TO THE EDITOR:
Following the successful example of imatinib in chronic myeloid leukemia, personalized oncology is often considered as the choice of optimal targeted therapies fitted to the mutational landscape of the cancer. However, this paradigm is far from being generalized, and randomized clinical trials of other drugs have failed to demonstrate major clinical improvement (if any) with this approach.1-3 However, the generation of big data in medicine raises new hope for personalized therapy by integrating numerous clinical and biological data to refine outcome prediction. A very promising demonstration of this “knowledge bank approach” recently came from the study reported by Gerstung et al that was based on the analysis of >1500 acute myeloid leukemia (AML) patients from 3 randomized clinical trials.4 In this article, the authors demonstrated the efficacy of their model to improve overall survival prediction compared with current classifications. More interestingly, the knowledge bank approach is able to predict the different causes of mortality in AML patients (ie, nonremission death, relapse death, nonrelapse death), as well as how allogeneic stem cell transplantation (ASCT) might impact these probabilities for each patient. As stated by the authors, the use of this scoring system might profoundly change patient care, especially with regard to ASCT. However, although the accuracy of this prognostic system has been confirmed for overall survival in an independent cohort from The Cancer Genome Atlas, the ability of the algorithm to predict the different causes of mortality has yet to be validated. Moreover, among the different caveats that might challenge the relevance of this scoring system in real life, the heterogeneity of high-throughput sequencing data generation and analysis is a major concern. For example, the filtering strategy used by Gerstung et al is only applicable in the context of a centralized laboratory analyzing large cohorts of patients and not for routine analysis.
To evaluate the potential of the knowledge bank approach in routine clinical practice, we have studied a retrospective cohort of AML patients who were treated with high-dose chemotherapy in a tertiary care center. These patients were fully characterized at the time of diagnosis at the clinical, biological, cytogenetic, and molecular levels.
Between 2005 and 2017, we identified 155 patients (71 women and 84 men; median age, 56.5 years; range, 20-74 years) who received an induction therapy regimen for AML containing cytarabine and anthracycline. ASCT was performed in 70 patients, during the first complete remission (n = 54) or after relapse (n = 16). The overall survival curve of the cohort estimated by the Kaplan-Meier method was in the usual range of what is observed in AML patients treated with intensive therapy (supplemental Figure 1, available on the Blood Web site). The median follow-up of living patients (n = 89 at the time of the study) was 27.9 months. The Gerstung et al algorithm includes 10 clinical, 26 cytogenetic, and 58 molecular parameters. Given the retrospective nature of the study, there were missing data (supplemental Figure 2; supplemental Table 2), which could have diminished the performance of the algorithm. Among the 1550 potential clinical annotations for the entire cohort (10 parameters in 155 patients), 6 were missing. Nine patients lacked full cytogenetic information because of karyotyping failure. Among the 8990 potential molecular annotations (58 parameters in 155 patients), 1821 (20.3%) were missing. Most of these (1240 [68.1%]) corresponded to genes not included in any version of the panel used in our laboratory (ATRX, CBLB, CUX1, GNAS, KDM5A, MLL5, MLL3, SF3A1, and U2AF2); the others were missing in a fraction of the patients depending on the version of the panel used. Importantly, no data were lacking for the 6 genes having the greatest impact on prognosis (TP53, CEBPA, NPM1, FLT3, DNMT3A, and MLL). Details of the sequencing technique used and the bioinformatic pipelines, as well as the methodology used to calculate the different probabilities of survival for each patient according to the knowledge bank approach, are provided in supplemental Methods.
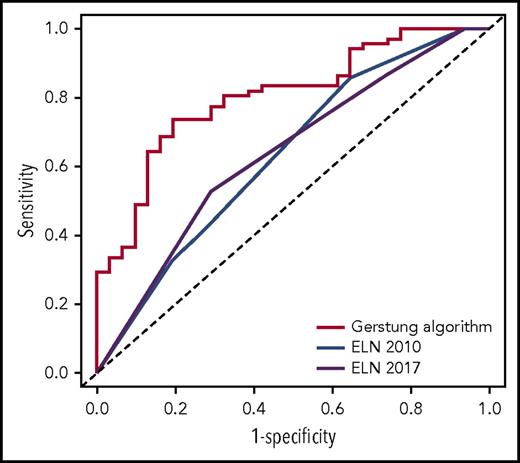
We first assessed the potential of the knowledge bank approach to predict overall survival at 3 years by comparing algorithm prediction with observed outcome. For this analysis, we did not use the information about ASCT in predicting survival with the Gerstung et al algorithm, which would have artificially favored the knowledge bank approach. The performance of the knowledge bank approach to identify the patients who died during the first 3 years (area under the receiver operating characteristic [ROC] curve [AUC], AUCknowledge databank = 0.804; 95% confidence interval [CI], 0.715-0.893) was greater than the European LeukemiaNet (ELN) 20105 (AUCELN 2010 = 0.632; 95% CI, 0.517-0.747; P = .004) and ELN 20176 (AUCELN 2017 = 0.638; 95% CI, 0.527-0.748; P = .005) classifications, which represent the current standard for establishing AML prognosis (Figure 1).
ROC curves showing the performance of the knowledge bank approach and the ELN 2010 and ELN 2017 classifications to identify patients who will die in the first 3 years after AML diagnosis.
ROC curves showing the performance of the knowledge bank approach and the ELN 2010 and ELN 2017 classifications to identify patients who will die in the first 3 years after AML diagnosis.
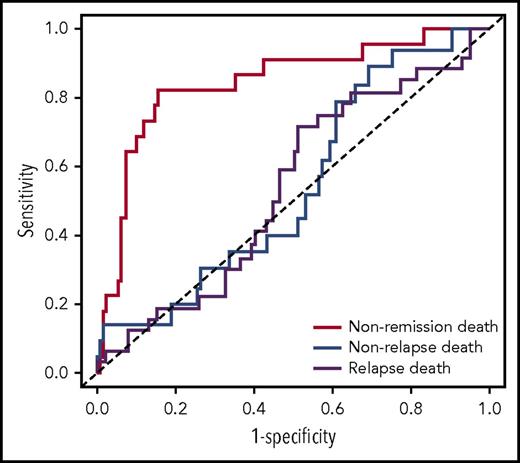
We then tested the strength of the knowledge bank approach to predict more subtle outcomes, such as nonremission death, relapse death, and nonrelapse death. The outcome of the 96 patients not censored at 3 years is presented in supplemental Table 1. ASCT was used to calculate the risk for relapse death and nonrelapse death among the patients having achieved complete remission. The knowledge bank approach effectively predicted nonremission death (AUCknowledge databank = 0.844; 95% CI, 0.746-0.941) and relapse death (AUCknowledge databank = 0.694; 95% CI, 0.567-0.820), but did not predict nonrelapse death (AUCknowledge databank = 0.581; 95% CI, 0.433-0.730) (Figure 2
ROC curves showing the performance of the knowledge bank approach to predict nonremission death, relapse death, and nonrelapse death.
ROC curves showing the performance of the knowledge bank approach to predict nonremission death, relapse death, and nonrelapse death.
).
Taken together, this study confirms the strength of the knowledge bank approach to improve the prognostic evaluation of patients with AML. Interestingly, this approach outperforms the 3-year overall survival prediction achieved by state-of-the-art classifications and is able to predict the probability of nonremission death and relapse death. However, the present cohort was too small to assess the capacity of this approach to evaluate the benefits of ASCT, which is the ultimate objective of AML classifications, because only 6 patients younger than 60 years of age and having achieved first complete remission had a 10% predicted increase in overall survival with ASCT according to the knowledge bank approach. It is also of note that the excellent results with the knowledge bank approach were confirmed herein, despite many caveats, such as the lack of 20% of molecular data. The robustness of the algorithm to missing data was indeed predicted by Gerstung et al, because most of the prognostic information at the molecular level is supported by a small set of genes and because the algorithm is capable of imputing missing data. This observation suggests that simplification of the algorithm might be possible without losing much information. However, given the improvement in sequencing technologies and the diminution of the sequencing cost, it is quite feasible to sequence all of the genes required to calculate the algorithm in a real-life setting, to capture as much prognostic information as possible. Moreover, the knowledge bank approach seems robust, despite many differences between the pipelines used to generate and analyze the high-throughput sequencing data, which was a potential major limitation to the generalization of this algorithm. A prospective validation of the knowledge bank approach is now warranted.
The online version of this article contains a data supplement.
Acknowledgment
The authors thank Philip Robinson (direction de la recherche clinique et de l'innovation, Hospices Civils de Lyon) for help with English editing.
Authorship
Contribution: S. Huet, F.S., and P.S. analyzed the data and wrote the manuscript. E.P., X.T., and G.S. were involved in the clinical care of the patients. C.L., B.G., L.C., P.S., and S. Huet collected data. K.C., C.C., I.M., S. Hayette, I.T., and T.S. were involved in the diagnostic procedure.
Conflict of interest disclosure: The authors declare no competing financial interests.
Correspondence: Pierre Sujobert, Hospices Civils de Lyon, Centre Hospitalier Lyon Sud, Laboratoire d’Hématologie, 165 Chemin du Grand Revoyet, 69495 Pierre-Bénite Cedex, France; e-mail: pierre.sujobert@chu-lyon.fr.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal