Background: Some previous studies in Ph+ ALL suggested that T315I mutations of the ABL1 KD are present in many pts prior to tyrosine kinase inhibitor (TKI) treatment. However, these reports used RT-PCR to generate complementary DNA (cDNA) prior to analysis, which may introduce random errors and lead to false positive results. We hypothesized that ultrasensitive DS of genomic DNA (gDNA) would more accurately detect low-level pretreatment ABL1 mutations in Ph+ ALL because it does not rely upon error prone RT-PCR.
Methods: DS compares the nucleotide sequences of each strand of double-stranded molecules, improving the accuracy of conventional next-generation sequencing by more than 10,000-fold. DS of exons 4-10 of ABL1 was performed to an average molecular depth of >10,000x. A mixture of known ABL1 variants at varying low variant allelic frequencies (VAFs) versus a negative control were used to determine sensitivity and specificity of DS.
DS of ABL1 was performed on 64 pts with newly diagnosed Ph+ ALL prior to receiving frontline hyper-CVAD plus a TKI (imatinib, n=5; dasatinib, n=38; ponatinib, n=21). DS was also performed on cDNA samples from RT-PCR to assess the RT-induced error rate. On RNA from relapse samples, the KD (codons 221 through 500) of BCR-ABL1 was sequenced by the Sanger method, using a nested PCR approach, with a detection limit of 10-20%.
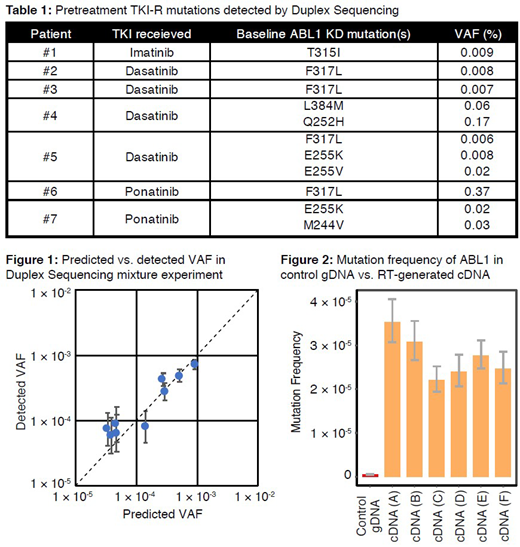
Results: A total of 115 pretreatment ABL1 mutations were detected by DS, with ≥1 mutation detected in 47/64 pts (73%). These mutations were generally present at very low levels (median VAF 0.008% [range, 0.004%-0.649%]). Eleven TKI resistance (TKI-R) mutations (10% of all detected mutations) were identified by DS in 7 pts (Table 1). Eighteen pts (28%) relapsed; however, none of the pts with TKI-R mutations relapsed, despite 5 pts receiving a TKI at least intermediately resistant to the detected mutation. Using Sanger sequencing, TKI-R mutations were detected at relapse in 9 pts (T315I, n=6; F317I, V229L and V338G, n=1 each). None of these TKI-R mutations was detected by DS in pretreatment samples. Together, these results suggest that low-level pretreatment ABL1 KD mutations are not clinically meaningful in Ph+ ALL.
To validate the DS methodology, we examined its sensitivity and specificity using 6 gDNA samples containing ABL1 KD mutations (9 different SNVs, including 6 TKI-R) mixed with control gDNA from a healthy young individual for predicted VAFs of 1/250 to 1/25,000. Mutations were identified by DS with 100% sensitivity down to VAF <5.6x10-5, with high precision (r2=0.93 for detected vs. predicted VAF; Figure 1). None of the spiked-in mutations were detected in the negative control (100% specificity).
In the control samples, the background mutation frequency of ABL1 was ~5x10-7. Using a null model of random distribution of mutations in normal samples at this frequency, there was no significant difference in the proportion of TKI-R mutations detected with DS versus the expected background frequency, suggesting that there is no biased detection of TKI-R mutations by DS.
To assess whether cDNA may be susceptible to artefactual mutations induced by RT steps, DS was performed on cDNA from 6 pts in whom no pretreatment ABL1 KD mutation was detected by DS. cDNA generated by RT had a 50-100x increased mutation frequency relative to the normal gDNA samples (Figure 2). False TKI-R mutations were observed in all cDNA samples (ranging from 11-16 TKI-R variants per sample). Overall, the fraction of TKI-R variants relative to total variants detected was significantly higher in all 6 cDNA samples compared to the negative control.
Conclusion: DS had 100% sensitivity and specificity for detection of TKI-R mutations in ABL1. In contrast, RT-generated cDNA increased mutation frequency by 50- to 100-fold, suggesting that RT-based methods may falsely assess the presence of TKI-R mutations. Prior to TKI treatment, DS detected ABL1 mutations at a rate similar to healthy controls, and even in cases when a low-level TKI-R mutation was detected by DS, there was no association with relapse. Together, these findings suggest that pretreatment ABL1 mutations in Ph+ ALL are due to random age-related mutagenesis and do not represent clinically meaningful subclones that contribute to relapse. The superior accuracy of DS compared to RT-PCR has broad implications in the detection of low-level mutations across cancers, which may allow for early interventions to target emerging resistant subclones.
Short:Takeda Oncology: Consultancy, Research Funding; AstraZeneca: Consultancy; Amgen: Honoraria. Kantarjian:Ariad: Research Funding; AbbVie: Honoraria, Research Funding; Takeda: Honoraria; Agios: Honoraria, Research Funding; Amgen: Honoraria, Research Funding; Cyclacel: Research Funding; Daiichi-Sankyo: Research Funding; Astex: Research Funding; Jazz Pharma: Research Funding; Pfizer: Honoraria, Research Funding; Immunogen: Research Funding; BMS: Research Funding; Actinium: Honoraria, Membership on an entity's Board of Directors or advisory committees; Novartis: Research Funding. Sasaki:Otsuka: Honoraria; Pfizer: Consultancy. Ravandi:Macrogenix: Consultancy, Research Funding; Cyclacel LTD: Research Funding; Menarini Ricerche: Research Funding; Selvita: Research Funding; Xencor: Consultancy, Research Funding; Amgen: Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding. Cortes:Astellas Pharma: Consultancy, Honoraria, Research Funding; Immunogen: Consultancy, Honoraria, Research Funding; Sun Pharma: Research Funding; Merus: Consultancy, Honoraria, Research Funding; BiolineRx: Consultancy; Bristol-Myers Squibb: Consultancy, Research Funding; Biopath Holdings: Consultancy, Honoraria; Takeda: Consultancy, Research Funding; Pfizer: Consultancy, Honoraria, Research Funding; Jazz Pharmaceuticals: Consultancy, Research Funding; Forma Therapeutics: Consultancy, Honoraria, Research Funding; Novartis: Consultancy, Honoraria, Research Funding; Daiichi Sankyo: Consultancy, Honoraria, Research Funding. Konopleva:Reata Pharmaceuticals: Equity Ownership, Patents & Royalties; Astra Zeneca: Research Funding; Agios: Research Funding; Genentech: Honoraria, Research Funding; Ascentage: Research Funding; Kisoji: Consultancy, Honoraria; Calithera: Research Funding; Stemline Therapeutics: Consultancy, Honoraria, Research Funding; Forty-Seven: Consultancy, Honoraria; Eli Lilly: Research Funding; AbbVie: Consultancy, Honoraria, Research Funding; Cellectis: Research Funding; Amgen: Consultancy, Honoraria; Ablynx: Research Funding; F. Hoffman La-Roche: Consultancy, Honoraria, Research Funding. Garcia-Manero:Novartis: Research Funding; Amphivena: Consultancy, Research Funding; Helsinn: Research Funding; AbbVie: Research Funding; Celgene: Consultancy, Research Funding; Astex: Consultancy, Research Funding; Onconova: Research Funding; H3 Biomedicine: Research Funding; Merck: Research Funding. Pratt:TwinStrand Biosciences: Employment. Williams:TwinStrand Biosciences: Employment. Valentine:TwinStrand Biosciences: Employment. Salk:TwinStrand Biosciences: Employment. Radich:TwinStrand Biosciences: Research Funding; Novartis: Other: RNA Sequencing. Jabbour:Pfizer: Consultancy, Research Funding; AbbVie: Consultancy, Research Funding; Takeda: Consultancy, Research Funding; Amgen: Consultancy, Research Funding; Adaptive: Consultancy, Research Funding; BMS: Consultancy, Research Funding; Cyclacel LTD: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal