INTRODUCTION: CXCL9, CXCL10 and CXCL11 (CXCL9-10-11) are closely related cytokines that specifically bind to their receptor CXCR3. They act inducing chemotaxis, proliferation and/or cytotoxicity of CD4+ Th1 and cytotoxic T cells, which express CXCR3. Although the CXCL9-10-11/CXCR3 axis promotes immune activation, their pro- or anti-tumor effects in chronic lymphocytic leukemia (CLL) remain controversial. The aims of this study are: 1. To investigate serum levels of CXCL9-10-11 and the protein expression of their receptor CXCR3, as well as Th1 and cytotoxic gene expression signatures and protein expression of the cytotoxic molecules granzyme B and perforin in peripheral blood (PB) CD4+ T cells of controls, CLL-like monoclonal B-cell lymphocytosis (MBL) and CLL Binet stage A patients. 2. To assess the correlations between all previous parameters. 3. To evaluate Th1, cytotoxic and PD1+ T cell populations during disease progression.
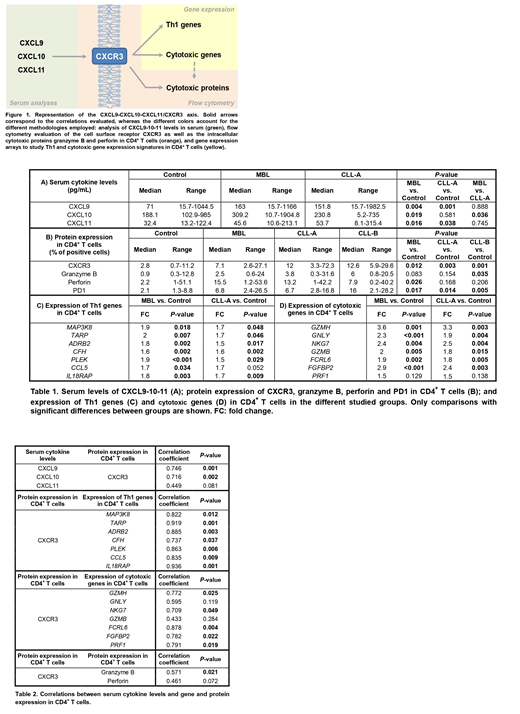
METHODS: Samples from 52 MBL subjects, 61 untreated CLL patients (Binet stage A/B [CLL-A/CLL-B]: 53/8) and 31 age-matched controls were employed. Serum levels (pg/mL) of CXCL9-10-11 were measured in 24 controls, 41 MBL and 44 CLL-A patients using Human CXCL9/MIG Quantikine ELISA Kit (R&D Systems) and U-PLEX Platform (Meso Scale Discovery). In addition, cryopreserved PB mononuclear cells from 8 controls, 11 MBL, 10 CLL-A and 8 CLL-B were studied by flow cytometry. Anti-CD3, anti-CD4, anti-granzyme B, anti-perforin, anti-CXCR3 and anti-PD1 antibodies, FVS510 and Fixation/Permeabilization Kit were used for cell staining (BD Biosciences). Protein expression of CXCR3, granzyme B, perforin and PD1 (measured as percentage of positive cells in PB CD4+ T cells) was assessed using FACSCanto II cytometer (BD Biosciences). In addition, purified CD4+ cells from PB (purity≥90%) were isolated by immunomagnetic methods (Miltenyi Biotec) to analyze gene expression in 9 controls, 13 MBL and 14 CLL-A patients. Extracted RNA (RIN>7) was hybridized to GeneChip Human Gene 2.0 ST arrays (Affymetrix). Differential gene expression was evaluated with linear models in R, and genes with P-value<0.05 and |FC|>1.5 were considered differentially expressed. Linear regression and Pearson correlations were calculated to evaluate the relationship between the different components of the CXCL9-10-11/CXCR3 axis (Figure 1). P-values<0.05 were considered significant.
RESULTS: MBL subjects showed significantly increased CXCL9-10-11 serum levels as well as CXCR3 protein expression, Th1 and cytotoxic gene expression and perforin protein expression in CD4+ T cells compared to controls. A similar trend was obtained for CLL-A versus controls, although the differences for CXCL10 and cytotoxic proteins did not achieve statistical significance despite their increased levels (Table 1). CXCL9 and CXCL10 serum levels were significantly and strongly correlated with CXCR3 protein expression in CD4+ T cells (r=0.746, P-value=0.001 and r=0.716, P-value=0.002, respectively). Very strong positive and significant correlations were also detected between CXCR3 protein expression and Th1 gene expression in CD4+ T cells (correlation coefficients >0.8 for 6/7 genes). Significant positive correlations were also observed between CXCR3 protein expression and cytotoxic genes as well as granzyme B protein (Table 2). Protein expression of CXCR3 and cytotoxic molecules were similarly increased in the different stages of the disease. However, CLL-B patients displayed an increased percentage of CD4+ T cells expressing PD1 (around 7% in MBL and CLL-A versus 16% in CLL-B), although significance was not achieved (Table 1).
CONCLUSIONS: 1. The increased levels of the different components of the CXCL9-10-11/CXCR3 axis in MBL and CLL-A, together with the strong correlations observed, point to an important activation of this molecular pathway in the first stages of the disease. 2. Correlations between CXCR3 and Th1/cytotoxic genes/proteins suggest that the increased Th1/cytotoxic features of CD4+ T cells in MBL and CLL-A are triggered by CXCL9-10-11/CXCR3 stimulation, and might be considered as a potential target for CLL immunotherapy. 3. The lower percentage of PD1+ CD4+ T cells in MBL/CLL-A may allow efficient effector Th1/cytotoxic responses at these stages of the disease.
ACKNOWLEDGEMENTS. PI11/01621, PI15/00437, 2017/SGR437, Fundació La Caixa, Fundación Española de Hematología y Hemoterapia (FEHH).
Gimeno:Abbvie: Speakers Bureau; JANSSEN: Consultancy, Speakers Bureau. Rai:Genentech/Roche: Membership on an entity's Board of Directors or advisory committees; Pharmacyctics: Membership on an entity's Board of Directors or advisory committees; Astra Zeneca: Membership on an entity's Board of Directors or advisory committees; Cellectis: Membership on an entity's Board of Directors or advisory committees. Abrisqueta:Celgene: Consultancy, Honoraria; Janssen: Consultancy, Honoraria, Other: Travel, Accommodations, expenses, Speakers Bureau; Abbvie: Consultancy, Honoraria, Other: Travel, Accommodations, expenses, Speakers Bureau; Roche: Consultancy, Honoraria, Other: Travel, Accommodations, expenses, Speakers Bureau. Bosch:Acerta: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding, Speakers Bureau; Kyte: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding, Speakers Bureau; Takeda: Honoraria, Research Funding; Novartis: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding, Speakers Bureau; AstraZeneca: Honoraria, Research Funding; F. Hoffmann-La Roche Ltd/Genentech, Inc.: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding, Speakers Bureau; Celgene: Honoraria, Research Funding; AbbVie: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding, Speakers Bureau; Janssen: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding, Speakers Bureau.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal