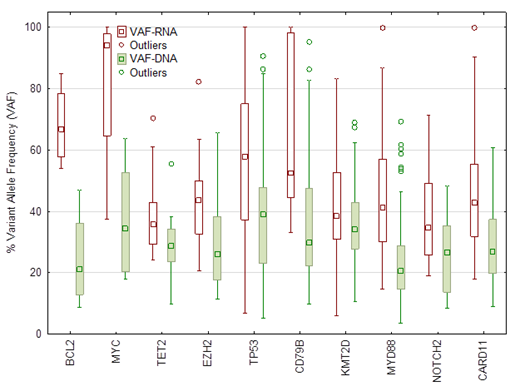
Cellular RNA levels are tightly regulated by very complex nuclear and cytoplasmic processes. The regulation of mutant mRNA in cancer cells is rarely studied. We explored the effects of mutations on mRNA levels in patients with diffuse large B-cell lymphoma (DLBCL). Using next generation sequencing (NGS) and variant allele frequency (VAF) of mutant RNA, we compared relative mutant mRNA or variant allele frequency (RNA-VAF) with variant allele frequency of mutant DNA (DNA-VAF) in the same samples from patients with DLBCL. Methods: RNA and DNA were extracted from 427 FFPE samples from patients with DLBCL. We sequenced the DNA using 177 gene panel and the RNA using 1408 gene panel. The DNA sequencing is based on Single Primer Extension (SPE) library preparation with unique molecular identifier (UMI) (Qiagen, Germantown, MD). The RNA sequencing is based on hybrid capture. Sequencing data of DNA is analyzed using the DRAGEN Platform. Sequence duplicates were removed before calculating VAF. The RNA sequencing data is analyzed using Illumina basespace. RNA VAF is calculated also after removing duplicates using Isaac variant caller. Only mutations detected by both DNA and RNA variant callers are compared. Results: A total of 1770 mutations were detected using the DNA panel and 2207 mutations were detected using the larger RNA sequencing panel. We focused on the most commonly mutated genes that included in both DNA and RNA panels and compared the VAF of the same mutations between DNA and RNA. The selected genes are: KMT2D, NOTCH2, CARD11, MYC, MYD88, EZH2, TP53, CD79B, BCL2, and TET2. The overall VAF in the RNA was significantly higher (P<0.00001) (median:43.9%, minimum: 6%, maximum: 100%) as compared with that of the DNA (median: 28.8%, minimum: 3.5%, maximum: 95%). When each gene is considered individual, all genes showed significantly higher VAF in RNA as compared with DNA. As expected some mutations were detected in DNA, but not in in RNA and vice versa. However, the number of mutations detected in these 10 genes using DNA sequencing was significantly (P= 0.0001) higher (#658) as compared with mutations detected in RNA (#471). Most of the missed mutations by RNA were termination mutations. The most striking RNA-missed mutations were in NOTCH2. The DNA testing showed 81 mutations, while the RNA testing listed only 19 mutations. Almost all NOTCH2 mutations missed by RNA sequencing wer Pro6ArgfsTer27, which leads to early termination of mRNA (loss of function). When we looked at overall NOTCH2 mRNA levels, NOTCH2 mRNA was significantly higher (P=0.002, Kruskal-Wallis ANOVA) in samples with NOTCH2 mutation detected in both DNA and RNA as compared with mutations detected in DNA only. The NOTCH2 mRNA levels were also lower in samples with mutations detected in DNA only (P=0.046) as compared with wild-type NOTCH2. Conclusion: This data suggests that stability of mutant mRNA is significantly higher for most mutations and most genes. However, there are exceptions, especially when the mutations are termination at early amino acid. NOTCH2 pro6ArgfsTer27 mutation is an example of early termination of transcription, which leads to significant instability and reduction in NOTCH2 mRNA levels acting as a tumor suppressor, while other mutations in the gene lead to over expression and more oncogenenic function. This data suggests that molecular profiling of cancer should include evaluating RNA mutations and expression levels and not all mutations detected in a gene are the same. Furthermore, increased stability of most mutant mRNA may have some implication on methods used for detected minimal residual disease.
Albitar:Genomic Testing Ccoperative: Employment, Equity Ownership. Tam:Takeda: Consultancy; Paragon Genomics: Consultancy. Hsi:Abbvie: Research Funding; Jazz: Consultancy; Eli Lilly: Research Funding; Cleveland Clinic&Abbvie Biotherapeutics Inc: Patents & Royalties: US8,603,477 B2. Piris:Nanostring: Membership on an entity's Board of Directors or advisory committees; Kyowa Kirin: Membership on an entity's Board of Directors or advisory committees; Kura: Research Funding; Millenium/Takeda: Membership on an entity's Board of Directors or advisory committees, Other: Lecture Fees, Research Funding; Celgene: Membership on an entity's Board of Directors or advisory committees; Gilead: Membership on an entity's Board of Directors or advisory committees, Research Funding; Jansen: Membership on an entity's Board of Directors or advisory committees, Other: Lecture Fees. Kantarjian:Agios: Honoraria, Research Funding; Daiichi-Sankyo: Research Funding; Pfizer: Honoraria, Research Funding; AbbVie: Honoraria, Research Funding; Immunogen: Research Funding; Novartis: Research Funding; Jazz Pharma: Research Funding; Actinium: Honoraria, Membership on an entity's Board of Directors or advisory committees; Amgen: Honoraria, Research Funding; Astex: Research Funding; Takeda: Honoraria; BMS: Research Funding; Cyclacel: Research Funding; Ariad: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal