exchange factor activity and small GTPase activity gene ontology pathways. Proximity ligation assay (PLA) detection of endogenous (D) Yap1/Cdc42 and (E) Yap1/Cdc42-GTP interactions in HSC. The detected dimers are pseudo-colored in red. Nuclei are counterstained with DAPI and merged images are shown in the right micrographs. Scale bar is 5 µm. (F) Frequency of HSC in which PLA signal depicted in panels D and E was found in relation or not with polarization. *P < .05 and **P < .01 between Vav1Cre;Wt and Vav1Cre;ScribbleΔ/Δ; ##P < .01 between Cdc42 and Cdc42-GTP frequencies. (G) Cartoon representing the overall polarization status between Scribble, Yap1, and Cdc42 in Wt cells emphasizing the loss of copolarization and asymmetry in ScribbleΔ/Δ HSC.
Active Cdc42 is a crucial regulator in HSC aging and specifically Cdc42 allocation accurately predicts asymmetric potential and fate.1,2,45,46 We observed a significant (∼60%) decrease in Cdc42-GTP (active) levels after induced deletion of Scribble in BM progenitors through activated effector PAK pull-down experiments (supplemental Figure 3E-F). To determine whether the changes in Cdc42 activity corresponded with the scaffolding ability of Scribble over Yap1, we analyzed protein interactions between Scribble and Cdc42/Cdc42-GTP, as well as for Yap1 and Cdc42/Cdc42-GTP through PLA. We report interactions of polarized Scribble with Cdc42 and Cdc42-GTP47,48 for 80% of HSC that were completely lost upon deficiency of Scribble (supplemental Figure 3G-I). The Yap1/Cdc42 and Yap1/Cdc42-GTP complexes were present in the majority of HSC (∼60% of HSC) in which both proteins complex in an asymmetric polarized manner (Figure 4D-F). Upon loss of Scribble, the frequency of HSC which displayed a copolarized state were significantly reduced by one-half, whereas HSC acquired a nonpolar interaction state or even no PLA signal, indicating a loss of proximity between Yap1 and Cdc42/Cdc42-GTP (Figure 4D-F). Taken together, distinct cellular polarization states were identified in which Scribble is required for polarized cytoplasmic distribution of Yap1 with Cdc42 in HSC (Figure 4G).
HSC Scribble deficiency results in enhanced long-term self-renewal capacity
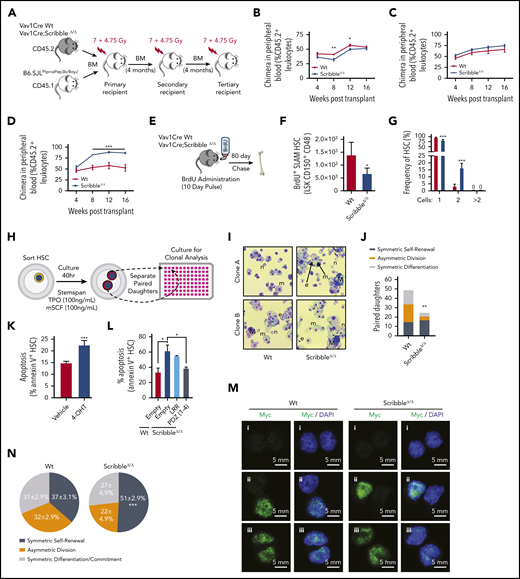
Despite the Scribble-dependent changes in the polarization of Yap1 with Cdc42 in HSC, hematopoietic-specific, constitutive ScribbleΔ/Δ HSC did not display any significant changes in PB white blood cell, neutrophil, and reticulocyte counts under basal hematopoietic activity (data not shown), similar to a previous report.13 We next examined HSC fitness with a serial competitive repopulation assay (Figure 5A). Analysis of the output hematopoiesis revealed a decreased reconstitution potential from ScribbleΔ/Δ BM during early stages (weeks 4-12) in primary transplant recipients (Figure 5B). However, the long-term repopulation of ScribbleΔ/Δ HSC was unchanged in PB (Figure 5B) and BM (supplemental Figure 4A-B). Additionally, we observed a significant expansion of HSC/P populations in the BM of these primary recipients (supplemental Figure 4B). A competitive advantage of ScribbleΔ/Δ HSC becomes apparent and statistically significant in subsequent tertiary recipients (Figure 5C-D; supplemental Figure 4C-D), which phenocopies the effect of conditional deletion of Lgl1.12 Chimera levels varied dramatically between Wt tertiary recipients where we observed a myeloid lineage bias consistent with aged hematopoiesis.49-51 However, the BM content of ScribbleΔ/Δ chimeric mice was 16-fold higher in lymphoid primed multipotent progenitors (supplemental Figure 4D) with increased multilineage differentiation potential in the PB, spleen, and BM (supplemental Figure 4E-G). Spleen weights of these animals were similar (supplemental Figure 4H) after tertiary reconstitution and we detected no evidence of myeloproliferation. Thus, serial competitive transplantation reveals that ScribbleΔ/Δ HSC have increased long-term repopulation ability with persistent self-renewal and maintained multilineage differentiation potential. These results are at odds with the expected organism-based HSC aging in which a loss of self-renewal and a myeloid lineage bias would be expected,52 suggesting that a process other than HSC aging is at play.
ScribbleΔ/Δ hematopoietic reconstitution develops a competitive advantage when serially transplanted by maintaining self-renewal divisions. (A) Schematic representing a serial competitive repopulation assay (CRA). Equal amounts of BM from CD45.2+ Vav1Cre Wt and Scribble null mice mixed with congenic CD45.1+ B6.SJLPtprcaPep3b/BoyJ competitor cells were transplanted at a 1:1 ratio into lethally irradiated B6.SJLPtprcaPep3b/BoyJ recipients. (B-D) PB chimera (CD45.2+ leukocytes) of (B) primary, (C) secondary, and (D) tertiary recipients. (E) Schematic representing a long-term BrdU incorporation assay in which animals freely imbibed water containing BrdU (1 mg/mL) for 10 days. Animals were euthanized after 80 days post-BrdU administration to quantify quiescence within hematopoietic stem and progenitor (HSC/P) populations determined by BrdU retention (BD Pharmingen intracellular staining kit: anti-BrdU, Alexa 488). (F) Absolute BrdU retaining (BrdU+) HSC assessed by FACS analysis of BM from mice as described in panel E. (G) Division of sorted and individually deposited HSC depicting the averages and standard deviations of the relative number (%) of wells containing the indicated number of cells after 20 hours in culture. Four independent experiments, n > 450 HSC. (H) Schematic of an in vitro paired daughter cell assay to assess fate decisions among individually sorted HSC. (I) Representative cytospin images of paired daughters. m, macrophage; n, neutrophil; e, erythrocyte; and M, megakaryocyte. (J) Absolute number of paired daughter cells analyzed for division modality, assessed as the presence or absence of full multilineage differentiation potential among individual paired daughter clones. n of paired daughter separations >200. Chi-squared analysis. (K) Cell death analysis using Annexin V and 7-AAD staining on Rosa26Cre;Scribblefl/fl HSC after 40 hours of culture with 4-OH tamoxifen. (L) Cell death analysis using Annexin V staining in Lin− Sca-1+ c-kit+ BM cells isolated from Vav1Cre;Wt or Vav1Cre;ScribbleΔ/Δ mice transduced with EMPTY (Ef1α-IRES-RFP) lentivirus or Scribble structure-function mutants as indicated. (M) Immunofluorescence depicting fate determinant allocation of Myc and the corresponding HSC division mode in Wt and ScribbleΔ/Δ paired daughter HSC (nocodazole, 10 nM for 24 hours). Low Myc expression in (i) paired daughter cells represents symmetric self-renewal, (ii) high and low Myc expression between the 2 daughters represents an asymmetric division, whereas (iii) high Myc expression in both cells is indicative of symmetric commitment. (N) Quantification of fate determinant allocation and division mode among Wt and ScribbleΔ/Δ paired daughter HSC. *P < .05, **P < .01, and ***P < .001.
ScribbleΔ/Δ hematopoietic reconstitution develops a competitive advantage when serially transplanted by maintaining self-renewal divisions. (A) Schematic representing a serial competitive repopulation assay (CRA). Equal amounts of BM from CD45.2+ Vav1Cre Wt and Scribble null mice mixed with congenic CD45.1+ B6.SJLPtprcaPep3b/BoyJ competitor cells were transplanted at a 1:1 ratio into lethally irradiated B6.SJLPtprcaPep3b/BoyJ recipients. (B-D) PB chimera (CD45.2+ leukocytes) of (B) primary, (C) secondary, and (D) tertiary recipients. (E) Schematic representing a long-term BrdU incorporation assay in which animals freely imbibed water containing BrdU (1 mg/mL) for 10 days. Animals were euthanized after 80 days post-BrdU administration to quantify quiescence within hematopoietic stem and progenitor (HSC/P) populations determined by BrdU retention (BD Pharmingen intracellular staining kit: anti-BrdU, Alexa 488). (F) Absolute BrdU retaining (BrdU+) HSC assessed by FACS analysis of BM from mice as described in panel E. (G) Division of sorted and individually deposited HSC depicting the averages and standard deviations of the relative number (%) of wells containing the indicated number of cells after 20 hours in culture. Four independent experiments, n > 450 HSC. (H) Schematic of an in vitro paired daughter cell assay to assess fate decisions among individually sorted HSC. (I) Representative cytospin images of paired daughters. m, macrophage; n, neutrophil; e, erythrocyte; and M, megakaryocyte. (J) Absolute number of paired daughter cells analyzed for division modality, assessed as the presence or absence of full multilineage differentiation potential among individual paired daughter clones. n of paired daughter separations >200. Chi-squared analysis. (K) Cell death analysis using Annexin V and 7-AAD staining on Rosa26Cre;Scribblefl/fl HSC after 40 hours of culture with 4-OH tamoxifen. (L) Cell death analysis using Annexin V staining in Lin− Sca-1+ c-kit+ BM cells isolated from Vav1Cre;Wt or Vav1Cre;ScribbleΔ/Δ mice transduced with EMPTY (Ef1α-IRES-RFP) lentivirus or Scribble structure-function mutants as indicated. (M) Immunofluorescence depicting fate determinant allocation of Myc and the corresponding HSC division mode in Wt and ScribbleΔ/Δ paired daughter HSC (nocodazole, 10 nM for 24 hours). Low Myc expression in (i) paired daughter cells represents symmetric self-renewal, (ii) high and low Myc expression between the 2 daughters represents an asymmetric division, whereas (iii) high Myc expression in both cells is indicative of symmetric commitment. (N) Quantification of fate determinant allocation and division mode among Wt and ScribbleΔ/Δ paired daughter HSC. *P < .05, **P < .01, and ***P < .001.
We recapitulated these in vivo findings using stroma-dependent cobblestone area-forming cell (CAFC) frequency analysis (supplemental Figure 4I). ScribbleΔ/Δ BM contains a reduced frequency (∼60%) in the number of day 7 CAFC, which parallels the lag in early reconstitution after primary transplant, whereas the frequency of late (days 28-35) CAFC (corresponding to HSC) is three- to fivefold increased over Wt BM (supplemental Figure 4J), indicating enhanced self-renewal potential. Taken together, these experiments provide evidence that polarized Scribble plays a functional role in hematopoiesis, regulating HSC self-renewal abilities and overall fitness upon serial BM transplantation. Importantly, expression of a transcriptionally competent, gain-of-function Yap1 mutant (Yap1 S112A) in HSC (supplemental Figure 4K-L) did not alter either survival when challenged with serial myeloablative stress (supplemental Figure 4M) or HSC competition repopulation abilities, complementary with a previous report27 (supplemental Figure 4N). These data indicate that the functional phenotypes associated with ScribbleΔ/Δ HSC is not driven by active Yap1-dependent nuclear functions.
Scribble deficiency decreases survival of nascent HSC daughter cells while sparing the frequency of low Myc, symmetric self-renewal divisions
Similar to the loss of quiescence observed when both Yap1/Taz are deleted in HSC (Figure 1C), ScribbleΔ/Δ HSC showed a decrease in the BM content of G0 HSC and a concomitant increase in the content of cycling HSC in the S/G2 and M phases (supplemental Figure 5A). This loss of quiescence became more apparent when we analyzed 5-bromodeoxycytidine (BrdU) incorporation and retention in vivo53 (Figure 5E). ScribbleΔ/Δ mice displayed a 50% reduction in the content of BrdU-retaining (also called “dormant”53 ) BM HSC and HSC/P (Figure 5F; supplemental Figure 5B).
As a method to assess division kinetics at the single-cell level, we sorted individual HSC into single wells and monitored their divisional history up to 40 hours in vitro. Consistent with our in vivo bulk HSC findings, roughly 20% of ScribbleΔ/Δ HSC have already undergone 1 cellular division (yielding 2 cells within the same well) within 20 hours, whereas such an event for Wt HSC is nearly unobservable54,55 (Figure 5G; supplemental Figure 5C). By 40 hours of culture, both Wt and ScribbleΔ/Δ HSC have reached similar levels of primary divisions (supplemental Figure 5D). Clonal analysis of these paired daughter cells revealed altered division modalities (fate decisions) among ScribbleΔ/Δ HSC (Figure 5H-I; supplemental Figure 5E). ScribbleΔ/Δ HSC were enriched in self-renewing divisions (Figure 5J; supplemental Figure 5F). Scribble depletion correlates with increased apoptosis of HSC upon induced, tamoxifen-driven in vitro deletion of Scribble in Rosa26CreERTi2;ScribbleΔ/Δ BM HSC (Figure 5K). Furthermore, when dividing cells were analyzed by time-lapse cinematography at times between 20 and 50 hours of culture in medium containing a permeable activated caspase 3/7 dependent fluorescent dye, we observed the frequent presence of apoptotic events in Scribble-deficient daughter cells (∼25%) compared with Wt (∼3%) daughter cells (supplemental Videos 1 and 2). Notably, the PDZ domain of Scribble introduced into ScribbleΔ/Δ stem and progenitor cells suffices to successfully restore survival of HSC/P to levels comparable to those of Wt HSC/P transduced with the control empty vector (Figure 5L). However, reconstitution with the LRR containing mutant fails to revert the apoptosis levels seen in the ScribbleΔ/Δ cells (Figure 5L).
Finally, we analyzed the fate allocation, as assessed by Myc activation and translocation,31 within paired daughter cells before the observed apoptosis. Analysis of nuclear Myc in paired daughter cells revealed that ScribbleΔ/Δ HSC indeed have increased symmetric self-renewal divisions in vivo, at the expense of asymmetric divisions (Figure 5M-N; supplemental Figure 5G). Altogether, these results support the concept of in vivo selection of self-renewing HSC clones in hematopoietic-specific Scribble deficiency. Collectively, our data prove that Scribble controls asymmetric division potential and fate through the PDZ mediated scaffolding of cytosolic Yap1 with Cdc42-GTP.
Additional deficiency of Scribble restores HSC fitness of Yap1/TazΔ/Δ HSC and associates with Rac activation
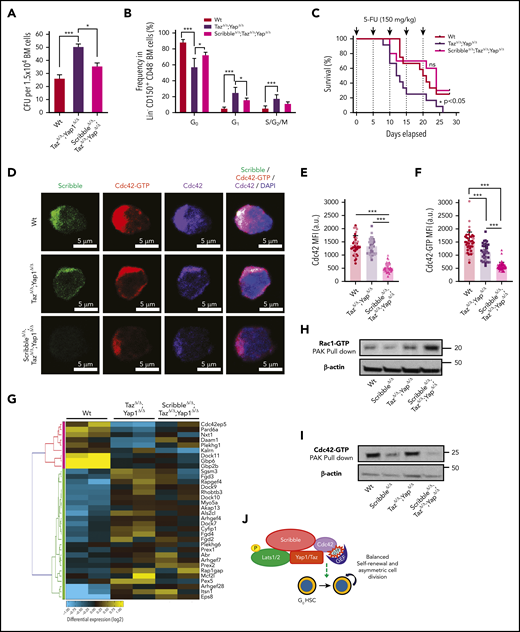
Given that the loss of Yap1/Taz severely diminishes HSC quiescence and survival after 5-FU (Figure 1) and that we have correlated such phenotypes with a Scribble-dependent cytosolic sequestration of Yap1 to modulate Cdc42-GTP activity, we wondered if the loss of all 3 proteins, Yap1, Taz, and Scribble, would result in further decreased quiescence with increased sensitivity to 5-FU. To our surprise, the triple deficiency significantly restored the CFU formation (Figure 6A) and the level of HSC quiescence (Figure 6B) observed in the double Yap1/TazΔ/Δ scenario to Wt levels, and as a result, restored survival following serial administration of 5-FU (median survival of 12.5 vs 24 days, P < .05; Figure 6C). These data indicate that the additional deficiency of Scribble in Yap1/Taz-mutant HSC restores quiescence and HSC fitness.
Deficiency of Scribble restores fitness of Yap/TazΔ/Δ HSC and associates with Rac activation. (A) Number of colony-forming units (CFU) from BM cells. (B) Cell-cycle analysis of Lin− CD48− CD150+ HSC harvested from the BM of Mx1Cre Wt, TazΔ/Δ;Yap1Δ/Δ, and ScribbleΔ/Δ;TazΔ/Δ;Yap1Δ/Δ mice 1 week after poly I:C. Stages of cell cycle were assessed by FACS analysis with incorporation of DNA binding Hoechst 33342 (2 mg/mL) and nucleotide binding PyroninY (0.25 mg/mL). (C) Kaplan-Meier survival analysis after serial myeloablation with 5-fluorouracil (150 mg/kg) 5 days apart. (D) Immunofluorescence of HSC showing Cdc42 expression and localization along with active Cdc42-GTP expression and localization. Nuclei are counterstained with DAPI and merged images are shown in the right micrographs. Scale bar 5 µm. (E) Quantification of Cdc42 expression measured by MFI. (F) Quantification of Cdc42-GTP expression measured by MFI. (G) Heat map depicting genes within the Rho guanyl nucleotide exchange factor activity and small GTPase activity gene ontology pathways showing differential regulation between Mx1Cre Wt, TazΔ/Δ;Yap1Δ/Δ, and ScribbleΔ/Δ;TazΔ/Δ;Yap1Δ/Δ HSC. (H) Rac1/Cdc42 activation PAK pulldown on lineage-depleted (Lin−) BM cells. (I) Cdc42 effector PAK pull-down from on Lin− BM cells. (J) Cartoon depicting a complete picture of the Scribble polarity complex and its components in relation with HSC self-renewal and cell-cycle entry. *P < .05 and ***P < .001.
Deficiency of Scribble restores fitness of Yap/TazΔ/Δ HSC and associates with Rac activation. (A) Number of colony-forming units (CFU) from BM cells. (B) Cell-cycle analysis of Lin− CD48− CD150+ HSC harvested from the BM of Mx1Cre Wt, TazΔ/Δ;Yap1Δ/Δ, and ScribbleΔ/Δ;TazΔ/Δ;Yap1Δ/Δ mice 1 week after poly I:C. Stages of cell cycle were assessed by FACS analysis with incorporation of DNA binding Hoechst 33342 (2 mg/mL) and nucleotide binding PyroninY (0.25 mg/mL). (C) Kaplan-Meier survival analysis after serial myeloablation with 5-fluorouracil (150 mg/kg) 5 days apart. (D) Immunofluorescence of HSC showing Cdc42 expression and localization along with active Cdc42-GTP expression and localization. Nuclei are counterstained with DAPI and merged images are shown in the right micrographs. Scale bar 5 µm. (E) Quantification of Cdc42 expression measured by MFI. (F) Quantification of Cdc42-GTP expression measured by MFI. (G) Heat map depicting genes within the Rho guanyl nucleotide exchange factor activity and small GTPase activity gene ontology pathways showing differential regulation between Mx1Cre Wt, TazΔ/Δ;Yap1Δ/Δ, and ScribbleΔ/Δ;TazΔ/Δ;Yap1Δ/Δ HSC. (H) Rac1/Cdc42 activation PAK pulldown on lineage-depleted (Lin−) BM cells. (I) Cdc42 effector PAK pull-down from on Lin− BM cells. (J) Cartoon depicting a complete picture of the Scribble polarity complex and its components in relation with HSC self-renewal and cell-cycle entry. *P < .05 and ***P < .001.
To determine whether Cdc42 expression and/or activation were modified by the triple deficiency of Scribble/Yap1/Taz, we performed single-cell analysis of the localization of Cdc42 and Cdc42-GTP. Immunofluorescence revealed that the combined deficiency of Yap1/Taz reduces the levels of Cdc42-GTP by ∼25% in HSC without affecting the expression level of Cdc42 (Figure 6D-F). Our data confirmed that the triple deficiency of Yap1, Taz, and Scribble resulted in further decrease (∼60%) in active Cdc42 expression with a comparable decrease of the overall Cdc42 protein (Figure 6D-F), indicating that the additional deficiency of Scribble results also in loss of Cdc42 protein rather than exclusively loss of its activation.
Because Cdc42 activity was not restored like the function of triple-deficient HSC, we compared the transcriptome of HSC from Mx1Cre;Wt, Mx1Cre;Yap1Δ/Δ;TazΔ/Δ, and Mx1Cre;ScribbleΔ/Δ;Yap1Δ/Δ;TazΔ/Δ to determine whether specific transcriptional programs are modulated in triple-deficient HSC, leading to the overall neutralizing functional effect (supplemental Figure 6A). By applying a filter on the same set of genes found differentially expressed in Mx1Cre;ScribbleΔ/Δ (supplemental Figure 3A-C) and Mx1Cre;Yap1Δ/Δ;TazΔ/Δ (Figure 4A-C), we identified that the Cdc42-specific GEFs, such as Fgd4, were largely neutralized in Mx1Cre;ScribbleΔ/Δ;Yap1Δ/Δ;TazΔ/Δ HSC (Figure 6G), and there was an upregulation of the expression of GEFs with positive activity on Rac proteins, specifically Prex142 (supplemental Figure 6B). Consistent with this upregulation in guanine nucleotide exchange factors specific for Rac1, we observed an approximate twofold upregulation of active Rac (Rac-GTP; Figure 6H) and ∼75% inhibition of Cdc42 activation (Cdc42-GTP; Figure 6I) in Mx1Cre;ScribbleΔ/Δ;Yap1Δ/Δ;TazΔ/Δ stem and progenitor cells. Increased Rac1 levels have recently been reported to increase HSC repopulating capacity both in vitro and in vivo,56 supporting the thesis that the increased activation of Rac in triple-deficient HSC is involved in the restoration of quiescence and animal survival after serial administration of 5-FU, confirming the complex interplay of the Rho GTPase subfamily of proteins in the control of HSC activity.
Discussion
In this report, we have identified that the combined deficiency of Yap1/Taz results in loss of HSC fitness evidenced by decreased competitive repopulation ability in serial transplantations and survival upon multiple cycles of 5-FU chemotherapy.57-59 This effect may be independent of transcriptional regulation by Yap1 because the expression of an active mutant of Yap1 does not alter the hematopoietic reconstitution. Rather, the role of Yap1/Taz in HSC seems to relate to their effect on polarity-based cell division and differentiation decisions controlled by the polarity master regulator Cdc42. However, the fact that the canonical signaling pathway downstream of Cdc42, aPKC/Par3/Par6, does not impair hematopoiesis11 suggests that a mechanism of Yap1 regulation via direct phosphorylation by PKCζ18 is dispensable in HSC where an alternative pathway may control HSC fitness. Our data provide evidence for a functional complex containing Scribble, Yap1, p-Lats, and Cdc42/Cdc42-GTP in dividing HSC that controls quiescence, fate, and fitness (Figure 6J).
Scribble deficiency in HSC results in loss of function of HSC activity in primary recipients of competitive grafts but results in hematopoietic gain of function in tertiary recipient animals, phenocopying the effects of Lgl1 deficiency.12 The positive regulatory role of Scribble in tertiary recipients seems to depend on a Lgl1-independent pathway as the LRR domains of Scribble (responsible for binding Lgl1) are largely dispensable for overall Yap1 polarization. In this report, we identified Scribble as a Yap1/Lats1 polarizing scaffold molecule in quiescent HSC. Upon forced division, deletion of Scribble in HSC disrupts the polarized cytosolic placement of Yap1 and increases apoptosis of daughter cells while impairing asymmetric cell divisions and maintaining symmetric self-renewal divisions with low levels of Myc activation. The survival of dividing HSC is restored by expression of the PDZ domain-containing mutant of Scribble. As a result, even after tertiary transplantation in which HSC aging may be at play, Scribble-deficient hematopoiesis seems to maintain its self-renewing HSC activity. Mechanistically, Scribble is required to retain cytosolic Yap1 polarization, whereas cytosolic Yap1 is required to maintain progenitor colony formation. The amino-terminus LRR domain of Scribble is required for polarized localization of the upstream negative regulator of Yap1, phosphorylated (Thr1079) Lats1; however, this domain could not rescue Yap1 polarization and function alone. We also identified that cytosolic Scribble and Yap1/Taz are required for activation of Cdc42 and the combined loss of Scribble, Yap1/Taz results in compensatory upregulated transcriptional expression of Rac-specific GEF, Rac activation, and HSC fitness restoration. The gene regulatory network associated with the transcriptional upregulation of specific GEFs in Scribble/Yap1/Taz triple-deficient HSC remains unknown. The fact that the phenotypes identified with Yap1/Taz and Scribble deficiency require a combination of inflammation and either transplantation or chemotherapy suggest that this system is active upon stress conditions. Specific inflammatory stressors may result in activation signaling pathways that require the expression of the complex Yap1-Taz/Scribble/Cdc42.
The roles of Cdc42 and Rac in HSC self-renewal are well established. Gene-targeted deletion of Cdc42 in mice leads to an increase in the HSC/P population in the BM because of enhanced cell-cycle progression, leading to HSC exhaustion and impaired long-term engraftment potential.4 Rac GTPases play a crucial role in homing, migration, interaction with the microenvironment, and long-term engraftment potential of HSC/Ps.60-63 Gain of function of Rac has been recently associated with increased HSC repopulating capacity.56 Similarly, the gain of function of RhoA prevents asymmetric distribution of activated p38 MAPK of regenerating HSC in the context of transplantation.7 Our data strongly indicates that the deficiency of Yap1/Taz and Scribble commonly result in changes in the transcriptome of GEFs for Cdc42, Rac, or RhoA. These changes are consistent with compensatory upregulation of the expression of Cdc42 GEFs and modest upregulation of Rac GEFs in both Yap1/Taz and ScribbleΔ/Δ HSC. Overall, the triple deficiency of Scribble/Yap1/Taz results in upregulation of the Rac-specific GEF Prex1 and restored expression of the Cdc42-specific GEF Fgd4 to Wt levels. A GST-PAK pulldown for Rac confirmed that the triple deficiency of Scribble/Yap1/Taz does result in increased Rac activity and decreased Cdc42 activity. Activated Rac-mediated HSC quiescence would explain the restoration of hematopoietic fitness of Scribble/Yap1/Taz deficient mice undergoing serial 5-FU administration (supplemental Figure 6C).
Our data provide the first genetic demonstration that Hippo-regulated Yap1 controls HSC quiescence and fitness, whereas the combined Scribble/Yap1/Taz complex controls Cdc42 activity and HSC fate through survival of asymmetrically dividing HSC.
The data reported in this article have been deposited in the Gene Expression Omnibus database (accession number GSE151465).
Data that is not required publicly can be obtained from M.J.A. and J.A.C.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported in part by grants from the National Institutes of Health National Heart, Lung, and Blood Institute F31 HL132468 (M.J.A.) and R01 HL147536 (J.A.C.); National Cancer Institute R01 CA193350 (Y.Z.), R01 CA192642 and R01 CA218254 (M.T.D.-M.), R01 CA211794 (J.M.), and P30 CA030199 (J.M.); and National Institute of Diabetes and Digestive and Kidney Diseases R01 DK104814 (Y.Z.) and R01 DK108743 (J.M.); and the Albert J Ryan Foundation and Cardell Fellowship for Excellence in Graduate Research (M.J.A.).
Authorship
Contribution: M.J.A., R.C.N., S.H., A.M.W., and B.B. performed experiments; M.J.A. and J.A.C. analyzed data and wrote the manuscript; M.J.A. and J.A.C. designed experiments; M.-D.F., M.X., Q.R.L., H.G., Y.Z., M.T.D.-M., and J.M. provided critical reagents, insightful views and critiques and edited the manuscript; and J.A.C. led the group and supervised M.J.A.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jose A. Cancelas, Division of Experimental Hematology and Cancer Biology, Cincinnati Children’s Hospital Medical Center, Hoxworth Blood Center, University of Cincinnati Academic Health Center, 3130 Highland Ave., Cincinnati, OH, 45267; e-mail: jose.cancelas@uc.edu; jose.cancelas@cchmc.org.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal