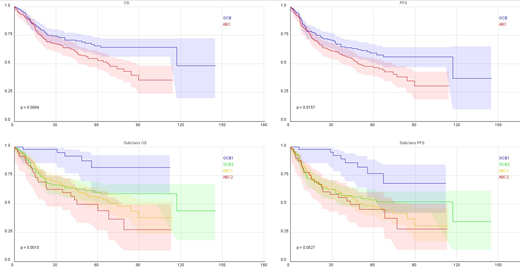
Introduction: Targeted RNA sequencing using Next Generation Sequencing (NGS) has significant advantages over transcriptome sequencing. In addition to information on mutations, fusion and alternative splicing, RNA quantification using targeted RNA sequencing is sensitive, reproducible and provides better dynamic range. We used targeted RNA sequencing for RNA profiling of diffuse large B-cell lymphoma (DLBCL) and explored its utility in the sub-classification of DLBC to ABC and GCB. Method: RNA extracted from 441 FFPE lymphnode samples with DLBC lymphoma and sequenced targeting 1408 genes. These cases were previously subclassified as ABC vs GCB using expression profiling and immunohistochemistry. We first normalized RNA expression data to PAX5 expression, then we tried to narrow down important markers using univariate significance tests. Setting the cutoff for false discovery rate at 0.0001, 48 variables remained significant, including 46 RNA levels and two genes (MYD88 and EZH2) mutation status. Using 60% of samples as training set, we used multiple statistical approaches for classification. Deep learning emerged as the best approach. We used autoencoder with 5 hidden layers and developed a model for classification of ABC vs GCB. To further improve on classification, we divided patients in each subgroup based on survival using simple tree model. In this tree model, expression level of CD58 emerged as a powerful prognostic marker for the ABC group and RLTPR expression in the GCB group. Results: Using probability of scoring developed based on deep learning and logestic regression, approximately 30% of the cases had a score between 0.5 and 0.75. For the remaining 70% of patients, the ABC vs GCB classification showed sensitivity and specificity of 96% and 97% for the testing set. We also applied the same approach to 60 independent cases classified using NanoString (Lymph2Cx). Our model showed sensitivity and specificity of 96% and 97% in the NanoString independent cases. Using the tree model for further classification of the ABC and GCB classes, CD58 mRNA levels separated the ABC group into two subgroups (ABC1 and ABC2) and RLTPR mRNA separated the GCB into two subgroups (GCB1 and GCB2) with significant difference in overall survival (P=0.0010) and progression-free survival (PFS) (P=0.0027). Conclusion: Targeted RNA sequencing is very reliable and practical for the subclassification of DLBCL and can provide clinical-grade reproducible test for prognostically subclassification of DLBCL.
Albitar:Genomic Testing Ccoperative: Employment, Equity Ownership. De Dios:Genomic Testing Ccoperative: Employment. Tam:Takeda: Consultancy; Paragon Genomics: Consultancy. Hsi:Abbvie: Research Funding; Eli Lilly: Research Funding; Cleveland Clinic&Abbvie Biotherapeutics Inc: Patents & Royalties: US8,603,477 B2; Jazz: Consultancy. Ferreri:Roche: Research Funding; Celgene: Consultancy, Research Funding; Novartis: Consultancy; Kite: Consultancy. Piris:Millenium/Takeda: Membership on an entity's Board of Directors or advisory committees, Other: Lecture Fees, Research Funding; Celgene: Membership on an entity's Board of Directors or advisory committees; Gilead: Membership on an entity's Board of Directors or advisory committees, Research Funding; Jansen: Membership on an entity's Board of Directors or advisory committees, Other: Lecture Fees; Nanostring: Membership on an entity's Board of Directors or advisory committees; Kyowa Kirin: Membership on an entity's Board of Directors or advisory committees; Kura: Research Funding. Kantarjian:Ariad: Research Funding; Agios: Honoraria, Research Funding; Daiichi-Sankyo: Research Funding; Novartis: Research Funding; BMS: Research Funding; Takeda: Honoraria; Actinium: Honoraria, Membership on an entity's Board of Directors or advisory committees; AbbVie: Honoraria, Research Funding; Jazz Pharma: Research Funding; Cyclacel: Research Funding; Immunogen: Research Funding; Amgen: Honoraria, Research Funding; Pfizer: Honoraria, Research Funding; Astex: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal