Key Points
Next-generation sequencing of HLA identified completely different predisposing and protective factors for iTTP in the Japanese and whites.
In silico analysis suggested that the shared ADAMTS13 peptide may bind HLA-DR proteins encoded by different DRB1 alleles.
Abstract
Immune-mediated thrombotic thrombocytopenic purpura (iTTP) is a rare autoimmune disorder caused by neutralizing anti-ADAMTS13 autoantibodies. In white individuals, HLA allele DRB1*11 is a predisposing factor for iTTP, whereas DRB1*04 is a protective factor. However, the role of HLA in Asians is unclear. In this study, we analyzed 10 HLA loci using next-generation sequencing in 52 Japanese patients with iTTP, and the allele frequency in the iTTP group was compared with that in a Japanese control group. We identified the following HLA alleles as predisposing factors for iTTP in the Japanese population: DRB1*08:03 (odds ratio [OR], 3.06; corrected P [Pc] = .005), DRB3/4/5*blank (OR, 2.3; Pc = .007), DQA1*01:03 (OR, 2.25; Pc = .006), and DQB1*06:01 (OR,: 2.41; Pc = .003). The estimated haplotype consisting of these 4 alleles was significantly more frequent in the iTTP group than in the control group (30.8% vs 6.0%; Pc < .001). DRB1*15:01 and DRB5*01:01 were weak protective factors for iTTP (OR, 0.23; Pc = .076; and OR, 0.23, Pc = .034, respectively). On the other hand, DRB1*11 and DRB1*04 were not associated with iTTP in the Japanese. These findings indicated that predisposing and protective factors for iTTP differ between Japanese and white individuals. HLA-DR molecules encoded by DRB1*08:03 and DRB1*11:01 have different peptide-binding motifs, but interestingly, bound to the shared ADAMTS13 peptide in an in silico prediction model.
Introduction
Thrombotic thrombocytopenic purpura (TTP) is a rare and devastating disease resulting from a severe deficiency in the von Willebrand factor–cleaving protease ADAMTS13.1-3 This condition leads to life-threatening microvascular thrombi in systemic organs that are caused by an imbalance between ADAMTS13 and von Willebrand factor. There are 2 types of TTP: congenital and acquired. Congenital TTP is caused by mutations in the ADAMTS13 gene. Acquired immune-mediated TTP (iTTP) is caused by the development of ADAMTS13 autoantibodies, which inhibit the activity of ADAMTS13 or enhance its clearance from blood.
Generally, the pathogenesis of an autoimmune disorder is considered multifactorial, and the HLA system plays an important role as a genetic risk factor. Several HLA alleles have been found to increase the risk of several diseases, including rheumatoid arthritis,4 systemic lupus erythematosus,5 and chronic thyroiditis.6 As for iTTP, HLA-DRB1*11 and HLA-DRB1*04 are predisposing and protective factors in white individuals, respectively.7-9 A Dutch group reported that the presentation of the core sequence FINVAPHAR in the CUB2 domain of ADAMTS13 on HLA-DRB1*11 contributes to the onset of iTTP by stimulating CD4+ T cells.10,11 In addition, an Italian group used immunochip analysis to identify the susceptibility loci for iTTP in the HLA region. They reported that the single-nucleotide polymorphism (SNP) rs6903608 increases the risk of iTTP development by 2.6-fold in whites.12 This SNP is located in the intron between HLA-DRA and HLA-DRB5 (6p21.32).13,14
In Japan, there have been no large-scale HLA analyses of iTTP. Therefore, to identify the alleles that increase iTTP susceptibility in the genetically isolated Japanese population, we performed high-resolution HLA typing using next-generation sequencing (NGS), which enabled us to obtain 6-digit resolution of individual HLA alleles. In addition, we analyzed the frequency of rs6903608 in members of the same cohort who had iTTP. NGS identified the unique HLA alleles associated with iTTP development in the Japanese. Furthermore, we performed an in silico analysis to clarify how this specific allele found in Japanese patients contributes to iTTP development.
Materials and methods
Patients
The diagnosis of iTTP was based on the diagnostic and treatment guidelines for TTP 2017 in Japan.15 This study enrolled patients with thrombocytopenia and hemolytic anemia who also exhibited decreased levels of ADAMTS13 activity (below 10% of healthy controls) and the presence of anti-ADAMTS13 inhibitor [at least 0.5 Bethesda units (BU)/mL]. Fifty-two patients with iTTP from 19 institutions across Japan were eligible and underwent HLA typing by NGS. Clinical outcomes of iTTP patients were evaluated for cumulative rates of clinical response and refractoriness according to a previously published consensus on the standardization of terminology in TTP.16 The median follow-up period was 967 days (range, 129-6852), and no patients died of TTP (2 died of lung cancer). This study was approved by the ethics committees of Nara Medical University and each referring institute. Written informed consent was obtained from all patients.
Samples and measurements
Whole blood collected in EDTA at enrollment was used for HLA typing. In addition, a questionnaire was submitted to Nara Medical University regarding patients’ routine laboratory test results and their treatments and clinical outcomes in the acute phase of iTTP. Laboratory tests assessed platelet counts and the levels of ADAMTS13 activity, anti-ADAMTS13 functional inhibitor, hemoglobin, lactate dehydrogenase (LDH), total bilirubin, and serum creatine. Both ADAMTS13 activity and an anti-ADAMTS13 functional inhibitor were measured by ADAMTS13-act-enzyme-linked immunosorbent assay.17
HLA typing by NGS
All subjects were genotyped for HLA-A, -B, and -C, -DRB1, -DRB3/4/5, -DQA1, -DQB1, -DPA1, and -DPB1 by NGS. HLA typing was performed based on the protocols of commercially available kits (Scisco Genetics, Seattle, WA) with Illumina MiSeq technology.18 The sequencing involved consecutive PCRs incorporating bar codes to track individual samples, followed by NGS using the MiSeq platform. Four-digit resolution of HLA alleles was used for analysis because their amino acid sequences, rather than nucleotide sequences, regulate antigen-specific immune responses. We compared allele frequencies at each locus in iTTP patients with those in a healthy Japanese population as reported by Nakajima et al in 2001.19 Haplotypes were constructed based on genotyping results using the expectation-maximization algorithm.20 Because the allele frequency at HLA-DPA1 was unavailable in the report of Nakajima et al, the remaining 10 loci were analyzed.
Genotyping of rs6903608
An Italian group reported that HLA variant rs6903608 is a risk factor for developing iTTP in whites.12 We genotyped rs6903608 in Japanese patients by using a SNP TaqMan assay (Thermo Fisher Scientific, Carlsbad, CA).21 According to the manufacturer’s instructions, 4 ng of each template DNA sample, 5 µL of TaqMan Genotyping Master Mix (Thermo Fisher Scientific), and 0.5 µL of TaqMan Genotyping Assay Mix (rs6903608, Thermo Fisher Scientific) were mixed and applied to a Step One Plus Real-Time PCR System (Thermo Fisher Scientific). A couple of alleles were amplified automatically, and their genotypes were predicted from allelic discrimination plots by StepOne software. In this study, we compared the allele frequency of rs6903608 between Japanese patients with iTTP and healthy Japanese controls, with data on the latter reported in jMorp, a Japanese multiomics reference panel (https://jmorp.megabank.tohoku.ac.jp/).22
Comparison of MHC class II pocket motifs in terms of HLA supertypes
We compared major histocompatibility complex (MHC) class II pocket motifs and HLA supertypes of predisposing alleles between Japanese and white individuals. MHC molecules can be classified into supertypes based on structural features and peptide specificity; this classification is highly important in the field of vaccine development.23,24 Peptide specificity depends on the 9 pocket motifs of MHC molecules, and of those motifs, those in the 1, 4, 6, 7, and 9 positions are considered to be key anchors.25 The “motif viewer” in the online analysis system NetMHCIIpan, version 3.2 (https://services.healthtech.dtu.dk/),26 was used to visually compare the pocket motifs of MHC class II molecules encoded by predisposing alleles.
Prediction of peptide binding with an in silico assay
To identify the interaction between ADAMTS13-derived peptides and HLA-DR molecules derived from susceptible DRB1 alleles identified in Japanese patients, an in silico peptide-binding assay was performed using NetMHCIIpan, version 3.2.26 NetMHCIIpan is constructed from an extended data set, obtained from the Immune Epitope Database, regarding quantitative binding affinities between MHC molecules, including HLA-DR, -DQ, and -DP, and various peptides. The amino acid sequence of the human ADAMTS13 molecule was obtained from a previous report.27 We calculated the binding affinities between HLA-DR molecules encoded by TTP-related DRB1 alleles and 9-amino-acid core peptides from the signaling peptide region to the CUB2 domain region. Strong and weak binders were defined for calculated percentage rank (%rank) values below 2% and 10%, respectively. %rank indicates the percentage rank of the predicted affinity compared with a set of 200 000 random natural peptides. This measure is not affected by the inherent bias of certain molecules toward higher or lower mean predicted affinities.
Statistical analysis
The HLA allele frequencies in patients with iTTP and controls were compared by Fisher’s exact test, and the Bonferroni correction was applied to the observed P-values. We calculated the corrected P-value (Pc) by multiplying it by the number of alleles in each locus whose frequency in the control group was more than 1%. Pairs of clinical laboratory parameters were compared by using the Mann-Whitney U test. Statistical analyses were carried out with EZR.28 We considered values of P < .05 to be statistically significant.
Results
Characteristics of Japanese patients with iTTP
The characteristics of patients with iTTP are shown in supplemental Table 1 (available on the Blood Web site). The male/female ratio in this group was 22:30, and the median age was 56.5 years (range, 1-81). Based on the underlying disease, iTTP patients were classified as having primary TTP (n = 38), autoimmune disease (n = 12), malignancy (n = 1), or acute pancreatitis (n = 1). Patients with autoimmune diseases included 6 with systemic lupus erythematosus, 3 with mixed connective tissue disease, 2 with Sjögren syndrome, and 1 with primary biliary cholangitis. Only 1 patient with iTTP had a medical history of gastric cancer, which was treated with chemotherapy. Plasma ADAMTS13 activity was below 3% in all cases, and the median value of functional antibody was 2.5 BU/mL (range, 0.5-113 BU/mL). Both severe thrombocytopenia (median platelet count, 11.5 × 109/L) and hemolytic anemia (median hemoglobin level, 8.0 g/dL; median LDH level, 968 U/L) were also seen in most patients. Most patients with iTTP did not show severe renal impairment (median serum creatinine: 0.88 mg/dL). Cardiac troponin results were available in only 20 of 52 patients; troponin was positive in 11 (55%) of the 20 analyzed patients. However, no patients developed cardiac events, including acute myocardial infarction and angina pectoris. On the other hand, 35 patients (67%) had neurological findings ranging from headache to coma, including neurological dysfunction, convulsion, and clouding of consciousness.
During the acute phase of TTP, 49 of 52 patients received a combination regimen of plasma exchange (PEX) with fresh frozen plasma and corticosteroids. Additional immunosuppressants were administered to several patients, as follows: rituximab in 24 patients, cyclosporin A in 6, cyclophosphamide in 5, vincristine in 3, and mycophenolate mofetil in 1. The median number of PEX treatments was 11 (range, 3-42), and 19 of 49 patients who underwent PEX were judged as having refractory TTP, as defined by the consensus on the standardization of terminology in TTP.16
Comparison of allele frequencies between iTTP and healthy individuals in Japan
The allele frequencies in iTTP patients and controls are shown in Table 1. Regarding HLA class II alleles (DRB1, DRB3/4/5, DQA1, and DQB1), the following were predisposing factors for iTTP in the Japanese population: DRB1*08:03 (odds ratio [OR], 3.06; observed P = 2.86 × 10−4; Pc = .005), DRB3/4/5*blank (OR, 2.3; observed P = 9.13 × 10−4; Pc = .007), DQA1*01:03 (OR, 2.25; observed P = 8.94 × 10−4; Pc = .006), and DQB1*06:01 (OR, 2.41; observed P = 2.49 × 10−4; Pc = .003). The estimated haplotype consisting of these 4 alleles was more frequent in the iTTP group than in the control group (30.8% vs 6.0%; Pc < .001).
Comparison of allele frequency between iTTP patients and controls in Japan
| HLA type . | Allele frequency in iTTP patients (%) n = 52 . | Allele frequency in controls (%) n = 516 . | P (observed) . | P (corrected) . | OR (95% CI) . |
|---|---|---|---|---|---|
| DRB1 | |||||
| DRB1*08:03 | 17.3 | 6.4 | 2.86 × 10−4 | .005 | 3.06 (1.63-5.51) |
| DRB1*15:01 | 2.9 | 11.6 | 4.22 × 10−3 | .076 | 0.23 (0.05-0.70) |
| DRB3/4/5 | |||||
| Blank | 27.9 | 14.3 | 9.13 × 10−4 | .007 | 2.30 (1.40-3.72) |
| DRB5*01:01 | 2.9 | 11.6 | 4.22 × 10−3 | .034 | 0.23 (0.05-0.70) |
| DQA1 | |||||
| DQA1*01:03 | 29.8 | 15.9 | 8.94 × 10−4 | .006 | 2.25 (1.38-3.60) |
| DQB1 | |||||
| DQB1*06:01 | 29.8 | 14.9 | 2.49 × 10−4 | .003 | 2.41 (1.48-3.88) |
| HLA type . | Allele frequency in iTTP patients (%) n = 52 . | Allele frequency in controls (%) n = 516 . | P (observed) . | P (corrected) . | OR (95% CI) . |
|---|---|---|---|---|---|
| DRB1 | |||||
| DRB1*08:03 | 17.3 | 6.4 | 2.86 × 10−4 | .005 | 3.06 (1.63-5.51) |
| DRB1*15:01 | 2.9 | 11.6 | 4.22 × 10−3 | .076 | 0.23 (0.05-0.70) |
| DRB3/4/5 | |||||
| Blank | 27.9 | 14.3 | 9.13 × 10−4 | .007 | 2.30 (1.40-3.72) |
| DRB5*01:01 | 2.9 | 11.6 | 4.22 × 10−3 | .034 | 0.23 (0.05-0.70) |
| DQA1 | |||||
| DQA1*01:03 | 29.8 | 15.9 | 8.94 × 10−4 | .006 | 2.25 (1.38-3.60) |
| DQB1 | |||||
| DQB1*06:01 | 29.8 | 14.9 | 2.49 × 10−4 | .003 | 2.41 (1.48-3.88) |
No disease-related alleles were identified in the A, B, C, or DPB1 loci.
By contrast, DRB1*15:01 and DRB5*01:01 were associated with a reduced risk of iTTP (OR, 0.23; observed P = 4.22 × 10−3; Pc = .076; and OR, 0.23; observed P = 4.22 × 10−3; Pc = .034, respectively). There is strong linkage disequilibrium between DRB1*15:01 and DRB5*01:01.29 Inconsistent Pc values for these alleles can be explained by differences in the number of alleles used for multiplication. Because the Bonferroni correction was reported to underestimate the results of statistical analyses,30 the DRB1*15:01-DRB5*01:01 haplotype was regarded as a protective factor for iTTP. The positive rate of this haplotype in patients with a medical history of autoimmune disease was similar to that in patients with primary iTTP, and no specific TTP-related alleles were identified in patients with autoimmune disease.
Among the HLA-A, -B, -C, and -DPB1 loci, no significant differences were found between the iTTP and control groups. In Japanese individuals, HLA-DRB1*11 was not identified as a predisposing factor for iTTP, and HLA-DRB1*04 was not associated with protection for iTTP. All allele frequencies are shown in supplemental Table 2. HLA analysis has been performed to investigate the association between HLA loci and disease development.
Clinical manifestations of predisposing haplotypes
In this study, 16 of 52 patients had the predisposing haplotype DRB1*08:03-DRB3/4/5*blank-DQA1*01:03-DQB1*06:01. Of these 52 patients, 14 were heterozygotes and 2 were homozygotes. To investigate the association between DRB1*08:03 and clinical parameters in patients with iTTP, we compared blood test results in the acute phase between patients with and without this haplotype (positive and negative groups, respectively), as shown in Table 2. Interestingly, the median elevations of both LDH and total bilirubin in the positive group were significantly smaller than those in the negative group (LDH, 742 vs 1138 U/L; P < .017; total bilirubin, 2.25 vs 3.3 mg/dL; P < .015, respectively). On the other hand, no significant differences were found between the 2 groups in the levels of ADAMTS13 activity, anti-ADATMS13 inhibitor, platelet count, hemoglobin, or serum creatinine. Regarding treatment response, the number of PEX procedures was not significantly different between the 2 groups (median, 6.5 vs 11.0; P = .187), but was slightly lower in the positive group than in the negative group. The rate of refractory cases was not significantly different between the 2 groups.
The effects of HLA-DRB1*08:03 on laboratory test results in patients with iTTP in the acute phase
| . | HLA-DRB1*08:03 . | . | |
|---|---|---|---|
| . | Positive (n = 16) . | Negative (n = 36) . | P . |
| ADAMTS13 activity (%) | <0.5 (<0.5-<0.5) | <0.5 (<0.5-<0.5) | .766 |
| ADAMTS13 inhibitor (BU/mL) | 3.15 (1.78-4.65) | 2.0 (1.0-3.5) | .254 |
| Platelet count (109/L) | 15.5 (7.0-32.3) | 11.0 (9.0-14.0) | .211 |
| Hemoglobin (g/dL) | 7.95 (6.93-9.63) | 8.0 (7.2-9.3) | .873 |
| LDH (U/L) | 742 (509-965) | 1138 (725-1598) | .017 |
| Total bilirubin (mg/dL) | 2.25 (1.2-2.43) | 3.3 (2.2-4.9) | .015 |
| Serum creatine (mg/dL) | 0.8 (0.68-0.9) | 1.0 (0.7-1.1) | .103 |
| . | HLA-DRB1*08:03 . | . | |
|---|---|---|---|
| . | Positive (n = 16) . | Negative (n = 36) . | P . |
| ADAMTS13 activity (%) | <0.5 (<0.5-<0.5) | <0.5 (<0.5-<0.5) | .766 |
| ADAMTS13 inhibitor (BU/mL) | 3.15 (1.78-4.65) | 2.0 (1.0-3.5) | .254 |
| Platelet count (109/L) | 15.5 (7.0-32.3) | 11.0 (9.0-14.0) | .211 |
| Hemoglobin (g/dL) | 7.95 (6.93-9.63) | 8.0 (7.2-9.3) | .873 |
| LDH (U/L) | 742 (509-965) | 1138 (725-1598) | .017 |
| Total bilirubin (mg/dL) | 2.25 (1.2-2.43) | 3.3 (2.2-4.9) | .015 |
| Serum creatine (mg/dL) | 0.8 (0.68-0.9) | 1.0 (0.7-1.1) | .103 |
Median (25%-75%).
Genotyping of rs6903608 in Japanese patients with iTTP
In the rs6903608 SNP, the original C is replaced with T. In the healthy Japanese population, the allele frequencies of C and T have been reported to be 28.9% and 71.1%, respectively.22 In our SNP analysis, the genotype frequencies were C/C, 9.6%; C/T, 40.3%; and T/T, 50.0%; and the allele frequencies were C, 29.8%, and T, 70.2%. Because there were no significant differences in allele frequency between iTTP patients and healthy volunteers, we concluded that, unlike in whites, rs6903608 is not a genetic risk factor for development of iTTP in the Japanese.
Comparison of pocket motifs of HLA-DR molecules and MHC-peptide binding predictions in an in silico analysis
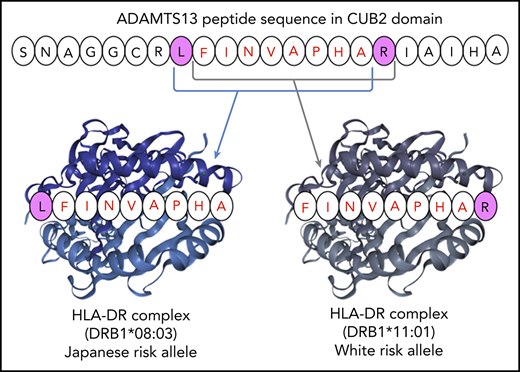
The pocket motifs of HLA-DR proteins encoded by DRB1*08:03 and DRB1*11:01 alleles were predicted using the motif viewer in NetMHCIIpan, version 3.2. As shown in Figure 1, the specificity of each pocket presented by Seq2Logo31 was dissimilar between the 2 HLA-DR molecules. Pocket motifs 1, 4, 6, 7, and 9 were particularly different from one another; these play an essential role in binding to peptides for antigen presentation.25 These results suggest that DRB1 alleles are categorized into different supertypes, and the fact that HLA-DR molecules are encoded by different DRB1 alleles indicates that peptide presentation differs among activating T cells. Next, we predicted HLA-DR molecule-peptide binding with NetMHCIIpan, version 3.2. Five core peptides (9 amino acids in length), shown in Table 3, were extracted as strongly binding peptides. Among them, LFINVAPHA in the CUB2 domain was the strongest binder to HLA-DR molecules encoded by DRB1*08:03 in in silico analysis. Surprisingly, the CUB2 peptide FINVAPHAR, which was previously reported to have high affinity to HLA-DR molecules encoded by DRB1*11:01 in whites,10 did not show any binding affinity to molecules encoded by DRB1*08:03 (%rank >10%). However, an amino acid shifted by 1 peptide, LFINVAPHA, was predicted to be a very strong binder to HLA-DR molecules encoded by DRB1*08:03 in Japanese patients with iTTP. We found that HLA-DR proteins encoded by DRB1*08:03 in Japanese patients with iTTP may recognize the shared ADAMTS13 peptide, as is the case with HLA-DR molecules encoded by DRB1*11:01 in white patients with iTTP.
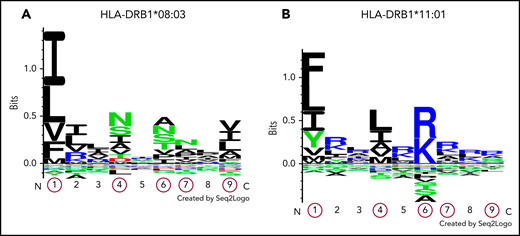
Logos displaying the binding motifs for HLA-DR molecules encoded by DRB1*08:03 and DRB1*11:01. The numbers shown on the x-axis indicate different pocket motifs. Each letter represents the abbreviation of an amino acid. Basic, acidic, and neutral amino acids are shown in blue, red, and green, respectively. The height of each column of letters is equal to the information content at the given position in the binding motif. The relative height of each letter within each column reflects the percentile of the frequency of each amino acid at that pocket. Pocket motifs 1, 4, 6, 7, and 9, circled in red, play essential roles in peptide binding after antigen presentation. This figure shows that the peptide-binding motif of DR molecules encoded by HLA-DRB1*08:03 (A) may differ from that of DR molecules encoded by HLA-DRB1*11:01 (B).
Logos displaying the binding motifs for HLA-DR molecules encoded by DRB1*08:03 and DRB1*11:01. The numbers shown on the x-axis indicate different pocket motifs. Each letter represents the abbreviation of an amino acid. Basic, acidic, and neutral amino acids are shown in blue, red, and green, respectively. The height of each column of letters is equal to the information content at the given position in the binding motif. The relative height of each letter within each column reflects the percentile of the frequency of each amino acid at that pocket. Pocket motifs 1, 4, 6, 7, and 9, circled in red, play essential roles in peptide binding after antigen presentation. This figure shows that the peptide-binding motif of DR molecules encoded by HLA-DRB1*08:03 (A) may differ from that of DR molecules encoded by HLA-DRB1*11:01 (B).
HLA-DR molecules encoded by DRB1*08:03 peptide binding assay for the whole ADAMTS13 molecule in silico
| . | ADAMTS13 peptide . | Domain . | Sequence (core sequence) . | %rank . | Predicted binding . |
|---|---|---|---|---|---|
| 1 | 101-115 | Metalloprotease | ERYVLTNLNIGAELL | 1.9 | Strong binder |
| 2 | 570-584 | Spacer | YVTFLTVTPNLTSVY | 1.1 | Strong binder |
| 3 | 914-928 | Thrombospondin-1 motif 5 | ELRFLCMDSALRVPV | 1.7 | Strong binder |
| 4 | 1324-1338 | CUB2 | GCRLFINVAPHARIA | 0.3 | Strong binder |
| 5 | 1334-1348 | CUB2 | HARIAIHALATNMGA | 1.6 | Strong binder |
| . | ADAMTS13 peptide . | Domain . | Sequence (core sequence) . | %rank . | Predicted binding . |
|---|---|---|---|---|---|
| 1 | 101-115 | Metalloprotease | ERYVLTNLNIGAELL | 1.9 | Strong binder |
| 2 | 570-584 | Spacer | YVTFLTVTPNLTSVY | 1.1 | Strong binder |
| 3 | 914-928 | Thrombospondin-1 motif 5 | ELRFLCMDSALRVPV | 1.7 | Strong binder |
| 4 | 1324-1338 | CUB2 | GCRLFINVAPHARIA | 0.3 | Strong binder |
| 5 | 1334-1348 | CUB2 | HARIAIHALATNMGA | 1.6 | Strong binder |
The underlined bold sequences indicate core sequence which is composed of 9 amino acids peptides and recognized by 9 pocket motifs of HLA-DR molecules.
Discussion
This is the first study in which NGS was used to analyze HLA in patients with iTTP. HLA typing at 4-digit resolution in Japanese patients showed that haplotype DRB1*08:03-DRB3/4/5*blank-DQA1*01:03-DQB1*06:01, which was not identified in white patients, was a significant predisposing factor in Japanese patients with iTTP, and the haplotype DRB1*15:01-DRB5*01:01 was a protective factor in this population. Among these associated alleles, DRB1*08:03 showed the highest OR. Of interest, DRB1*08:03 is unique to individuals with east Asian ancestry.32
The clinical manifestations associated with this predisposing haplotype were investigated based on laboratory findings obtained during the acute phase (Table 2). The levels of ADAMTS13 activity, anti-ADATMS13 inhibitor, platelet count, hemoglobin, and serum creatinine were not different between patients with and without the haplotype, whereas the haplotype-positive group showed mild elevations of LDH and total bilirubin. The clinical response was almost the same between the 2 groups. These results suggest that iTTP patients with DRB1*08:03 may develop milder TTP than those without it, but the difference is not caused by that haplotype.
Considering the diversity of HLA types among different regions and ethnicities, HLA typing data in nonwhites is needed, to reveal the mechanisms of iTTP development for various genetic backgrounds. Martino et al33 reported that African and Caribbean black patients and healthy black controls showed no significant differences in the allele frequencies of HLA types that are related to disease in whites, including DRB1*11, DRB1*04, and DQB1*03. However, the protective allele DRB1*04 was much less common in black controls than in white controls.
An analysis of data from studies conducted in western countries identified several HLA-DR and -DQ alleles as predisposing or protective factors for iTTP, as shown in Table 4.34 Among whites, DRB1*11 is a well-known and extensively analyzed phenotype associated with iTTP.7-9 DRB3*, DQB1*02:02, DRB1*15-DQB1*06, and DRB1*11-DQB1*03 were also shown to be associated with iTTP susceptibility.9,35 By contrast, DRB1*04, DRB1*13-DQB1*06, and DRB1*07-DQB1*02 are thought to be protective factors against iTTP.9,35 Considering the linkage disequilibrium between DR and DQ alleles, the pathogenesis of iTTP may involve not only the DR allele, but also the DQ allele. On the other hand, in an immunochip analysis, an Italian group found that a common variant, rs6903608, was associated with a 2.6-fold increase in the risk of iTTP development.12 However, DRB1*11 was not identified as a genetic risk for iTTP in the Italian study. The discrepancy between our results and theirs may have been caused by the linkage disequilibrium between rs6903608 and DRB1*11:01.13 To date, the effect of this SNP has not been analyzed in Japanese patients with iTTP. Our results indicated that the prevalence of rs6903608 in the Japanese was not different between iTTP patients and healthy volunteers. This may be because DRB1*11:01 was not identified as a predisposing allele in Japanese patients with iTTP.
HLA alleles associated with iTTP
| Allele . | Effect . | Race . | Reference . |
|---|---|---|---|
| DRB1*11 | Predisposing | White | 7 |
| White | 8 | ||
| White | 9 | ||
| DRB3* | Predisposing | White | 7 |
| DQB1*02:02 | Predisposing | White | 9 |
| DRB1*11-DQB1*03 | Predisposing | White | 34 |
| DRB1*15-DQB1*06 | Predisposing | White | 34 |
| DRB1*08:03-DRB3/4/5*blank-DQA1*01:03-DQB1*06:01 | Predisposing | Japanese | Present study |
| DRB1*04 | Protective | White | 7 |
| White | 8 | ||
| DRB1*07-DQB1*02 | Protective | White | 34 |
| DRB1*13-DQB1*06 | Protective | White | 34 |
| DRB1*15:01-DRB5*01:01 | Protective | Japanese | Present study |
| Allele . | Effect . | Race . | Reference . |
|---|---|---|---|
| DRB1*11 | Predisposing | White | 7 |
| White | 8 | ||
| White | 9 | ||
| DRB3* | Predisposing | White | 7 |
| DQB1*02:02 | Predisposing | White | 9 |
| DRB1*11-DQB1*03 | Predisposing | White | 34 |
| DRB1*15-DQB1*06 | Predisposing | White | 34 |
| DRB1*08:03-DRB3/4/5*blank-DQA1*01:03-DQB1*06:01 | Predisposing | Japanese | Present study |
| DRB1*04 | Protective | White | 7 |
| White | 8 | ||
| DRB1*07-DQB1*02 | Protective | White | 34 |
| DRB1*13-DQB1*06 | Protective | White | 34 |
| DRB1*15:01-DRB5*01:01 | Protective | Japanese | Present study |
In the immune system, antigen-presenting cells display specific antigenic peptides in conjunction with HLA class II complexes; this activates antigen-specific CD4+ T cells that elicit and propagate B-cell responses, resulting in the induction of plasma cells that produce specific antibodies.36,37 A recent analysis of dendritic cells pulsed with complexes of HLA class II and ADAMTS13 peptides suggest that the CUB2 domain–derived peptide FINVAPHAR is preferentially presented by the HLA-DR protein encoded by HLA-DRB1*11.11 The mimicry between pathogen-derived peptides and this ADAMTS13-derived peptide may contribute to iTTP pathogenesis. To confirm whether HLA-DR molecules constructed with the predisposing allele DRB1*08:03 in the Japanese could bind to ADAMTS13 peptides like FINVAPHAR in whites, we performed an analysis of MHC peptide in silico with the NetMHCIIpan system. We analyzed only the DRB1 protein because, in contrast to the DPA and DQA proteins, paired DRA1 protein has a nonpolymorphic (invariant) binding site.26,38 Therefore, the various DRB1 alleles should largely account for the peptide-binding affinities of both the HLA-DRB1 and HLA-DR molecules, even though HLA-DR molecules are composed of α and β chains encoded by DRA1 and DRB1, respectively. As DRB1*11 and DRB1*08 have been reported to be members of a different HLA supertype,39 our results indicated that HLA-DR had a different binding motif based on sequence logos (Figure 1). Initially, it seemed that the peptide-binding motifs of DR molecules encoded by HLA-DRB1*08:03 could be different from that of HLA-DRB1*11:01; however, a search of all ADAMTS13 peptides showed that HLA-DR molecules encoded by DRB1*08:03 bound LFINVAPHA (%rank, 0.3) but not FINVAPHAR (%rank, 13.0), the former peptide being shifted by only 1 amino acid. By contrast, those encoded by DRB1*11:01 showed a weak binding affinity to LFINVAPHA (%rank, 5.0) compared with the previously reported peptide FINVAPHAR (%rank, 1.1). This discovery demonstrated that HLA-DR molecules encoded by the specific allele DRB1*08:03 found in Japanese patients with iTTP could bind to the same CUB2 domain–derived peptides LFINVAPHAR as HLA-DR molecules encoded by the DRB1*11:01 allele and that the HLA-DR molecules associated with the 2 alleles displayed different binding motifs. Furthermore, both Japanese and white individuals may develop iTTP through the activation of CD4+ T cells recognizing the common ADAMTS13 peptide in the context of DRB1 alleles encoding different HLA-DR proteins. In an in silico assay, the binding affinity to LFINVAPHAR by the HLA-DR complexes encoded by DRB1*08:03 was higher than that of the HLA-DR molecules encoded by DRB1*11:01 (%rank, 0.3 vs 1.1, respectively). Therefore, the second LFINVAPHAR-binding HLA-DR protein consisting of DRB1*11:01 could be a genetic risk factor for iTTP in whites because DRB1*08:03 is an extremely rare alleles in that population. Our finding may contribute to defining the precise mechanism of iTTP development.
This study had several limitations. First, the number of patients in our HLA analysis was smaller than that in a previous study from a western country.8 Second, our samples included several patients with secondary iTTP (eg, caused by autoimmune diseases). Although the effect of underlying autoimmune diseases on iTTP-related alleles was unclear, the allele frequencies of predisposing and protective haplotypes in primary and secondary iTTP were almost identical (data not shown). Finally, our hypothesis that HLA-DR molecules encoded by DRB1*08:03 can bind to the CUB2-derived peptide LFINVAPHA is based only on the results of in silico analysis. Further analysis using an in vitro MHC-peptide binding assay may confirm our findings.
We identified specific disease-related HLA alleles (haplotypes) in Japanese patients with iTTP that had not reported in western countries. These results were inconsistent with those reported in a white population, possibly because the Japanese population has historically been genetically isolated.
For original data, please e-mail the corresponding author.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by research grants from the Ministry of Health, Labour, and Welfare of Japan (M.K., M. Murata, and M. Matsumoto).
Authorship
Contribution: K.S. designed the study concept, analyzed the data, performed the experiments and wrote the manuscript; M.K. and K. Hosomichi provided advice for statistical and MHC-peptide analyses; H.T. performed the HLA analysis; A.H., H.U., K.N., T.O., M.H., M. Matsui, K.I., A.O., K. Okuhiro, Y. Yamashita, M.I., H.K., N.T., N.K., T. Matsukawa, H.S., K. Ohshiro, K. Hayashi, Y.U., T. Mushino, Y.O., and Y. Yamada collected samples for analysis; M. Murata designed the study and analyzed the data; and M. Matsumoto designed and directed the study and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Masanori Matsumoto, Nara Medical University, 840 Shijocho, Kashihara, Nara 634-8522, Japan; e-mail: mmatsumo@naramed-u.ac.jp.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal