INTRODUCTION: Classic Hodgkin lymphoma (cHL) is uniquely characterized by an extensively dominant microenvironment composed primarily of different types of non-cancerous immune cells with a rare population (~1%) of tumor cells. Detailed characterization of these cellular components and their spatial relationship is crucial to understand crosstalk and therapeutic targeting in the cellular ecosystem of the tumor microenvironment (TME).
METHODS: In this study, we performed high dimensional and spatial profiling of immune cells in the TME of cHL. Single cell RNA sequencing (scRNA-seq) was performed with the 10x Genomics platform on cell suspensions collected from lymph nodes of 22 cHL patients, including 12 of nodular sclerosis subtype, 9 of mixed cellularity subtype and 1 of lymphocyte-rich subtype, with 5 reactive lymph nodes (RLNs) serving as normal controls. Illumina sequencing (HiSeq 2500) was performed to yield single-cell expression profiles for 127,786 cells. We also performed multicolor IHC and imaging mass cytometry (IMC) on TMA slides from the same patients.
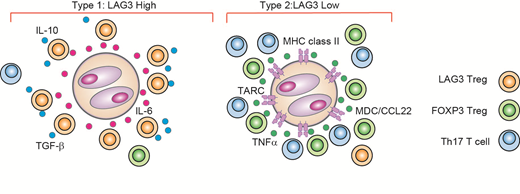
RESULTS: Unsupervised clustering using PhenoGraph identified 22 cell clusters including 12 T cell clusters, 7 B cell clusters and 1 macrophage cluster. While most immune cell populations were common between cHL and RLN, we observed an enrichment of cells from cHL in all 3 regulatory T cell (Treg) clusters. The most cHL-enriched cluster was characterized by high expression of LAG3, in addition to common Treg markers such as IL2RA (CD25) and TNFRSF18 (GITR), but lacked expression of FOXP3, consistent with a type 1 regulatory (Tr1) T cell population.
LAG3+ T cells in cHL had high expression of immune-suppressive cytokines IL-10 and TGF-b . In vitro exposure of T cells to cHL cell line supernatant induced significantly higher levels of LAG3 in naïve T cells compared to co-culture with other lymphoma cell line supernatant or medium only. CD4+ LAG3+ T cells isolated by FACS also suppressed the proliferation of responder CD4+ T cells when co-cultured in vitro. Additionally, Luminex analysis revealed that cHL cell lines secrete substantial amounts of cytokines and chemokines that can promote Tr1 cell differentiation (e.g. IL-6).
Our scRNA-seq analysis revealed that LAG3 expression was significantly higher in cHL cases with loss of major histocompatibility class II (MHC-II) expression on HRS cells as compared to MHC-II positive cases (P = 0.019), but was not correlated with EBV status or histological subtype. Strikingly, LAG3 was identified as the most up-regulated gene in cells from MHC-II negative cases compared to MHC-II positive cases.
Topological analysis using multicolor IHC and IMC revealed that in MHC-II negative cases, HRS cells were surrounded by LAG3+ T cells. In these cases, the density of LAG3+ T cells in HRS cell-rich regions was significantly increased, and the average distance between an HRS cell and its closest LAG3+ T cell neighbor was significantly shorter. These associations were confirmed in an independent cohort of 166 cHL patients.
Finally, we observed a trend towards an inferior disease-specific survival (DSS; P = 0.072) and overall survival (OS; P = 0.12) in cases with an increased number of LAG3+ T cells. A high proportion of LAG3+ T cells (> 20%) was identified as an independent prognostic factor for DSS by multivariate Cox regression.
CONCLUSIONS: Our results reveal a diverse TME composition with inflammatory and immunosuppressive cellular components that are linked to MHC class II expression status on HRS cells (Figure). Unprecedented transcriptional and spatial profiling at the single cell level has established the pathogenic importance of HRS cell-induced CD4+ LAG3+ T cells as a mediator of immunosuppression in cHL, with potential implications for novel therapeutic approaches.
Savage:Seattle Genetics, Inc.: Consultancy, Honoraria, Research Funding; BMS, Merck, Novartis, Verastem, Abbvie, Servier, and Seattle Genetics: Consultancy, Honoraria. Scott:Roche/Genentech: Research Funding; Celgene: Consultancy; Janssen: Consultancy, Research Funding; NanoString: Patents & Royalties: Named inventor on a patent licensed to NanoSting [Institution], Research Funding. Steidl:Bristol-Myers Squibb: Research Funding; Nanostring: Patents & Royalties: Filed patent on behalf of BC Cancer; Roche: Consultancy; Seattle Genetics: Consultancy; Bayer: Consultancy; Juno Therapeutics: Consultancy; Tioma: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal