Key Points
PHF19 is positively correlated with poorer clinic outcomes of MM and essential for MM growth in vitro and in vivo.
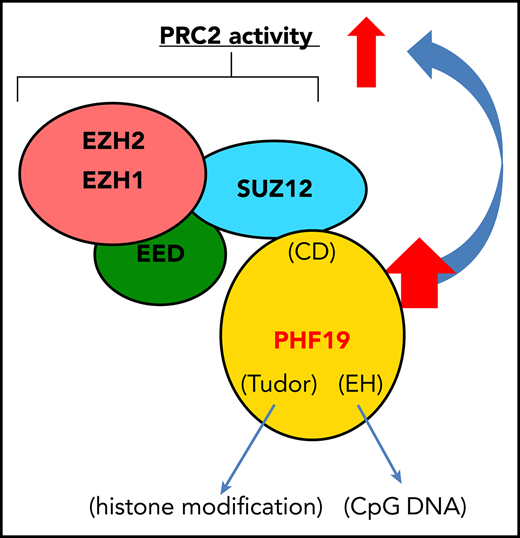
PHF19 potentiates tumorigenicity via PRC2 activation and H3K27me3 spreading, rendering sensitivity of MM cells to PRC2 inhibitors.
Abstract
Dysregulation of polycomb repressive complex 2 (PRC2) promotes oncogenesis partly through its enzymatic function for inducing trimethylation of histone H3 lysine 27 (H3K27me3). However, it remains to be determined how PRC2 activity is regulated in normal and diseased settings. We here report a PRC2-associated cofactor, PHD finger protein 19 (PHF19; also known as polycomb-like 3), as a crucial mediator of tumorigenicity in multiple myeloma (MM). Overexpression and/or genomic amplification of PHF19 is found associated with malignant progression of MM and plasma cell leukemia, correlating to worse treatment outcomes. Using various MM models, we demonstrated a critical requirement of PHF19 for tumor growth in vitro and in vivo. Mechanistically, PHF19-mediated oncogenic effect relies on its PRC2-interacting and chromatin-binding functions. Chromatin immunoprecipitation followed by sequencing profiling showed a critical role for PHF19 in maintaining the H3K27me3 landscape. PHF19 depletion led to loss of broad H3K27me3 domains, possibly due to impaired H3K27me3 spreading from cytosine guanine dinucleotide islands, which is reminiscent to the reported effect of an “onco”-histone mutation, H3K27 to methionine (H3K27M). RNA-sequencing–based transcriptome profiling in MM lines also demonstrated a requirement of PHF19 for optimal silencing of PRC2 targets, which include cell cycle inhibitors and interferon-JAK-STAT signaling genes critically involved in tumor suppression. Correlation studies using patient sample data sets further support a clinical relevance of the PHF19-regulated pathways. Lastly, we show that MM cells are generally sensitive to PRC2 inhibitors. Collectively, this study demonstrates that PHF19 promotes MM tumorigenesis through enhancing H3K27me3 deposition and PRC2’s gene-regulatory functions, lending support for PRC2 blockade as a means for MM therapeutics.
Introduction
Polycomb repressive complex 2 (PRC2) plays pivotal roles in both normal and malignant development.1,,-4 Biochemically, PRC2 forms a delicate multimeric core structure5 and utilizes an enzymatic subunit, either enhancer of Zeste homolog 2 (EZH2) or a related EZH1 methyltransferase, to catalyze methylation of histone H3 lysine 27 (H3K27). H3K27 trimethylation (H3K27me3) is believed to elicit transcriptional silencing effects via recruiting downstream readers and effectors, thereby modulating gene-expression programs crucial for development, differentiation, and cell fate determination.2,4,6,7 Previous studies also documented important roles for various PRC2-interacting factors, including JARID2,8,-10 polycomb-like (comprising 3 family members: PHF1/PCL1, MTF2/PCL2, and PHF19/PCL3)11,,,-15 and RNAs,16,17 in regulating the genomic targeting and/or enzymatic activities of PRC2 under different biological contexts.6 Mutation and deregulation of the PRC2-encoding genes are frequent in cancer.4,18 Deep sequencing of patient samples has identified recurrent gain-of-function and loss-of-function mutations of EZH2 in B-cell lymphoma and myeloid neoplasms, respectively.19,-21 These mutations were subsequently demonstrated to promote oncogenesis using relevant models.4,22,-24 However, it remains to be defined whether deregulation of various PRC2-associated partners is also crucially involved in malignant development.
Here, we report that PHF19, a polycomb-like member of PRC2 cofactors, acts as a critical mediator of tumorigenesis in multiple myeloma (MM), a common malignancy of plasma cells. MM and plasma cell leukemia (PCL), a more aggressive form of MM, develop from clinically insidious stages such as monoclonal gammopathy of uncertain significance through a step-wise progression, which often involves acquisition of both genetic and epigenetic alterations to facilitate generation of full-blown tumors.25,,,,-30 We find overexpression and genomic gain of PHF19 associated with malignant progression of MM and PCL. There is a marked correlation between higher expression of PHF19 and worse outcomes of MM patients in several clinical trial studies. Using loss-of-function approaches, we demonstrate essential roles of PHF19 in promoting MM tumor growth both in vitro and in the xenografted animal models. Mechanistically, the oncogenic function of PHF19 depends on a C-terminal domain that mediates physical interaction with PRC2, as well as the N-terminal regions known to bind chromatin. Suppressing PHF19 expression in MM cells not only leads to the globally decreased H3K27me3 but also, importantly, results in the derepression of PRC2 target genes. Notably, PHF19 depletion leads to loss of broad H3K27me3 domains, possibly due to impaired spreading of H3K27me3 from cytosine guanine dinucleotide island (CGI) elements, whereas a majority of CGI-bound H3K27me3 peaks are found retained. Transcriptome profiling data obtained from both MM cell lines and primary patient samples further reveal a positive correlation between PHF19 and the silencing of cell cycle inhibitors and interferon-JAK-STAT signaling genes. Further, we show that the enforced expression of STAT1, a gene downstream of interferon-JAK signaling, or treatment with PRC2 inhibitors, suppressed MM growth. Taken together, this study describes a previously unexplored yet critical oncogenic pathway in MM in which PHF19 overexpression enhances broad H3K27me3 domain formation and PRC2 activities to promote malignant progression and transformation. Despite recent improvement in MM therapeutics, targeting the PHF19-PRC2 complex shall expand the current anti-MM arsenal, especially for those refractory cases.
Methods
Cell lines and tissue culture
Human MM lines used in the study are MM1.S (American Tissue Culture Collection [ATCC], CRL-2974), KMS11 (a gift of K.C. Anderson), L-363 (DSMZ, ACC49), U266 (ATCC, TIB-196), NCI-H929 (ATCC, CRL-9068), and RPMI-8226 (ATCC, CRM-CCL-155). Luciferase-labeled L-363 and U266 lines were previously described.31 Other lines include K562 (ATCC, CRL-243), Raji (ATCC, CCL-86), 293T (ATCC, CRL-3216), and NIH-3T3 (ATCC, CRL-165). These lines were cultured according to the vendor’s specifications.
Virus preparation, infection, and stable cell generation
Virus was prepared with the packaging system in 293T cells and used for the infection of cells, following by drug selection as previously described.13,32,33 Details of the used short hairpin RNA (shRNA), single guide RNA (sgRNA), and complementary DNAs (cDNAs) are described in supplemental Methods (available on the Blood Web site).
Western blot and coIP
Mass spectrometry analysis
For quantification of histone modifications, total histones were isolated from cells using an acidic extraction protocol and subjected to mass-spectrometry–based measurement.33,-35 Purification and identification of PHF19-associated complexes from cells stably expressing a Flag-tagged PHF19 were performed as before.13
ChIP-seq and RNA-seq
Chromatin immunoprecipitation followed by sequencing (ChIP-seq) was carried out as before13,32,33,36 and ChIP samples submitted to the University of North Carolina (UNC) Chapel Hill High-Throughput Sequencing Facility for preparation of multiplexed libraries. RNA preparation and RNA-sequencing (RNA-seq) library construction using TruSeq RNA Library Preparation Kit-v2 (Illumina, RS-122-2002) were previously described.13,32,33,36
GSEA
Gene set enrichment analysis (GSEA) was performed with the downloaded GSEA software (www.broadinstitute.org/gsea) by exploring the Molecular Signatures Database (www.broadinstitute.org/gsea/msigdb/annotate.jsp).
Human tumor line xenografted models
All animal experiments were approved by and conducted in accord with guidelines of Institutional Animal Care and Use Committee at UNC. Nonobese diabetic/severe combined immunodeficiency/IL2Rgamma-null mice (NSG; JAX Laboratory) were maintained by the Animal Studies Core, UNC Lineberger Cancer Center. Each recipient mouse was inoculated with 1 million luciferase-labeled MM cells suspended in phosphate-buffered saline via tail-vein injection. In vivo MM growth was monitored via weekly chemiluminescence imaging of mice following intraperitoneal injection with d-luciferin. In vivo treatment with UNC1999 was performed using the same protocol described before.33
Statistical analysis
Results of quantification were presented in the format of average ± standard deviation of 3 independent experiments. For survival analysis, the log-rank (Mantel-Cox) test was performed. All other analyses used the Student t test.
Additional methods
Additional methods are provided in supplemental Methods.
Results
PHF19 overexpression underlies malignant progression of MM and PCL, correlating with poorer clinical outcomes of patients
Using the Cancer Cell Line Encyclopedia transcriptome data set,37 we found that PHF19 expression is significantly higher in B-cell–derived malignancies, including MM, relative to other blood cancer subtypes and solid tumors (Figure 1A; supplemental Figure 1A). Next, we examined the publicly available data sets of MM patient samples and found the elevated expression and/or genomic gains of PHF19 correlated significantly with disease progression from normal or clinically insidious stages to malignant MM and PCL (Figure 1B-D; supplemental Figure 1B). Genomic gain of PHF19, located at the chromosome 9q33, may be partly due to trisomy 9 (data not shown), an early event of MM progression,38 and the degree of PHF19 gain was found comparable to that of BRD4 (Figure 1D), an oncogene previously shown to be amplified in MM.39 Elevated expression of PHF19 was also found associated with relapse of MM (Figure 1E). Importantly, by exploring multiple clinical trial studies of primary MM patients with outcome data,40,41 we consistently identified PHF19 among the top-ranked transcripts showing the most significant correlation to poorer prognosis (supplemental Figure 1C and supplemental Table 1); MM patients with higher PHF19 expression in tumor generally displayed a significantly worse outcome in clinical trials, including an advanced Total Therapy 6 treatment regimen40 (Figure 1F-G; supplemental Figure 1D). Together, these observations strongly indicate a role of PHF19 in promoting malignant progression and relapse of MM, which was not studied before.
Overexpression of PHF19 is correlated with malignant progression and adverse clinical outcomes of MM patients. (A) PHF19 expression in MM, relative to other indicated cancers, according to the Cancer Cell Line Encyclopedia data set.37 AML, acute myeloid leukemia; T-ALL, T-cell acute lymphoblastic leukemia. *P < .05; **P < .01; ***P < .001; ****P < .0001. (B-C) PHF19 expression in samples from normal controls or patients diagnosed with monoclonal gammopathy of undetermined significance (MGUS), smoldering MM (SMM), MM, or PCL in the indicated data sets. (D) Copy-number variation of the indicated gene in MM patients (National Center for Biotechnology Information Gene Expression Omnibus data set GSE21349). (E) PHF19 expression in MM patients, either at baseline or after disease relapse, based on the indicated data set. (F-G) Kaplan-Meier survival curve for PHF19 expression in the Mulligan et al cohort of relapsed MM patients (F; n = 264) receiving the trial of bortezomib41 and the Heuck et al cohort of relapsed MM patients (G; n = 55) receiving the Total Therapy 6 (TT6) regimen.40 NS, not significant.
Overexpression of PHF19 is correlated with malignant progression and adverse clinical outcomes of MM patients. (A) PHF19 expression in MM, relative to other indicated cancers, according to the Cancer Cell Line Encyclopedia data set.37 AML, acute myeloid leukemia; T-ALL, T-cell acute lymphoblastic leukemia. *P < .05; **P < .01; ***P < .001; ****P < .0001. (B-C) PHF19 expression in samples from normal controls or patients diagnosed with monoclonal gammopathy of undetermined significance (MGUS), smoldering MM (SMM), MM, or PCL in the indicated data sets. (D) Copy-number variation of the indicated gene in MM patients (National Center for Biotechnology Information Gene Expression Omnibus data set GSE21349). (E) PHF19 expression in MM patients, either at baseline or after disease relapse, based on the indicated data set. (F-G) Kaplan-Meier survival curve for PHF19 expression in the Mulligan et al cohort of relapsed MM patients (F; n = 264) receiving the trial of bortezomib41 and the Heuck et al cohort of relapsed MM patients (G; n = 55) receiving the Total Therapy 6 (TT6) regimen.40 NS, not significant.
PHF19 is essential for MM cell proliferation in vitro
The PHF19 gene encodes different protein isoforms: PHF19, a long isoform known to interact with PRC2,11,12 and PHF19S, a short isoform only containing the N-terminal domains of PHF1942 (Figure 2A). Both isoforms are expressed in MM cell lines and patient samples, with their relative expression correlating positively with one another (supplemental Figure 2A-B). To determine potential MM-promoting functions of PHF19, we stably transduced independent shRNA, which targets either PHF19 alone or both PHF19 and PHF19S isoforms (Figure 2A, red lines), into a set of MM and/or PCL cell lines that cover different genetic subtypes of MM such as t(4;14) and t(14;16)30 (supplemental Figure 2C). PHF19 shRNAs had the expected knockdown (KD) effect on PHF19 isoforms (Figure 2B-C), which also led to the severely impaired growth of each tested MM/PCL line (Figure 2D-I). To rule out off-target effect of shRNA, we further rescued PHF19-KD cells with a PHF19 cDNA carrying “silent” mutations at shRNA-targeted regions without affecting the encoded protein sequence. We found that rescue with the long isoform PHF19, but not the short PHF19S, fully rescued proliferation defects seen in different KD lines (Figure 2J-L; supplemental Figure 2D-G). Additionally, the PHF19-dependent MM cell growth was confirmed using CRISPR/Cas9-mediated PHF19 knockout (Figure 2M-O). Using soft agar–based colony formation, a surrogate assay for scoring MM-initiating stem cells, we found that PHF19 KD suppressed colony-forming capacities of KMS11 and RPMI-8226 cells, a defect again readily rescued by the long isoform of PHF19, but not PHF19S (Figure 2P-R). We also noticed that MM cells after rescue of KD expressed PHF19 at a significantly higher level than parental cells (supplemental Figure 2D) and also showed higher rates of proliferation (Figure 2K-L; supplemental Figure 2F-G), indicating a correlation between PHF19 expression and degree of MM aggressiveness. Further, compared with control, ectopic expression of PHF19 and not PHF19S significantly promoted colony formation of NIH-3T3 fibroblast cells in the soft agar assays (Figure 2S). Together, these results demonstrate a critical requirement of PHF19 for sustaining MM growth in vitro. PHF19 KD in 2 non-MM cell lines that express PHF19 at comparable levels, including K562 myeloid leukemia cells and Raji lymphoma cells (supplemental Figure 2A), did not affect cell growth (Figure 2T), indicating a unique involvement of PHF19 in MM tumorigenesis.
PHF19 is essential for MM cell growth in vitro. (A) Domain structure of PHF19 isoforms. Red lines indicate the used shRNAs. (B-C) Immunoblots (B) and reverse transcription (RT) followed by qPCR (C) of PHF19 after shRNA-induced KD in L-363 cells. (D-I) Growth of L-363 (D), MM1.S (E), RPMI-8226 (F), NCI-H929 (G), KMS11 (H), and U266 (I) cells after transduction of the indicated shRNA relative to empty vector (EV). (J) Immunoblots of the indicated Flag-tagged PHF19 isoform used for rescuing PHF19 KD in L-363 cells. (K-L) Growth of PHF19-KD L-363 (K) or MM1.S (L) cells after rescue with either EV or the indicated PHF19 isoform, compared with parental WT cells. (M-O) Growth (M) of the indicated MM cells stably transduced with a PHF19-targeting sgRNA (sg) and doxycycline (Dox)-inducible Cas9, following treatment with doxycycline relative to mock. Cas9/sg-mediated genomic editing of PHF19 was verified by immunoblotting (N) and sequencing (O). (P-S) Colony-formation assays using KMS11 (P-Q; representative colony images shown in panel P), RMPI-8226 (R), or NIH-3T3 cells (S) after stable transduction of the indicated shRNA and/or gene. (T) Growth of K562 or Raji cells after transduction of the indicated shRNA relative to mock. KO, knockout; PAM, protospacer adjacent motif.
PHF19 is essential for MM cell growth in vitro. (A) Domain structure of PHF19 isoforms. Red lines indicate the used shRNAs. (B-C) Immunoblots (B) and reverse transcription (RT) followed by qPCR (C) of PHF19 after shRNA-induced KD in L-363 cells. (D-I) Growth of L-363 (D), MM1.S (E), RPMI-8226 (F), NCI-H929 (G), KMS11 (H), and U266 (I) cells after transduction of the indicated shRNA relative to empty vector (EV). (J) Immunoblots of the indicated Flag-tagged PHF19 isoform used for rescuing PHF19 KD in L-363 cells. (K-L) Growth of PHF19-KD L-363 (K) or MM1.S (L) cells after rescue with either EV or the indicated PHF19 isoform, compared with parental WT cells. (M-O) Growth (M) of the indicated MM cells stably transduced with a PHF19-targeting sgRNA (sg) and doxycycline (Dox)-inducible Cas9, following treatment with doxycycline relative to mock. Cas9/sg-mediated genomic editing of PHF19 was verified by immunoblotting (N) and sequencing (O). (P-S) Colony-formation assays using KMS11 (P-Q; representative colony images shown in panel P), RMPI-8226 (R), or NIH-3T3 cells (S) after stable transduction of the indicated shRNA and/or gene. (T) Growth of K562 or Raji cells after transduction of the indicated shRNA relative to mock. KO, knockout; PAM, protospacer adjacent motif.
PHF19 is required for MM tumorigenesis in vivo
Next, we aimed to determine whether PHF19 is required for MM tumorigenesis in vivo. To this end, we generated 2 independent xenografted models via tail vein injection of the luciferase-labeled MM1.S (Figure 3A-D) or L-363 cells (Figure 3E-G) into NSG mice. In these xenografts, PHF19 KD was achieved using different approaches: doxycyline inducible (Figure 3A-D) or stable expression of shRNA (Figure 3E-G). Through chemiluminescence imaging (Figure 3B-C,F-G; panels of “vehicle”) and examination of bone marrow cells (Figure 3D; supplemental Figure 3A), we observed efficient bone infiltration of MM cells and subsequent malignant expansion in the bone of mice xenografted with mock-treated cells. Such a tumorigenic process was found significantly suppressed by PHF19 KD in both MM1.S (Figure 3A-D, Dox vs vehicle) and L-363 xenografted models (Figure 3E-G, sh1254 vs shEV). As a result, mice xenografted with the PHF19-KD MM cells displayed a significantly prolonged event-free survival compared with controls (Figure 3A,E, red vs black). At the same time, rescue of KD restored tumorigenesis in vivo with a significantly accelerated kinetics (Figure 3E-G, blue vs red). Together, the MM cell xenografted models substantiated an important role for PHF19 in malignant growth in vivo.
PHF19 is required for MM tumorigenesis in vivo. (A-C) Kaplan-Meier curve showing the event-free survival of NSG mice xenografted with the luciferase-labeled MM1.S cells that stably expressed a doxycyline-inducible, PHF19-specific shRNA (sh1254_Tet-on). Seven days after injection of cells via tail vein, mice were randomized into 2 cohorts, with one subjected to vehicle treatment as control and the other to doxycycline (Dox) as the PHF19-KD cohort, followed by weekly measurement of chemiluminescence signals (representative image and summary of signals shown in panels B and C, respectively). (D) Wright-Giemsa staining of the bone marrow samples prepared 8 weeks after xenograft of MM1.S cells into NSG mice, either mock treated (vehicle) or with PHF19 KD (Dox). Scale bar, 10 μM. (E-G) Kaplan-Meier curve for the event-free survival (E), representative image (F; 30 days postxenograft), and summary of chemiluminescence signals (G) of mice xenografted with the luciferase-labeled L-363 cells that stably expressed either empty vector (shEV), a PHF19-specific shRNA (sh1254), or sh1254 plus an sh1254-resistant PHF19 cDNA (sh1254+PHF19). n, cohort size.
PHF19 is required for MM tumorigenesis in vivo. (A-C) Kaplan-Meier curve showing the event-free survival of NSG mice xenografted with the luciferase-labeled MM1.S cells that stably expressed a doxycyline-inducible, PHF19-specific shRNA (sh1254_Tet-on). Seven days after injection of cells via tail vein, mice were randomized into 2 cohorts, with one subjected to vehicle treatment as control and the other to doxycycline (Dox) as the PHF19-KD cohort, followed by weekly measurement of chemiluminescence signals (representative image and summary of signals shown in panels B and C, respectively). (D) Wright-Giemsa staining of the bone marrow samples prepared 8 weeks after xenograft of MM1.S cells into NSG mice, either mock treated (vehicle) or with PHF19 KD (Dox). Scale bar, 10 μM. (E-G) Kaplan-Meier curve for the event-free survival (E), representative image (F; 30 days postxenograft), and summary of chemiluminescence signals (G) of mice xenografted with the luciferase-labeled L-363 cells that stably expressed either empty vector (shEV), a PHF19-specific shRNA (sh1254), or sh1254 plus an sh1254-resistant PHF19 cDNA (sh1254+PHF19). n, cohort size.
PHF19 facilitates broad H3K27me3 domain formation, possibly by promoting H3K27me3 spreading from CGIs
To dissect the molecular function of PHF19 in MM, we first performed biochemical pull-down of PHF19 followed by mass spectrometry–based identification and detected PRC2 core components such as EZH2 and SUZ12 as PHF19’s major partners (supplemental Figure 3B). To further assess such a PHF19-PRC2 interaction, we turned to mass spectrometry–based quantification of histone modifications and observed the significantly decreased H3K27me3 in L-363 cells with PHF19 KD relative to mock (Figure 4A, red vs black; regardless of H3 isoform). Expression and stability of PRC2 core components were unaffected by PHF19 KD in L-363 or NCI-H929 cells (supplemental Figure 3C-D). Post-KD of PHF19, we also observed an increase of H3K36me2 (Figure 4A), a histone modification antagonizing H3K27me3.25,43 Alterations of H3K27me3 and H3K36me2 due to PHF19-KD were corrected following PHF19 rescue (Figure 4A, gray vs red). By immunoblotting, we validated these alterations in L-363 and MM1.S cells after PHF19 KD (Figure 4B; supplemental Figure 3E).
PHF19 potentiates the PRC2-catalyzed H3K27me3 in MM. (A-B) Mass spectrometry (A) and immunoblotting (B) analyses of the indicated histone modifications in L-363 cells following mock treatment (shEV), PHF19 KD (sh1254), or PHF19-KD rescue (sh1254+PHF19). (C-E) Heatmap (C) and the averaged H3K27me3 ChIP-seq read densities (D-E) at the H3K27me3 peaks called in L-363 cells with mock treatment (shEV) or PHF19-KD (shPHF19), grouped into peaks at promoter (C, left, and D) and nonpromoter regions (C, right, and E). Shown on each row of heatmap is ChIP-seq read density across ±5 kb from the center of a called peak, with columns sorted by the H3K27me3 signals in mock-treated cells. Data shown here used reads sampled to the same sequencing depth. (F-G) Integrative Genomics Viewer views of H3K27me3 ChIP-seq read densities at genomic regions covering the CDKN1C-KCNQ1 (F) and STAT5B/5A/3 loci (G) in L-363 cells, either mock treated (shEV) or with PHF19-KD (shPHF19), and WT MM1.S and KMS11 cells. Bottom of the panels shows distribution of CGI elements (black bars). (H) Summary of the total number of H3K27me3 peaks, detected at genomic regions with (red) or without (blue) CGI, in L-363 cells stably expressed with shEV or shPHF19. (I) Venn diagram showing H3K27me3 peaks in WT KMS11 and L363 cells. (J) H3K27me3 ChIP-qPCR for the indicated gene promoter in L-363, MM1.S, and KMS11 cells stably transduced with shEV or shPHF19. Y-axis shows the average ± standard error (SE) of signals from 3 independent experiments after normalization to input. *P < .05; **P < .01.
PHF19 potentiates the PRC2-catalyzed H3K27me3 in MM. (A-B) Mass spectrometry (A) and immunoblotting (B) analyses of the indicated histone modifications in L-363 cells following mock treatment (shEV), PHF19 KD (sh1254), or PHF19-KD rescue (sh1254+PHF19). (C-E) Heatmap (C) and the averaged H3K27me3 ChIP-seq read densities (D-E) at the H3K27me3 peaks called in L-363 cells with mock treatment (shEV) or PHF19-KD (shPHF19), grouped into peaks at promoter (C, left, and D) and nonpromoter regions (C, right, and E). Shown on each row of heatmap is ChIP-seq read density across ±5 kb from the center of a called peak, with columns sorted by the H3K27me3 signals in mock-treated cells. Data shown here used reads sampled to the same sequencing depth. (F-G) Integrative Genomics Viewer views of H3K27me3 ChIP-seq read densities at genomic regions covering the CDKN1C-KCNQ1 (F) and STAT5B/5A/3 loci (G) in L-363 cells, either mock treated (shEV) or with PHF19-KD (shPHF19), and WT MM1.S and KMS11 cells. Bottom of the panels shows distribution of CGI elements (black bars). (H) Summary of the total number of H3K27me3 peaks, detected at genomic regions with (red) or without (blue) CGI, in L-363 cells stably expressed with shEV or shPHF19. (I) Venn diagram showing H3K27me3 peaks in WT KMS11 and L363 cells. (J) H3K27me3 ChIP-qPCR for the indicated gene promoter in L-363, MM1.S, and KMS11 cells stably transduced with shEV or shPHF19. Y-axis shows the average ± standard error (SE) of signals from 3 independent experiments after normalization to input. *P < .05; **P < .01.
To further dissect requirement of PHF19 for genome-wide deposition of H3K27me3 in MM, we performed H3K27me3 ChIP-seq in L363 cells before and after PHF19 KD (supplemental Figure 4A). We found that, compared with mock, PHF19 KD caused H3K27me3 loss or decrease at a majority (88%; 21 588 out of a total of 24 633 peaks) of the H3K27me3 peaks, irrespective of their locations at promoter or nonpromoter regions (Figure 4C-E; supplemental Figure 4B). Also note that there is a general loss of broad H3K27me3 domains after depletion of PHF19 in cells, as exemplified by the HOX gene clusters (supplemental Figure 4C-D, L363 shEV vs shPHF19), the cell cycle inhibitor CDKN1C (Figure 4F), and immune-related genes (STAT5A/5B, JAK1; Figure 4G; supplemental Figure 4E). In both wild-type (WT) and PHF19-KD cells, H3K27me3 demonstrated similar distributions among the promoter, intragenic, and intergenic regions (supplemental Figure 4F). However, only ∼15% of H3K27me3 peaks in WT cells were found at genomic regions with CGIs, a genomic element known to be crucial for initial recruitment of PRC2,44,-46 whereas the percentage of peaks with CGIs is relatively high (∼45%) in PHF19-KD cells due to disproportional loss of H3K27me3 at non-CGI regions (Figure 4H; also see black bars for CGIs in Figure 4F-G and supplemental Figure 4C-E). Such a drastic loss of H3K27me3 at non-CGI regions in PHF19-KD cells is consistent with the reported roles of polycomb-like proteins in stabilizing PRC2 at binding sites and/or promoting its distribution to nearby genomic regions.11,-13,46,-48 Furthermore, ∼60% of H3K27me3 peaks detected in MM1.S and KMS11 cells overlapped those in WT L-363 cells (Figure 4I; supplemental Figure 4G), as shown by those of known PRC2 targets (Figure 4F-G; supplemental Figure 4C-E). By ChIP followed by quantitative polymerase chain reaction (ChIP-qPCR), we validated a requirement of PHF19 for deposition of H3K27me3 at HOXB9, CDKN1A/C, and STAT5A in different MM lines (Figure 4J). Collectively, we show that in MM, PHF19 enhances the PRC2-mediated catalysis of H3K27me3, probably by promoting both the initial targeting of PRC2 and subsequent spreading of H3K27me3 to form broad repressive domains in the genome. Interestingly, loss of broad H3K27me3 domains seen after depletion of PHF19 is reminiscent to what was observed with an “onco”-histone mutation, H3K27 to methionine (H3K27M),49,50 thus showing that both positive and negative regulation of PRC2 activity may underlie tumorigenesis.
Interaction with PRC2 is essential for PHF19 to promote tumorigenesis in MM
To further define the role for PHF19-PRC2 interactions in MM oncogenesis, we performed coIP and found the C-terminal chromo-like domain (CD; Figure 2A) of PHF19 to be essential for the PHF19-PRC2 interaction (Figure 5A). Such a CD-dependent interaction with PRC2 was conserved in other polycomb-like proteins, PHF1 (Figure 5B) and MTF2 (supplemental Figure 5A). When transduced to MM cells, such a PRC2-interaction–defective mutant of PHF19 (PHF19ΔCD) exhibited comparable expression and nuclear localization relative to WT (supplemental Figure 5B; Figure 5C); however, unlike WT, PHF19ΔCD failed to rescue the slowed tumor growth caused by PHF19 KD, as assayed in liquid culture using MM1.S and L-363 cells (Figure 5D-E) or soft agar–based colony formation assay using KMS11 cells (Figure 5F). Relative to WT, PHF19ΔCD failed to sustain malignant tumorigenesis in the L-363 xenografted model, as measured by event-free survival (Figure 5G) and in vivo tumor growth (Figure 5H-I). Consistently, GSEA with multiple publicly available transcriptome data sets of primary MM samples identified positive correlations between higher PHF19 expression and PRC2-mediated gene silencing (Figure 5J-M; supplemental Figure 5C). These data collectively demonstrate that PHF19 relies on physical interaction with PRC2 to promote MM oncogenesis in cell line models and that such an interaction is clinically relevant.
Physical interaction with PRC2 is required for PHF19 to promote MM tumorigenesis. (A-B) CoIP for interaction of Flag-tagged PHF19 (A) or PHF1 (B), either WT or ΔCD, with endogenous EZH2 and SUZ12 in 293 cells. (C) Immunofluorescence of the indicated Flag-PHF19 in L-363 cells. Scale bar, 5 μM. (D-F) Growth of the indicated PHF19-KD MM cells after transduction of EV or the WT or ΔCD PHF19, relative to parental cells, in liquid culture (D-E) or soft agar–based (F) assays. (G-I) Kaplan-Meier curve of event-free survival (G), chemiluminescence signals (H), and representative imaging (I; 8 weeks after xenograft) of NSG mice xenografted with the PHF19-KD L-363 cells after rescue with EV or sh1254-resistant PHF19, either WT or ΔCD. (J-M) GSEA shows that higher PHF19 expression is negatively correlated with expression of genes directly bound by H3K27me3 (J) or repressed by EZH2 (K) or EED (L) and positively related to genes displaying positive correlation to EED (M) in 264 of MM patients, with the cohort divided to PHF19-high (top 50%) and PHF19-low (bottom 50%) based on a transcriptome data set (GSE9782).
Physical interaction with PRC2 is required for PHF19 to promote MM tumorigenesis. (A-B) CoIP for interaction of Flag-tagged PHF19 (A) or PHF1 (B), either WT or ΔCD, with endogenous EZH2 and SUZ12 in 293 cells. (C) Immunofluorescence of the indicated Flag-PHF19 in L-363 cells. Scale bar, 5 μM. (D-F) Growth of the indicated PHF19-KD MM cells after transduction of EV or the WT or ΔCD PHF19, relative to parental cells, in liquid culture (D-E) or soft agar–based (F) assays. (G-I) Kaplan-Meier curve of event-free survival (G), chemiluminescence signals (H), and representative imaging (I; 8 weeks after xenograft) of NSG mice xenografted with the PHF19-KD L-363 cells after rescue with EV or sh1254-resistant PHF19, either WT or ΔCD. (J-M) GSEA shows that higher PHF19 expression is negatively correlated with expression of genes directly bound by H3K27me3 (J) or repressed by EZH2 (K) or EED (L) and positively related to genes displaying positive correlation to EED (M) in 264 of MM patients, with the cohort divided to PHF19-high (top 50%) and PHF19-low (bottom 50%) based on a transcriptome data set (GSE9782).
Transcriptome analyses further delineated the PHF19-enforced gene pathways crucial for MM tumorigenesis
To delineate PHF19-mediated gene-regulatory networks in MM, we used RNA-seq to profile transcriptomes of MM1.S cells after PHF19 KD and rescue (supplemental Figure 6A-B) and identified 2366 genes that were derepressed due to PHF19 KD and then resilenced following PHF19 rescue (Figure 6A; supplemental Table 2). In agreement with the above correlational studies using patient data sets, GSEA using cell line RNA-seq data also showed positive relationship between PHF19 and PRC2-mediated silencing (Figure 6B-D; supplemental Figure 6C). PHF19 was also found to be positively correlated to transcripts associated with cell cycle progression or proliferation (Figure 6E-G; supplemental Figure 6D-E), providing an explanation for the observed role of PHF19 in MM growth. The same correlations also exist in transcriptome data sets of primary patients (Figure 6H-I; supplemental Figure 6F-G). Further, cell cycle progression analysis of MM1.S cells post-KD of PHF19 detected defects in the G1-S phase transition (Figure 6J). These results are consistent with ChIP-seq showing that cell cycle inhibitor genes are targets of PRC2 and H3K27me3 in MM (eg, CDKN1C; Figure 4F). Using multiple MM lines, we also validated suppressing effect of PHF19 on expression of cell cycle inhibitors (Figure 6K). Lower expression of CDKN1A is correlated with poorer survival of MM patients (supplemental Figure 6H), and nucleotide variations of the CDKN2A locus were reported to be associated with MM development in a genome-wide association study using a large cohort of clinical samples.51
Transcriptome profiling of MM cell lines and primary patient samples delineated the PHF19-enforced gene pathways crucial for MM tumorigenesis. (A) Heatmap showing relative expression of 2366 genes identified as both derepressed post-KD of PHF19 and re-repressed after rescue of PHF19-KD in MM1.S cells. Threshold of differential expression is adjusted DESeq P value (adj.p) of .01 and fold-change (FC) of 2 for transcripts with mean tag counts of ≥10. (B-G) GSEA reveals that, in MM1.S cells, PHF19 is correlated positively to the indicated gene sets showing positive association with EZH2 (B) or EED (C), negatively to the indicated EED-suppressed genes (D), and positively to genes related to cell cycle progression (E), E2F (F), and a proliferation phenotype (G). NES, normalized enrichment score. (H-I) GSEA using a MM patient sample transcriptome data set (GSE9782) shows that higher PHF19 expression is positively related to genes associated with cell cycle progression (H) or E2F (I). (J) Cell cycle progression of MM1.S cells after transduction of shEV (left) or shPHF19 (right). (K) RT-qPCR measures fold change in expression of the indicated cell cycle inhibitor gene in L-363, MM1.S, and NCI-H929 cells after transduction of shPHF19, relative to shEV. Y-axis shows average ± SE of values from three independent experiments. #, not examined due to gene deletion in cells. (L-M) GSEA reveals PHF19 KD positively correlated to gene sets related to the interferon signaling in MM1.S cells. (N) RT-qPCR measures fold change in expression of the indicated JAK/STAT signaling gene in L-363, MM1.S, and NCI-H929 cells after transduction of shPHF19 relative to shEV. (O) L-363 cell growth after transduction of STAT1 (red) or shPHF19 (red) relative to mock treated. Inset shows an immunoblot of the ectopically expressed STAT1.
Transcriptome profiling of MM cell lines and primary patient samples delineated the PHF19-enforced gene pathways crucial for MM tumorigenesis. (A) Heatmap showing relative expression of 2366 genes identified as both derepressed post-KD of PHF19 and re-repressed after rescue of PHF19-KD in MM1.S cells. Threshold of differential expression is adjusted DESeq P value (adj.p) of .01 and fold-change (FC) of 2 for transcripts with mean tag counts of ≥10. (B-G) GSEA reveals that, in MM1.S cells, PHF19 is correlated positively to the indicated gene sets showing positive association with EZH2 (B) or EED (C), negatively to the indicated EED-suppressed genes (D), and positively to genes related to cell cycle progression (E), E2F (F), and a proliferation phenotype (G). NES, normalized enrichment score. (H-I) GSEA using a MM patient sample transcriptome data set (GSE9782) shows that higher PHF19 expression is positively related to genes associated with cell cycle progression (H) or E2F (I). (J) Cell cycle progression of MM1.S cells after transduction of shEV (left) or shPHF19 (right). (K) RT-qPCR measures fold change in expression of the indicated cell cycle inhibitor gene in L-363, MM1.S, and NCI-H929 cells after transduction of shPHF19, relative to shEV. Y-axis shows average ± SE of values from three independent experiments. #, not examined due to gene deletion in cells. (L-M) GSEA reveals PHF19 KD positively correlated to gene sets related to the interferon signaling in MM1.S cells. (N) RT-qPCR measures fold change in expression of the indicated JAK/STAT signaling gene in L-363, MM1.S, and NCI-H929 cells after transduction of shPHF19 relative to shEV. (O) L-363 cell growth after transduction of STAT1 (red) or shPHF19 (red) relative to mock treated. Inset shows an immunoblot of the ectopically expressed STAT1.
In analysis of cell line and primary sample transcriptome data sets, we additionally identified a negative correlation between PHF19 and genes related to the interferon/JAK- and STAT1-related signaling pathway (Figure 6L-M; supplemental Figure 6I-N). ChIP-seq showed that genes related to this pathway such as JAK1 and STATs are directly bound by H3K27me3 in MM (Figure 4G; supplemental Figure 4E). By RT-qPCR, we confirmed the role of PHF19 in suppressing expression of the JAK-STAT signaling genes in multiple MM cell lines (Figure 6N). Furthermore, enforced expression of STAT1, an effector of interferon-JAK signaling,52 suppressed L363 cell growth (Figure 6O). Note that in Drosophila development, polycomb proteins were implicated for silencing of JAK-STAT signaling,53 and our studies of PHF19 in MM support a conservation of this pathway in human cancer.
Taken together, we demonstrate several cancer-related gene pathways that underlie the oncogenic effects of PHF19 in MM.
PHF19’s chromatin-binding domains are essential for MM tumorigenesis
Besides CD, a region essential for PHF19-PRC2 interactions (Figure 5A), PHF19 also harbors other functional motifs such as Tudor54 and extended homology (EH) domains, which were reported to bind histone modification or DNA.11,-13,46,47 To examine the role of these PHF19 motifs in MM oncogenesis, we used both deletion and point mutations to abolish the chromatin-binding activities harbored within these domains. The point mutations used were F74A of Tudor11,13 and substitution of a KKKK motif (either K333A or a compound KKKK-to-AAKA mutation) at EH.46 Comparable to WT, all of these deletion or point mutants showed similar nuclear localization and stable expression (Figure 7A; supplemental Figure 7A); however, none of them were able to rescue the slowed growth caused by PHF19-KD in L363 or MM1.S cells (Figure 7B-C). Unlike WT, the Tudor or EH mutants also failed to restore silencing of PRC2 targets such as CDKN1A/C, JAK1, and STAT1 (Figure 7D). Importantly, in the L363 cell xenografted model, these mutants were also found defective in restoring tumor growth relative to WT (Figure 7E-G; supplemental Figure 7B). These results thus show crucial requirements of PHF19’s chromatin-binding domains for MM tumorigenicity.
PHF19’s chromatin-associating domains are essential for PHF19-mediated gene-repressive and tumorigenic effects in MM. (A) Immunoblotting of the indicated Flag-PHF19 in L-363 cells. (B-C) Growth of the PHF19-KD L-363 (B) or MM1.S (C) cells after transduction of EV, WT PHF19, or the indicated Tudor or EH mutant relative to WT cells. (D) RT-qPCR for the indicated gene in the PHF19-KD L-363 cells after transduction of EV, WT PHF19, or the indicated mutant. Y-axis shows the average ± SE of signals from three independent experiments after normalization to β-actin. *P < .05; **P < .01; ***P < .001; ****P < .0001. (E-G) Representative chemiluminescence imaging (E; 8 weeks postxenograft), signal summary (F), and event-free survival (G) of NSG mice postxenograft of the PHF19-KD L-363 cells after rescue with EV, WT PHF19, or the indicated mutant (cohort size, n = 5). (H) Growth of the indicated MM cells treated with UNC1999, relative to dimethyl sulfoxide (DMSO), for 4, 8, 12, or 16 days. (I-K) Chemiluminescence imaging (I-J; with representative imaging 5 weeks postxenograft shown in panel J) and Kaplan-Meier curve showing event-free survival (K) of NSG mice after xenograft of the luciferase-labeled L-363 cells and subsequent treatment with UNC1999 or vehicle.
PHF19’s chromatin-associating domains are essential for PHF19-mediated gene-repressive and tumorigenic effects in MM. (A) Immunoblotting of the indicated Flag-PHF19 in L-363 cells. (B-C) Growth of the PHF19-KD L-363 (B) or MM1.S (C) cells after transduction of EV, WT PHF19, or the indicated Tudor or EH mutant relative to WT cells. (D) RT-qPCR for the indicated gene in the PHF19-KD L-363 cells after transduction of EV, WT PHF19, or the indicated mutant. Y-axis shows the average ± SE of signals from three independent experiments after normalization to β-actin. *P < .05; **P < .01; ***P < .001; ****P < .0001. (E-G) Representative chemiluminescence imaging (E; 8 weeks postxenograft), signal summary (F), and event-free survival (G) of NSG mice postxenograft of the PHF19-KD L-363 cells after rescue with EV, WT PHF19, or the indicated mutant (cohort size, n = 5). (H) Growth of the indicated MM cells treated with UNC1999, relative to dimethyl sulfoxide (DMSO), for 4, 8, 12, or 16 days. (I-K) Chemiluminescence imaging (I-J; with representative imaging 5 weeks postxenograft shown in panel J) and Kaplan-Meier curve showing event-free survival (K) of NSG mice after xenograft of the luciferase-labeled L-363 cells and subsequent treatment with UNC1999 or vehicle.
PRC2 inhibitors represent an attractive means for MM treatment
Our above results show that targeting PHF19-PRC2 complexes suppresses MM tumorigenicity. As there is a lack of inhibitor against PHF19, we turned to UNC1999, a dual inhibitor of EZH2 and EZH1 we previously reported.33,55 Indeed, multiple tested MM lines showed considerable sensitivity to UNC1999 in vitro (Figure 7H). Additionally, compared with mock, UNC1999 treatment significantly delayed tumor progression in vivo (Figure 7I-J) and prolonged survival of NSG mice xenografted with L-363 cells (Figure 7K). Thus, consistent with recent studies,56,,-59 targeting EZH2/1’s enzymatic activities by small molecules provides a strategy for treating PHF19-dependent MM.
Discussion
PRC2 and H3K27me3 represent a crucial epigenetic component for regulating gene-expression program and defining distinctive cellular states. PRC2 misregulation is increasingly appreciated as a central pathway of oncogenesis. The literature has documented various mechanisms that cause PRC2 deregulation in cancer, which at least include mutation of PRC2 genes20,60,,,-64 and misregulation of microRNA or histone demethylase that antagonizes PRC2.4,59,65 Recently, recurrent mutation of H3K27M, the PRC2 substrate, was detected in pediatric glioblastomas, resulting in the perturbed enzymatic functions and/or genomic distribution of PRC2.49,50,66,67 Further, growing evidence indicates that PRC2-interacting partners are implicated in oncogenesis. For example, PRC2-associated long non-coding RNAs were reported to promote oncogenesis.16 PHF1, a PHF19-related gene, is frequently involved in aberrant chromosomal translocations in sarcoma and soft tissue tumors.68,69 Dissecting the molecular basis underpinning PRC2 perturbation seen in disease is clinically important and shall shed light on mechanism-based therapeutics.
Here, we focused on PHF19 in MM, a plasma cell malignancy displaying considerable genetic heterogeneity. PHF19 overexpression is associated with MM progression. Using various tumor lines and xenografted models, we demonstrated an essential requirement of PHF19 for enforcing oncogenicity in MM. MM is generally characterized by an extraordinarily low mitotic rate,70 and PHF19 is required for cell cycle progression and colony-forming abilities of MM cells. Such oncogenic effects of PHF19 appear to exist across different genetic subtypes of MM (supplemental Figure 2C). Patients with higher PHF19 expression in tumors generally displayed the significantly worse outcomes in clinical trial studies (Figure 1F-G; supplemental Figure 1C-D). PHF19 may serve as a useful predictive or prognostic biomarker for MM. We favor a view that PHF19 enforces a more aggressive, proliferative phenotype during malignant transformation of plasma cells into MM that are already “hit” with initial lesions.
We also examined the mechanisms underlying MM-promoting effects of PHF19. Using a CD-deleted mutant of PHF19, we demonstrated that its oncogenic effect is dependent on association to PRC2. A recent study has reported direct interaction of SUZ12 with CD of PHF19.71 Our mass spectrometry– and ChIP-seq–based profiling shows that PHF19 is required for maintaining genomic patterns of PRC2-catalyzed H3K27me3. In particular, we demonstrated an essential role for PHF19 in H3K27me3 deposition, probably via spreading from CGIs, and broad H3K27me3 domain formation in the cancer genome. Interestingly, alteration in the H3K27me3 genomic landscape caused by PHF19 depletion is reminiscent to what was observed with H3K27M49,50 and enhancer of Zeste homologs inhibitory protein (EZHIP, also termed CXORF67),72 indicating a possible crosstalk among these PRC2-modulatory factors. In our hand, commercial PHF19 antibodies were unsuitable for ChIP-seq (data not shown). Also, beyond H3K27me3 that this study mainly profiled, future investigation is warranted to study into other potentially affected epigenetic pathways, including H3K27me3 readers and H3K36me2, and their crosstalk to PHF19-PRC2 complexes, ideally with clinical samples or patient-derived xenografts. For example, recent reports already suggested a rather complicated interplay among machineries that “write” or “read” different methylation states of H3K27 and H3K36 (such as H3K27me2 and H3K36me2).13,46,49,72,-74
Furthermore, transcriptome analysis of MM cells post-KD of PHF19 revealed the involvement of PHF19 for repression of PRC2 targets, including transcripts crucial for cell cycle progression and the interferon-JAK-STAT signaling. Mutagenesis of PHF19 revealed that its MM-promoting and gene-silencing functions rely not only on interaction with PRC2 but also on the conserved chromatin-binding domains (ie, Tudor and EH).11,-13,46,47 These observations favor a model that overexpression and/or amplification of PHF19 in MM enhances chromatin association of PHF19-PRC2 complexes, besides an H3K27me3-promoting effect seen with polycomb-like proteins.14,75 Together with gain-of-function mutation of EZH2 found in 10% to 20% of B-cell lymphomas, overexpression of PHF19 in MM as studied herein represents another mechanism by which a tumor gains the enhanced PRC2 activity in order to sustain malignant growth. In agreement, overexpression of EZH2 or mutation of KDM6A/UTX, an H3K27me3 demethylase, was reported in MM as well59,76 ; recently, a genome-wide association study identified JARID2, a gene encoding another crucial PRC2-interacting cofactor, as a susceptibility locus in MM.51 Therefore, multiple pathways operate in MM leading to PRC2 deregulation.
Despite recent development of proteasome inhibitors for MM treatment, relapse and drug resistance still occur, which often become unmanageable. It is worth noting that higher expression of PHF19 is associated with worse outcomes in clinical trials using US Food and Drug Administration–approved proteasome inhibitors (Figure 1F-G). Thus, targeting PHF19-PRC2 complexes may provide a new strategy to further improve MM therapeutics. This study and recent reports56,,-59 show that PRC2 inhibitors indeed suppress MM tumorigenesis. Because EZH2 is more widely expressed during development, we hypothesize that targeting PHF19 provides an attractive means for blocking PRC2 in a tumor- and tissue-specific manner. Toward this end, we show various PHF19 domains (Tudor, EH, and CD) to be functionally crucial for MM tumorigenesis, which shall represent valuable target sites for further development. Tudor belongs to a Royal family of chromatin “readers” that are potentially druggable.77 Additional investigation is needed for developing the PHF19-targeted strategies.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
ChIP-seq and RNA-seq datasets have been deposited in the Gene Expression Omnibus database (accession number GSE135890).
Acknowledgments
The authors thank the Animal Studies Core, Genomics Core, Bioinformatics Core, Small Animal Imaging Facility and Flow Cytometry Core of UNC at Chapel Hill for assistance. The authors also thank the members of the Wang Laboratory for technical support and discussion.
UNC facilities are supported, in part, by UNC Lineberger Comprehensive Cancer Center Core Support Grant P30-CA016086, and North Carolina Biotech Center Institutional Support Grant 2012-IDG-1006. This work was supported by National Institutes of Health (NIH)/National Cancer Institute (NCI) Grants R01-CA215284 and R01-CA211336 (to G.G.W.), and P01-CA196539 (to B.A.G); NIH/National Heart, Lung, and Blood Institute (NHLBI) Grant R01-HL133120 (to D.Z.); NIH/National Institute of General Medical Sciences (NIGMS) Grants R01-GM110174 (to B.A.G.), R01GM122749 (to J.J.), and P20GM121293, P20GM103625, and P20GM103429 (to A.J.T.); NIH/Office of the Director Grant S10OD018445 (to A.J.T.), NIH/National Center for Advancing Translational Sciences Grant UL1TR003107 (to A.J.T.); a Kimmel Scholar Award (to G.G.W.); grants from the When Everyone Survives Leukemia Research Foundation (to G.G.W.); the Gilead Sciences Research Scholars Program in Hematology/Oncology (to G.G.W.); and Gabrielle's Angel Foundation for Cancer Research (to G.G.W.). Z.R. is supported by a Department of Defense Career Development Award nested postdoctoral fellowship (W81XWH-14-10232; awarded to G.G.W.). G.G.W. is an American Society of Hematology Scholar in Basic Science, an American Cancer Society Research Scholar, and a Leukemia & Lymphoma Society Scholar.
Authorship
Contribution: Z.R. and J.H.A. performed tumor, genomics, and molecular experiments; Z.R., H.L., and Y.-H.T. analyzed various genomics data sets, with the assistance of J.H.A., P.W., and L.C. and guidance of D.Z. and G.G.W.; N.V.B., B.K., A.J.S., S.G.M., R.D.E., B.A.G., and A.J.T. performed mass spectrometry analysis of histones and protein complexes; Z.R., J.H.A., D.F.A., and L.C. performed various biochemical and genomics validation studies; R.W.J.G. and A.C.M. established MM models for in vivo imaging studies; A.M. and J.J. synthesized compounds and designed treatment; G.G.W. conceived and led the project; and Z.R. and G.G.W. generated figures and wrote the manuscript, with input from all authors.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for Z.R. is Division of Hematologic Malignancies, Johns Hopkins University School of Medicine, Baltimore, MD.
Correspondence: Greg Wang, UNC Lineberger Cancer Center, 450 West Dr, CB 7295, Chapel Hill, NC 27599-7295; e-mail: greg_wang@med.unc.edu.
REFERENCES
Author notes
Z.R., J.H.A., and H.L. contributed equally to this study.