Abstract
Improved personalized adjustment of primary therapy to the perceived risk of relapse by using new prognostic markers for treatment stratification may be beneficial to patients with acute lymphoblastic leukemia (ALL). Here, we review the advances that have shed light on the role of IKZF1 aberration as prognostic factor in pediatric ALL and summarize emerging concepts in this field. Continued research on the interplay of disease biology with exposure and response to treatment will be key to further improve treatment strategies.
Background
Pediatric acute lymphoblastic leukemia (ALL), the most common malignancy observed in children and adolescents, is characterized by broad clinical and biological heterogeneity that is largely sustained by a diverse background of disease-initiating and -maintaining recurrent structural and/or numerical genetic aberrations acquired by the leukemic clone.1-5 Despite this heterogeneity, overall treatment results in pediatric ALL are one of the true success stories in clinical oncology with current cure rates exceeding 85%.1,4 On modern clinical protocols, this is achieved by the application of risk-adapted therapy, reflecting the probability of treatment failure. For this purpose, prognostic factors are used to estimate an individual patient’s risk of relapse and to adjust the required treatment intensity by patient stratification into different therapeutic risk groups, for example, standard, intermediate, high, and very high risk.1,4,5 Unfortunately, despite all of these efforts, a significant proportion of patients with ALL still experiences relapse. Thus, further improved personalized adjustment of primary therapy to the perceived risk of ALL relapse by using new prognostic markers for treatment stratification is likely to be beneficial to patients and, therefore, of great interest to those involved in the diagnosis and treatment of pediatric ALL and beyond.
Technical advances in the “omics” field fueled multiple comprehensive exploratory studies, and a number of candidate prognostic markers for ALL risk stratification have been identified and published during the last 10 years.1-5 Caregivers are thus faced with an ever-growing body of literature on new prognostic markers, unfortunately, mostly full of uncertainties about their clinical relevance. Therefore, it does not come as a surprise that the majority of newly described genetic or genomic markers are not regularly used to support decision-making procedures on current clinical protocols for ALL. General reasons for this lack of translation include (1) large heterogeneity across studies with differently sized, selected, and treated patient populations; (2) methodological differences in marker assessment and uncertainties regarding assay procedures; (3) differences in reported end points and conflicting outcomes; and (4) differences in the statistical analyses used, as well as other issues. One obvious difficulty in the practical implementation of new prognostic markers for ALL is the limited clinical significance conferred by many of the new high-risk markers, making it difficult to justify exposure to more intensive and toxic treatments for the potential benefit of a minority of patients among those identified by the marker.
The genetics of IKZF1
The IKZF1 gene is located on chromosome band 7p12.2, consists of 8 exons, and codes for the transcription factor IKAROS with key regulatory functions in lymphopoiesis.6,7 IKAROS harbors 6 zinc fingers. Four of these are located in the DNA-binding domain encoded by exons 4 to 6 and are essential to maintain IKAROS’ tumor-suppressor function. The remaining 2 zinc fingers are encoded by exon 8 and mediate the dimerization of IKAROS either as a homodimer or with other transcription factors of its family (eg, AIOLOS and HELIOS).6-8 IKZF1 has remained in the spotlight of the leukemia field since 2008 when Mullighan et al first described that recurrent, mostly monoallelic, focal deletions affect the coding regions of IKZF1.9,10 These deletions involve either the whole gene or only parts of IKZF1 and are observed at an overall frequency of ∼15% in pediatric and 40% in adult ALL cases.2,11,12 Although some deletions (eg, those affecting the whole gene, intragenic deletions including exons 2 and/or 8, deletions of exon 1 and 5′ untranscribed regulatory regions) result in haploinsufficiency, other deletions, most commonly those affecting exons 4 to 7, lead to loss of the DNA-binding domain and generation of dominant-negative isoforms.2,9-12 The latter compromise the tumor-suppressor function of IKAROS translated from the remaining wild-type allele. The frequency of somatic point mutations in IKZF1 has been studied far less extensively. They are present to a much lower extent (Figure 1) and, as with deletions, their molecular consequence can be either haploinsufficiency (eg, truncating variants) or a dominant-negative effect (Figure 1).10,13-19 Recently, a new B-cell precursor (BCP) ALL subgroup characterized by the IKZF1 missense mutation p.Asn159Tyr (N159Y) affecting the DNA-binding domain was identified through a distinct gene-expression profile.17,18 Of interest, the specific gene-expression profile also differed from those BCP ALL cases with other known IKZF1 alterations. In addition, an increasing number of cases with fusion transcripts involving IKZF1 have been described (Figure 1).17-26 Besides the somatic alterations described earlier in this section, frequent intronic germline variants in IKZF1 (rs11978267 and rs4132601) have been identified in genome-wide association studies and consistently described to modestly modulate the risk of pediatric BCP ALL (Figure 1).27,28 Of major importance, Churchman et al recently characterized IKZF1 as a leukemia predisposition gene by reporting mostly adverse germline IKZF1 variation in familial pediatric ALL and 43 of 4963 (0.9%) unselected BCP ALL patients29 (Figure 1).
Genetic alterations of the IKZF1 gene at chromosome band 7p12.2 in pediatric ALL. Red boxes indicate the observed approximate frequencies of the different types of genetic aberrations: deletions (bottom left), gene fusions (bottom right), and somatic as well as germline single-nucleotide variants (top). The 2 intronic germline risk variants identified in genome-wide association studies are indicated by their Reference SNP cluster ID. Frequencies (percentages) of the most common specific deletions (DEL) within the group of IKZF1-deleted ALL are indicated in black. Chr, chromosome; UTR, untranslated region.
Genetic alterations of the IKZF1 gene at chromosome band 7p12.2 in pediatric ALL. Red boxes indicate the observed approximate frequencies of the different types of genetic aberrations: deletions (bottom left), gene fusions (bottom right), and somatic as well as germline single-nucleotide variants (top). The 2 intronic germline risk variants identified in genome-wide association studies are indicated by their Reference SNP cluster ID. Frequencies (percentages) of the most common specific deletions (DEL) within the group of IKZF1-deleted ALL are indicated in black. Chr, chromosome; UTR, untranslated region.
Most of the published data on IKZF1 deletions in pediatric ALL have been generated by multiplex ligation probe-dependent amplification (MLPA) analysis (Table 1).13,30-44 Alternative techniques include polymerase chain reaction (PCR) or array techniques.10,14,15,30,39,40,45,46 MLPA detects virtually all deletions targeting the coding sequence, and most large studies evaluating the prognostic effect of IKZF1 deletion in patient cohorts relied on this technique (Table 1). However, like array techniques, it cannot reliably detect deletions present in <25% of cells. Thus, MLPA may fail to detect IKZF1 deletions in samples showing a low leukemia burden or deletions that are limited to a leukemic subclone only. For recurrent intragenic deletions, this limitation can be overcome by quantitative PCR assays. This technique allows detection of intragenic IKZF1 deletions with a higher sensitivity than classical techniques and, furthermore, allows minimal residual disease (MRD) monitoring.47 However, it has to be emphasized that the prognostic impact of subclonal IKZF1 deletions has not been specifically evaluated yet and may differ from that of full clone deletions. Another debated issue relates to isolated IKZF1 exon 1 deletions and whether they should be considered in routine diagnosis or not. Their accurate detection can be complicated due to false MLPA positivity favored by a high guanine-cytosine content in this region. Molecular mapping of exon 1 deletions together with an adapted MLPA protocol with a reinforced denaturation step recently confirmed the frequency and impact of exon 1 deletions on IKZF1 transcription48 and suggest that these deletions should be considered for diagnosis when properly controlled. It is noteworthy that additional IKZF1 deletions targeting the 5′ regulatory regions have been described and are not detected by current MLPA protocols.9,48 IKZF1 sequence mutations were mainly analyzed by Sanger sequencing and targeted or nontargeted next-generation sequencing strategies whereas the majority of fusion genes have been identified in transcriptomic studies.10,13-26,29 The different qualities and sensitivities of these techniques need to be considered when comparing frequency and type of IKZF1 aberrations in the literature.
Summary of studies on the prognostic impact of IKZF1 aberration in treatment trials of pediatric ALL
| Trial . | Ref. . | Country . | Trial period . | No. of patients . | Type of ALL . | Frequency of aberrant IKZF1 (method of detection) . | EFS (DFS) . | OS . | Cumulative incidence of relapse . | Comments . |
|---|---|---|---|---|---|---|---|---|---|---|
| COG P9906, St. Jude Total XI, XII, XIII, XIV, XV, and Interfant-99 | 10 | US | COG P9906: 2000-2003 | COG P9906: 221 | High-risk BCP | 28.6% (SNP array, Sanger sequencing) | 25% vs 73% (at 5 y), P < .0001 | 73% vs 25% (at 5 y), P < .0001 | Groundbreaking first study demonstrating prognostic impact of IKZF1 deletion; includes 5 patients with IKZF1 point mutation in the COG cohort and 21 BCR-ABL1–rearranged cases in the St. Jude cohort | |
| St. Jude and Interfant: 1986-2007 | St. Jude and Interfant: 258 | BCP | 18.6% (SNP array) | 40% vs 72% (at 10 y), P < .001 | 48% vs 26% (at 10 y), P = .004 | |||||
| DCOG ALL9 | 13, 48 | The Netherlands | 1997-2000 | 131 | BCP | 13.0% (MLPA, Sanger sequencing) | 39% vs 89% (at 8 y), P < .001 | 56.0% vs 91.0% (at 8 y), P < .001 | Exemplifies the importance of IKZF1 deletion as a prognostic factor, especially in non–high-risk patients; included 2 BCR-ABL1–rearranged cases; subsequent study in related population demonstrated the prognostic power of integrated use of aberrant IKZF1 and MRD levels | |
| TPOG-ALL-93, TPOG-97-VHR, TPOG-ALL-2002 | 14 | Taiwan | 1995-2009 | 242 | BCP | 10.7% (PCR) | 15% vs 76% (at 10 y), P < .0001 | 38% vs 78% (at 10 y), P = .0016 | First study indicating poor prognostic effect of IKZF1 deletion in an Asian population | |
| DCOG ALL8, ALL9, ALL10, UKALL 97, 97/99, 2003 | 30 | The Netherlands, UK | 1997-2006 | DCOG: 34 | Down syndrome ALL | DCOG: 35.3% (CGH array, MLPA, Sanger sequencing) | DCOG: 45% vs 95% (at 6 y), P = .002 | DCOG: 66% vs 95% (at 6 y), P = .02 | DCOG: 37% vs 5% (at 6 y), P = .044 | Strong prognostic effect of IKZF1 deletion in Down syndrome ALL |
| UKALL: 85 | UKALL: 27.1% (MLPA) | UKALL: 21% vs 58% (at 6 y), P = .002 | UKALL: 15% vs 71% (at 6 y), P = .002 | UKALL: 37% vs 18% (at 6 y), P = .06 | ||||||
| Japan Childhood Leukemia Study ALL02 | 31 | Japan | 2002-2008 | 202 | BCP | 9.4% (MLPA) | 63% vs 89% (at 5 y),P = ..001 | 72% vs 90% (at 5 y),P = ..02 | Particularly strong prognostic effect of IKZF1 deletion in the NCI high-risk group | |
| JCCLSG ALL 2004 | 32 | Japan | 2004-2008 | 177 | BCP | 12.0% (MLPA) | 68% vs 85% (at 4 y), P = .04 | Prognostic effect of IKZF1 deletion more pronounced in high-risk patients | ||
| ALL-REZ BFM 2002 | 33 | Germany, Austria, Switzerland | 2002-2009 | 204 | BCP relapse | 33.3% (MLPA) | 30% vs 51% (at 5 y), P = .002 | 36% vs 60% (at 5 y), P = .001 | 41% vs 23% (at 5 y), P = .006 | Prognostic impact of IKZF1 deletion also in second-line treatment of relapsed ALL; one-quarter of IKZF1 deletions was acquired at relapse |
| AIEOP-BFM ALL 2000 | 34 | Germany | 1999-2005 | 694 | BCP and T ALL | 12.0% (MLPA) | 69% vs 85% (at 5 y), P < .0001 | 82% vs 92% (at 5 y), P = .003 | 21% vs 10% (at 5 y), P = .001 | Pronounced prognostic effect of IKZF1 deletion in the intermediate-risk group |
| AIEOP-BFM ALL 2000 | 35 | Italy | 2003-2005 | 410 | BCP | 13.2% (MLPA) | 70% vs 85% (at 5 y), P = .007 | 87% vs 93% (at 5 y), P = .100 | 24% vs 13% (at 5 y), P = .049 | Due to the modest effect of IKZF1 deletion observed, the relevance of IKZF1 deletion as a clinically useful stratification factor is debated |
| DCOG ALL8, ALL9, ALL10, COALL-97, COALL-03 | 36 | The Netherlands, Germany | 1991-2012 | 857 | BCP | 15.9% (MLPA) | 34% vs 13% (at 5 y), P = <.001 | Confirms and extends the strong prognostic effect of IKZF1 deletion; IKZF1 deletion remained predictive in intermediate risk patients on the MRD-guided DCOG ALL10 trial | ||
| EsPhALL and I-BFM SG studies (for pre-TKI assessment) | 37 | Austria, Czech Republic, France, Germany, The Netherlands, UK | 1995-2005 | Pre-TKI cohort: 84 EsPh-ALL cohort: 107 | BCR-ABL1–rearranged BCP | 66.0% (MLPA, CGH array, SNP array) | DFS pre-TKI: 30% vs 58% (at 4 y), P = .013 | Pre-TKI: 49% vs 75% (at 4 y), P = .075 | Pre-TKI: 57% vs 21% (at 4 y), P = .026 | IKZF1 deletion is demonstrated to be a poor prognostic marker in BCR-ABL1–rearranged pediatric ALL independent of imatinib treatment |
| DFS EsPhALL: 54% vs 63% (at 4 y), P = .168 | EsPhALL: 58% vs 83% (at 4 y), P = .070 | EsPhALL: 28% vs 31% (at 4 y), P = .817 | ||||||||
| DFS EsPhALL TKI-treated good-risk patients: 56% vs 75% (at 4 y), P = .051 | EsPhALL TKI-treated good-risk patients: 66% vs 100% (at 4 y), P = .039 | EsPhALL TKI-treated good-risk patients: 29% vs 25% (at 4 y), P = .249 | ||||||||
| UKALL trials ALL 97/99 and ALL 2003 | 38 | UK, Ireland | UKALL 97/99: 1997-2002 | UKALL 97/99: 864 | BCP | 13.4% (MLPA) | 56% vs 80% (at 5 y), P = <.001 | 70% vs 89% (at 5 y), P = <.001 | 40% vs 18% (at 5 y), P = <.001 | Integration of cytogenetic with genomic data including IKZF1 deletion refines risk groups in a clinically meaningful way |
| UKALL 2003: 2003-2011 | UKALL 2003: 782 | BCP | 11.1% (MLPA) | 80% vs 92% (at 5 y), P = <.001 | 86% vs 95% (at 5 y), P = <.001 | 16% vs 6% (at 5 y), P = .002 | ||||
| NOPHO ALL-1992, NOPHO ALL-2000, NOPHO ALL-2008 | 39 | Denmark, Finland, Norway, Sweden | 1992-2013 | 334 | BCP | 15.0% (SNP array, MLPA) | 60% vs 83% (at 10 y), P < .001 | 73% vs 89% (at 10 y), P = .001 | 35% vs 12% (at 10 y), P < .001 | Prognostic impact independent of WBC count and MRD; co-occurrence of PAR1 deletion increased prognostic impact of IKZF1 deletion |
| EORTC Children’s Leukemia Group study 58951 | 40 | Belgium, France, Portugal | 1998-2008 | 1223 | BCP | 14.6% (PCR, MLPA) | 68% vs 87% (at 5 y), P = <.001 | 87% vs 92% (at 5 y), P = .035 | IKZF1-aberrant BCP ALL benefited from vincristine-steroid pulses during maintenance treatment | |
| ALLR3 | 41 | UK, Ireland, The Netherlands, Australia, New Zealand | 2002-2013 | 222 | BCP relapse | 23.0% (MLPA) | 48% vs 53% (at 5 y), P = .30 | No prognostic impact of IKZF1 deletion at relapse | ||
| ANZCHOG ALL8 | 42 | Australia, New Zealand | 2002-2011 | 475 | BCP standard and intermediate risk | 10.5% (MLPA) | 53% vs 83% (at 7 y), P < .0001 | 78% vs 94% (at 7 y), P < .0001 | 41% vs 15% (at 7 y), P < .0001 | Strong prognostic effect of IKZF1 deletion in non–high-risk BCP ALL |
| ALL-BFM 95 | 45 | Germany | 1995-2000 | 655 | BCP intermediate risk | 12.2% (PCR) | 66% vs 82% (at 5 y), P = .001 | Vincristine-dexamethasone pulses during maintenance were not of benefit to IKZF1-deleted intermediate-risk patients | ||
| DFCI ALL Consortium Protocol 05-001 | 43 | US | 2005-2010 | 385 | BCP | 16.0% (MLPA) | 63% vs 88% (at 5 y), P < .001 | 79% vs 94% (at 5 y), P < .001 | 29% vs 8% (at 5 y), P < .001 | IKZF1 deletion was confirmed as an independent predictor of inferior outcome |
| Malaysia-Singapore ALL 2003, 2010 | 44 | Malaysia, Singapore | 2002-2017 | 665 | BCP | 15.9% (MLPA) | Without IKZF1 as high-risk criterion: 30% vs 8% (at 5 y), P < .001 | Demonstrates that intensifying treatment of IKZF1-deleted BCP ALL patients reduces risk of relapse | ||
| With IKZF1 as high-risk criterion: 14% vs 5% (at 5 y), P = .030 | ||||||||||
| COG Trial AALL0622 | 46 | US, Canada | 2008-2012 | 44 | BCR-ABL1–rearranged BCP | 56.8% (SNP array) | 52% vs 82% (at 5 y), P = .040 | 80% vs 100% (at 5 y), P = .040 | Confirms poor outcome for IKZF1-deleted BCR-ABL1–rearranged ALL on dasatinib-containing treatment regimen | |
| AIEOP-BFM ALL 2000 | 61 | Germany, Italy | 1999-2009 | 1408 | BCP | 14.6% (MLPA) | IKZF1-deleted, but not IKZF1plus: 77% vs 86% (at 5 y), P = .0005 | IKZF1-deleted, but not IKZF1plus: 89% vs 94% (at 5 y), P = .023 | IKZF1-deleted, but not IKZF1plus: 16% vs 11% (at 5 y), P = .045 | Definition of a poor MRD-dependent prognostic pattern termed IKZF1plus with IKZF1 deletions co-occurring with deletions in CDKN2A, CDKN2B, PAX5, or PAR1 in the absence of ERG deletion |
| IKZF1plus: 51% vs 86% (at 5 y), P < .0001 | IKZF1plus: 75% vs 94% (at 5 y), P < .0001 | IKZF1plus: 44% vs 11% (at 5 y), P < .0001 |
| Trial . | Ref. . | Country . | Trial period . | No. of patients . | Type of ALL . | Frequency of aberrant IKZF1 (method of detection) . | EFS (DFS) . | OS . | Cumulative incidence of relapse . | Comments . |
|---|---|---|---|---|---|---|---|---|---|---|
| COG P9906, St. Jude Total XI, XII, XIII, XIV, XV, and Interfant-99 | 10 | US | COG P9906: 2000-2003 | COG P9906: 221 | High-risk BCP | 28.6% (SNP array, Sanger sequencing) | 25% vs 73% (at 5 y), P < .0001 | 73% vs 25% (at 5 y), P < .0001 | Groundbreaking first study demonstrating prognostic impact of IKZF1 deletion; includes 5 patients with IKZF1 point mutation in the COG cohort and 21 BCR-ABL1–rearranged cases in the St. Jude cohort | |
| St. Jude and Interfant: 1986-2007 | St. Jude and Interfant: 258 | BCP | 18.6% (SNP array) | 40% vs 72% (at 10 y), P < .001 | 48% vs 26% (at 10 y), P = .004 | |||||
| DCOG ALL9 | 13, 48 | The Netherlands | 1997-2000 | 131 | BCP | 13.0% (MLPA, Sanger sequencing) | 39% vs 89% (at 8 y), P < .001 | 56.0% vs 91.0% (at 8 y), P < .001 | Exemplifies the importance of IKZF1 deletion as a prognostic factor, especially in non–high-risk patients; included 2 BCR-ABL1–rearranged cases; subsequent study in related population demonstrated the prognostic power of integrated use of aberrant IKZF1 and MRD levels | |
| TPOG-ALL-93, TPOG-97-VHR, TPOG-ALL-2002 | 14 | Taiwan | 1995-2009 | 242 | BCP | 10.7% (PCR) | 15% vs 76% (at 10 y), P < .0001 | 38% vs 78% (at 10 y), P = .0016 | First study indicating poor prognostic effect of IKZF1 deletion in an Asian population | |
| DCOG ALL8, ALL9, ALL10, UKALL 97, 97/99, 2003 | 30 | The Netherlands, UK | 1997-2006 | DCOG: 34 | Down syndrome ALL | DCOG: 35.3% (CGH array, MLPA, Sanger sequencing) | DCOG: 45% vs 95% (at 6 y), P = .002 | DCOG: 66% vs 95% (at 6 y), P = .02 | DCOG: 37% vs 5% (at 6 y), P = .044 | Strong prognostic effect of IKZF1 deletion in Down syndrome ALL |
| UKALL: 85 | UKALL: 27.1% (MLPA) | UKALL: 21% vs 58% (at 6 y), P = .002 | UKALL: 15% vs 71% (at 6 y), P = .002 | UKALL: 37% vs 18% (at 6 y), P = .06 | ||||||
| Japan Childhood Leukemia Study ALL02 | 31 | Japan | 2002-2008 | 202 | BCP | 9.4% (MLPA) | 63% vs 89% (at 5 y),P = ..001 | 72% vs 90% (at 5 y),P = ..02 | Particularly strong prognostic effect of IKZF1 deletion in the NCI high-risk group | |
| JCCLSG ALL 2004 | 32 | Japan | 2004-2008 | 177 | BCP | 12.0% (MLPA) | 68% vs 85% (at 4 y), P = .04 | Prognostic effect of IKZF1 deletion more pronounced in high-risk patients | ||
| ALL-REZ BFM 2002 | 33 | Germany, Austria, Switzerland | 2002-2009 | 204 | BCP relapse | 33.3% (MLPA) | 30% vs 51% (at 5 y), P = .002 | 36% vs 60% (at 5 y), P = .001 | 41% vs 23% (at 5 y), P = .006 | Prognostic impact of IKZF1 deletion also in second-line treatment of relapsed ALL; one-quarter of IKZF1 deletions was acquired at relapse |
| AIEOP-BFM ALL 2000 | 34 | Germany | 1999-2005 | 694 | BCP and T ALL | 12.0% (MLPA) | 69% vs 85% (at 5 y), P < .0001 | 82% vs 92% (at 5 y), P = .003 | 21% vs 10% (at 5 y), P = .001 | Pronounced prognostic effect of IKZF1 deletion in the intermediate-risk group |
| AIEOP-BFM ALL 2000 | 35 | Italy | 2003-2005 | 410 | BCP | 13.2% (MLPA) | 70% vs 85% (at 5 y), P = .007 | 87% vs 93% (at 5 y), P = .100 | 24% vs 13% (at 5 y), P = .049 | Due to the modest effect of IKZF1 deletion observed, the relevance of IKZF1 deletion as a clinically useful stratification factor is debated |
| DCOG ALL8, ALL9, ALL10, COALL-97, COALL-03 | 36 | The Netherlands, Germany | 1991-2012 | 857 | BCP | 15.9% (MLPA) | 34% vs 13% (at 5 y), P = <.001 | Confirms and extends the strong prognostic effect of IKZF1 deletion; IKZF1 deletion remained predictive in intermediate risk patients on the MRD-guided DCOG ALL10 trial | ||
| EsPhALL and I-BFM SG studies (for pre-TKI assessment) | 37 | Austria, Czech Republic, France, Germany, The Netherlands, UK | 1995-2005 | Pre-TKI cohort: 84 EsPh-ALL cohort: 107 | BCR-ABL1–rearranged BCP | 66.0% (MLPA, CGH array, SNP array) | DFS pre-TKI: 30% vs 58% (at 4 y), P = .013 | Pre-TKI: 49% vs 75% (at 4 y), P = .075 | Pre-TKI: 57% vs 21% (at 4 y), P = .026 | IKZF1 deletion is demonstrated to be a poor prognostic marker in BCR-ABL1–rearranged pediatric ALL independent of imatinib treatment |
| DFS EsPhALL: 54% vs 63% (at 4 y), P = .168 | EsPhALL: 58% vs 83% (at 4 y), P = .070 | EsPhALL: 28% vs 31% (at 4 y), P = .817 | ||||||||
| DFS EsPhALL TKI-treated good-risk patients: 56% vs 75% (at 4 y), P = .051 | EsPhALL TKI-treated good-risk patients: 66% vs 100% (at 4 y), P = .039 | EsPhALL TKI-treated good-risk patients: 29% vs 25% (at 4 y), P = .249 | ||||||||
| UKALL trials ALL 97/99 and ALL 2003 | 38 | UK, Ireland | UKALL 97/99: 1997-2002 | UKALL 97/99: 864 | BCP | 13.4% (MLPA) | 56% vs 80% (at 5 y), P = <.001 | 70% vs 89% (at 5 y), P = <.001 | 40% vs 18% (at 5 y), P = <.001 | Integration of cytogenetic with genomic data including IKZF1 deletion refines risk groups in a clinically meaningful way |
| UKALL 2003: 2003-2011 | UKALL 2003: 782 | BCP | 11.1% (MLPA) | 80% vs 92% (at 5 y), P = <.001 | 86% vs 95% (at 5 y), P = <.001 | 16% vs 6% (at 5 y), P = .002 | ||||
| NOPHO ALL-1992, NOPHO ALL-2000, NOPHO ALL-2008 | 39 | Denmark, Finland, Norway, Sweden | 1992-2013 | 334 | BCP | 15.0% (SNP array, MLPA) | 60% vs 83% (at 10 y), P < .001 | 73% vs 89% (at 10 y), P = .001 | 35% vs 12% (at 10 y), P < .001 | Prognostic impact independent of WBC count and MRD; co-occurrence of PAR1 deletion increased prognostic impact of IKZF1 deletion |
| EORTC Children’s Leukemia Group study 58951 | 40 | Belgium, France, Portugal | 1998-2008 | 1223 | BCP | 14.6% (PCR, MLPA) | 68% vs 87% (at 5 y), P = <.001 | 87% vs 92% (at 5 y), P = .035 | IKZF1-aberrant BCP ALL benefited from vincristine-steroid pulses during maintenance treatment | |
| ALLR3 | 41 | UK, Ireland, The Netherlands, Australia, New Zealand | 2002-2013 | 222 | BCP relapse | 23.0% (MLPA) | 48% vs 53% (at 5 y), P = .30 | No prognostic impact of IKZF1 deletion at relapse | ||
| ANZCHOG ALL8 | 42 | Australia, New Zealand | 2002-2011 | 475 | BCP standard and intermediate risk | 10.5% (MLPA) | 53% vs 83% (at 7 y), P < .0001 | 78% vs 94% (at 7 y), P < .0001 | 41% vs 15% (at 7 y), P < .0001 | Strong prognostic effect of IKZF1 deletion in non–high-risk BCP ALL |
| ALL-BFM 95 | 45 | Germany | 1995-2000 | 655 | BCP intermediate risk | 12.2% (PCR) | 66% vs 82% (at 5 y), P = .001 | Vincristine-dexamethasone pulses during maintenance were not of benefit to IKZF1-deleted intermediate-risk patients | ||
| DFCI ALL Consortium Protocol 05-001 | 43 | US | 2005-2010 | 385 | BCP | 16.0% (MLPA) | 63% vs 88% (at 5 y), P < .001 | 79% vs 94% (at 5 y), P < .001 | 29% vs 8% (at 5 y), P < .001 | IKZF1 deletion was confirmed as an independent predictor of inferior outcome |
| Malaysia-Singapore ALL 2003, 2010 | 44 | Malaysia, Singapore | 2002-2017 | 665 | BCP | 15.9% (MLPA) | Without IKZF1 as high-risk criterion: 30% vs 8% (at 5 y), P < .001 | Demonstrates that intensifying treatment of IKZF1-deleted BCP ALL patients reduces risk of relapse | ||
| With IKZF1 as high-risk criterion: 14% vs 5% (at 5 y), P = .030 | ||||||||||
| COG Trial AALL0622 | 46 | US, Canada | 2008-2012 | 44 | BCR-ABL1–rearranged BCP | 56.8% (SNP array) | 52% vs 82% (at 5 y), P = .040 | 80% vs 100% (at 5 y), P = .040 | Confirms poor outcome for IKZF1-deleted BCR-ABL1–rearranged ALL on dasatinib-containing treatment regimen | |
| AIEOP-BFM ALL 2000 | 61 | Germany, Italy | 1999-2009 | 1408 | BCP | 14.6% (MLPA) | IKZF1-deleted, but not IKZF1plus: 77% vs 86% (at 5 y), P = .0005 | IKZF1-deleted, but not IKZF1plus: 89% vs 94% (at 5 y), P = .023 | IKZF1-deleted, but not IKZF1plus: 16% vs 11% (at 5 y), P = .045 | Definition of a poor MRD-dependent prognostic pattern termed IKZF1plus with IKZF1 deletions co-occurring with deletions in CDKN2A, CDKN2B, PAX5, or PAR1 in the absence of ERG deletion |
| IKZF1plus: 51% vs 86% (at 5 y), P < .0001 | IKZF1plus: 75% vs 94% (at 5 y), P < .0001 | IKZF1plus: 44% vs 11% (at 5 y), P < .0001 |
AIEOP, Associazione Italiana di Ematologia e Oncologia Pediatrica; ALL-REZ, Acute Lymphoblastic Leukemia-Relapse Study; BFM, Berlin-Frankfurt-Münster; ALLR3, An International Collaborative Trial for Relapsed and Refractory Acute Lymphoblastic Leukaemia Combination Chemotherapy in Treating Young Patients With Relapsed or Refractory Acute Lymphoblastic Leukemia; ANZCHOG, Australian & New Zealand Children’s Haematology/Oncology Group; CGH, comparative genomic hybridization; COG, Children’s Oncology Group; DCOG, Dutch Childhood Oncology Group; DFCI, Dana-Farber Cancer Institute; DFS, disease-free survival; EFS, event-free survival; EORTC, European Organisation for Research and Treatment of Cancer; EsPHALL, Safety and Efficacy of Imatinib Added to Chemotherapy in Treatment of Ph+ Acute Lymphoblastic Leukemia in Children; I-BFM SG, International Berlin-Frankfurt-Münster Study Group; JCCLSG, Japanese Children's Cancer and Leukemia Study Group; MLPA, multiplex ligation probe-dependent amplification; MRD, minimal residual disease; NCI, National Cancer Institute; NOPHO, Nordic Society of Pediatric Hematology and Oncology; OS, overall survival; PCR, polymerase chain reaction; Ref., reference(s); Sanger sequencing, dye-terminator sequencing; SNP, single-nucleotide polymorphism; TKI, tyrosine kinase inhibitor; TPOG, Taiwan Pediatric Oncology Group; UK, United Kingdom; US, United States; VHR, very high risk; WBC, white blood cell.
IKZF1 as a prognostic factor in pediatric ALL
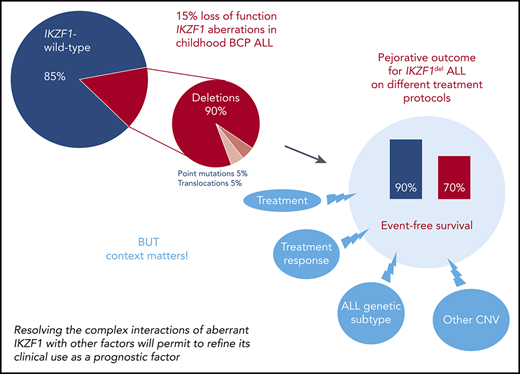
During the last 10 years, various studies have analyzed the clinical importance of IKZF1 aberration in pediatric ALL treated on different treatment protocols.10,13-16,30-46 The presence of IKZF1 deletions has been associated with older age at diagnosis, higher presenting white blood cell counts, and higher levels of MRD after induction and consolidation.10,31,32,34,35,43,44,49,50 Consequently, IKZF1 deletions are overrepresented in high-risk patients with pediatric ALL. Importantly, the distribution of IKZF1 aberrations within the subgroups of ALL is not homogeneous. Although approximately two-thirds of BCR-ABL1+ pediatric BCP ALL carry an IKZF1 deletion,9,37,44,46 the frequency in BCR-ABL1− BCP ALL subgroups is lower. It ranges from roughly 20% in the B-other group, which lacks the “classical” recurrent acquired genetic aberrations of ALL, but includes Philadelphia-like and BCR-ABL1–like BCP ALL characterized by an activated kinase signature, and 15% in high hyperdiploid ALL to <5% in ETV6-RUNX1-, TCF3- or KMT2A-rearranged ALL as well as T-cell ALL.10,31,34,35,38-40,44,51-53 Although very rare in BCR-ABL1− BCP ALL (∼1%), IKZF1 point mutations have been described in up to 10% of IKZF1 deletion-negative BCR-ABL1+ BCP ALL.10,13-16 One unifying and important feature of a majority of prognostic studies in pediatric BCP ALL is that the different types of IKZF1 deletions have been consistently linked to an unfavorable clinical outcome of frontline treatment (Table 1).10,13,14,30-46,54 It is worth mentioning that IKZF1 point mutations seem to have a similar impact on outcome.10,13-16 As a consequence, some international study groups on treatment of ALL early on included IKZF1 deletion status into their high-risk treatment-stratification strategies for BCP ALL patients whereas others did not follow this strategy because the prognostic effect of IKZF1 deletions alone was considered not sufficiently strong enough to justify exposure to the toxic side effects of high-risk or even very-high-risk therapy.38,53
Of significance, although it has been clearly demonstrated that IKZF1 aberrations exert an independent prognostic impact, some studies early on suggested the presence of effect modification or confounding by different levels of MRD, activated JAK-STAT signaling, or co-occurring deletions of the lymphoid transcriptional regulator gene BTG1 or the ets family transcription factor ERG.15,32,34,35,49,55-57 For example, integrated use of MRD and IKZF1 deletion status predicted 79% of relapses in a Dutch study.49 Such an epistatic effect of other genetic aberrations has been particularly documented for ERG deletions because IKZF1 deletion surprisingly does not affect the prognosis of BCP ALL when co-ocurring with ERG deletion.56,57 It is now known that ERG deletions almost exclusively occur in the recently described DUX4-rearranged BCP ALL subtype, which also bears IKZF1 deletions in >20% of the cases.19,58,59 In this respect, ERG deletion can be considered a surrogate marker for DUX4 rearrangements, although approximately one-third of DUX4-rearranged BCP ALL lacks an ERG deletion.60 So far, it is unclear whether the previously described prognostic effect of the ERG deletion56,57 is mediated by itself or the association with the DUX4 rearrangement. Of interest, there are hints that positivity for an ERG deletion also seems to confer an advantageous outcome within DUX4-rearranged BCP ALL.60 Hence, the interrelationships of ERG deletion and DUX4 rearrangements with IKZF1 deletions and their impact on outcome are complex and not fully understood. Recently, a large study of the International Berlin-Frankfurt-Münster (I-BFM) Study Group refined the prognostic strength of IKZF1 deletions in BCR-ABL1− pediatric BCP ALL by describing an extremely poor prognostic IKZF1 deletion-associated genetic aberration profile termed IKZF1plus.61 It is defined by IKZF1 deletions co-occurring with deletions in CDKN2A, CDKN2B, PAX5, or PAR1 in the absence of ERG deletion. Of interest, but unfortunately not yet understood, IKZF1plus exerts its strong prognostic impact only in patients still carrying measurable MRD of a leukemic cell load exceeding 10−4 after induction treatment. This simple to assess profile in combination with MRD analyses has already been implemented as a high-risk stratification criterion in the current frontline AIEOP-BFM ALL 2017 trial for treatment of ALL, and its prognostic value has been confirmed by others.62
Lastly, the prognostic importance of IKZF1 deletions for second-line treatment of BCP ALL is relatively underexamined in comparison with frontline protocols and, therefore, is less well understood. Despite differences in results of 2 published studies, 1 unifying conclusion is supportive of the idea that, similar to frontline therapy, the prognostic effect of IKZF1 deletions seem to be context-dependent.33,41
Overall, these observations indicate that the complex interplay of exposure and response to treatment with the underlying disease biology is the key driver determining the prognostic impact of IKZF1 aberrations. Thus, a greater understanding of the interrelationships between treatment, treatment response, and cooperating molecular lesions in IKZF1-aberrant ALL could help to develop improved treatments tailored to the different scenarios.
IKZF1 and mechanisms of treatment resistance
The suggested molecular mechanisms involved in drug resistance mediated by IKZF1 alteration in BCP ALL are complex and not completely understood. However, there is evidence that loss of IKAROS in normal BCP cells or BCR-ABL1+ BCP ALL leads to acquisition of a stem cell–like phenotype with increases in self-renewal, upregulation of focal adhesion kinase (FAK), cell-adhesion molecules, and drug resistance through formation of a de novo superenhancer landscape of collaborating master transcription regulators and B-cell transcription factors.63-65 Of interest, Churchman et al demonstrated reversal of such a phenotype in IKZF1-aberrant BCR-ABL1+ BCP ALL by treatment with retinoid receptor agonists.65 Retinoids induced selective expression of wild-type IKZF1 and initiated expression of IKZF1 target genes. Similar results were obtained through FAK inhibition and both treatments, retinoid receptor agonists and FAK inhibition, improved the sensitivity of BCR-ABL1+ BCP ALL to tyrosine kinase inhibitor therapy.65,66 In addition, there are intriguing data demonstrating that IKZF1 controls energy metabolism in BCP ALL with a direct link to glucocorticoid response.67,68 These important insights may allow the development of new specific therapeutic approaches targeting IKZF1-altered signaling networks in BCP ALL.
Perspective
Risk stratification and associated treatment strategies in trials on pediatric ALL are often not fully comparable. This may, at least in part, contribute to the partially different impact of aberrant IKZF1 on outcome observed in some of the published trials (Table 1). However, these complicating issues may also bear chances, if we engage into more detailed and careful comparisons of the different treatment backgrounds and their variant stratification strategies in context with IKZF1 aberrations and the relevant clinical end points. Following such strategies may allow us to nourish our knowledge on IKZF1 aberration with a better understanding of the differential response of affected patients to specific parts of the applied treatment protocols. Cooperative approaches incorporating well-characterized trial populations from large study groups will likely be key to success in this regard and should also answer many additional open questions such as the importance of a truly comprehensive assessment of the molecular spectrum of IKZF1 alterations (copy-number variation, nucleotide mutations, gene fusions), their prognostic importance in ALL subgroups, and the development of a full picture of the interplay of aberrant IKZF1 with cooperating genetic aberrations (eg, CDKN2A, PAX5, BTG1, JAK pathway aberrations, RAS pathway aberrations, other kinase activating lesions, ERG, subclonality of cooperating lesions).
Therefore, by continuing our research on IKZF1 in pediatric ALL, we will (1) be able to improve information for patients and/or their guardians regarding the risk of recurrence and final outcome of ALL; (2) very likely gain further insights into the biology of ALL; and (3) increase our ability to design and conduct clinical trials delivering optimized treatment strategies based on improved individual characterization of children and adolescents with ALL.
So, is IKZF1 aberration still a prognostic factor? As it turns out, the answer is: yes, more than ever!
Acknowledgments
The authors thank all participants and personnel involved in trials AIEOP-BFM ALL 2000, EORTC-CLG 58951, UKALL 97/99, and UKALL 2003. Particular thanks go to Gianni Cazzaniga for critical discussions. The authors also thank Anna Stengel and Stefanie Junk for their support regarding the development and adaptation of the Rscript for mutation plotting.
This work was supported by the ERA-NET TRANSCAN/European Commission under the 7th Framework Programme (FP7), Madeleine-Schickedanz-Kinderkrebsstiftung, Deutsche Krebshilfe, and Bloodwise blood cancer charity.
Authorship
Contribution: M.S., H.C., and A.V.M. designed and performed research, analyzed and discussed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Martin Stanulla, Department of Pediatric Hematology and Oncology, Hannover Medical School, Carl-Neuberg-Str 1, D-30625 Hannover, Germany; e-mail: stanulla.martin@mh-hannover.de.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal