Introduction: Cell-free circulating DNA (cfDNA) and its genetic mutations have been studied as a non-invasive tool to obtain clinical information in malignant lymphoma. We analysed serum cfDNA concentrations in lymphoma patients and investigated its gene mutation profile in diffuse large B-cell lymphoma (DLBCL) and follicular lymphoma (FL).
Methods: Serum samples from lymphoma patients diagnosed at our institution between 2016-2017 were obtained at the time of initial diagnosis. Serum samples of 10 healthy individuals were used as control. cfDNA was extracted using Maxwell RSC cfDNA Plasma Kit (Promega), and concentration was measured by Quantus Fluorometer (Promega). The serum concentration level in each subtype of lymphoma was compared to control using Mann-Whitney U test. Furthermore, progression free survival (PFS) was compared between those with and without elevated cfDNA concentrations using Kaplan-Meier method. We also assessed factors associated with elevated cfDNA. Using cfDNA, multiplex PCR was performed, then a sequence library constructed with an Ion Custom Amplicon panel. The panel for the sequence library was designed using the Ion AmpliSeq DesignerTM . Genomes were sequenced for the selected 121 target genes using the Ion ProtonTM System. Frequencies of each mutation were compared amongst DLBCL, FL, and DLBCL + FL using Kruskal-Wallis test.
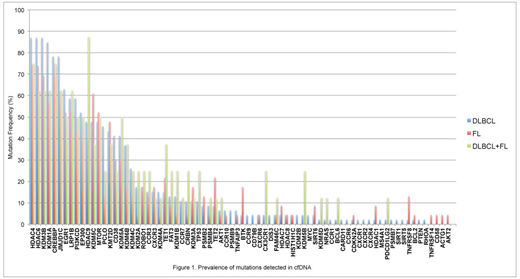
Results: We analysed 114 patients with newly diagnosed lymphoma (DLBCL n=63, DLBCL combined with FL n=8, DLBCL combined with extranodal marginal zone B cell lymphoma (MALT) n=1, FL n=20, FL/MALT n=1, Burkitt lymphoma (BL) n=1, MALT n=8, Mantle cell lymphoma (MCL) n=3, Low-grade B cell lymphoma n=2, Hodgkin lymphoma (HL) n=4, Peripheral T cell lymphoma (PTCL) n=3), as well as control (n=10). The median cfDNA concentration in all lymphoma patients was 55.8ng/ml (range 6.2-1220.0ng/ml), which was significantly higher than that for control (median 7.5ng/ml, range 4.4-14.1ng/ml; p<0.01). Elevated cfDNA concentration compared to control was observed in patients with DLBCL (median 61.7ng/ml, p<0.01), FL (52.3ng/ml, p<0.01), HL (116.0ng/ml, p=0.04) and MALT (37.4ng/ml, p=0.02). No significant differences were observed among different subtypes in our study. Elevated cfDNA (>40ng/ml) was significantly associated with shorter PFS in patients with DLBCL (PFS at 24 months; 100% vs. 79%, p=0.03). For this group of patients, B-symptoms (p=0.04), bulky masses (p=0.03) or non-germinal center B-cell like (GCB) subtype (p=0.02) were found to correlate with elevated cfDNA levels. We then analysed 77 cases with available genetic mutation data (DLBCL n=46, FL n=23, DLBCL + FL n=8). The frequencies of each mutation for each subtype are shown in Figure 1. Gene mutation profiles were similar in each group; CREBBP, EGR1, HDAC4, HDAC6, JMJD1C, and KDM3B being some of the most frequently mutated genes. PCLO and KDM1A mutations were more frequently found in DLBCL compared to FL.
Discussion and Conclusion: Serum cfDNA concentration is predictive of certain clinical characteristics and may be used as one of the predictors for survival outcomes in DLBCL. PCLO encodes protein that is part of the presynaptic cytoskeletal matrix and although its role in lymphoma is unclear, its mutation has previously been reported. KDM1A encodes histone demethylase, which has been widely discussed as a target for cancer therapy, yet its role in lymphoma is poorly understood. Further studies with a larger sample size and longer follow up period are warranted to better understand the role of cfDNA gene mutations in obtaining clinical information.
Mishima:Chugai-Roche Pharmaceuticals Co.,Ltd.: Consultancy. Yokoyama:Chugai-Roche Pharmaceuticals Co.,Ltd.: Consultancy. Nishimura:Celgene K.K.: Honoraria; Chugai-Roche Pharmaceuticals Co.,Ltd.: Consultancy. Hatake:Celgene K.K.: Research Funding; Janssen Pharmaceutical K.K.: Research Funding; Takeda Pharmaceutical Co.,Ltd.: Honoraria. Terui:Bristol-Myers Squibb, Celgene, Janssen, Takeda, MSD, Eisai, Ono, and Chugai-Roche Pharmaceuticals Co.,Ltd.: Honoraria; Bristol-Myers Squibb K.K.: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal