Key Points
FOXO1 is recurrently mutated in BL.
Nuclear FOXO1 promotes proliferation and survival in BL.
Abstract
Forkhead box class O1 (FOXO1) acts as a tumor suppressor in solid tumors. The oncogenic phosphoinositide-3-kinase (PI3K) pathway suppresses FOXO1 transcriptional activity by enforcing its nuclear exclusion upon AKT-mediated phosphorylation. We show here abundant nuclear expression of FOXO1 in Burkitt lymphoma (BL), a germinal center (GC) B-cell–derived lymphoma whose pathogenesis is linked to PI3K activation. Recurrent FOXO1 mutations, which prevent AKT targeting and lock the transcription factor in the nucleus, are used by BL to circumvent mutual exclusivity between PI3K and FOXO1 activation. Using genome editing in human and mouse lymphomas in which MYC and PI3K cooperate synergistically in tumor development, we demonstrate proproliferative and antiapoptotic activity of FOXO1 in BL and identify its nuclear localization as an oncogenic event in GC B-cell–derived lymphomagenesis.
Introduction
In T-cell–dependent antigen responses, B cells undergo proliferation and selection in germinal centers (GCs) of peripheral lymphoid tissues. Here, the entry and exit of B cells in the GC’s dark zone (DZ) and light zone (LZ) guide the fate of the activated cells,1 which mutate and switch their immunoglobulin genes.2 Both processes require DNA modifications that accidently lead to mutation or translocation of tumor-promoting genes, the major cause of (post) GC B-cell lymphomas.3 Burkitt lymphoma (BL) is a GC B-cell–derived tumor characterized by a c-MYC (MYC) translocation into the immunoglobulin locus.4 Although aberrant MYC expression promotes malignancies by induction of proliferation and other means, it also evokes increased apoptosis.5 Recently, we and others have identified phosphoinositide-3-kinase (PI3K) signaling as a potent prosurvival signal in BL pathogenesis.6,7
Alterations of this pathway are frequently detected in cancer,8 and a prominent mediator of the PI3K-signaling cascade is the conserved forkhead box class O (FOXO) gene family of transcription factors. In mammals, FOXO1, FOXO3, and FOXO4 are ubiquitously expressed and regulate their activity by posttranslational modifications.9 FOXO1 is critical for B-cell development,10 and in the GC reaction it instructs the transcriptional program of the DZ.11,12 As a general rule, PI3K activation and nuclear FOXO1 are mutually exclusive: PI3K-mediated AKT activation results in the phosphorylation of FOXO1 (at residues threonine 24 [T24], and serine 256 [S256] and 319 [S319]), mediating its interaction with 14-3-3 protein and the nuclear export of the complex.13,14 FOXO proteins regulate proapoptotic and antiproliferative genes, as well as antioxidant and DNA repair pathways.15,16 Thus, FOXO1 has tumor-suppressor function in various solid tumors and B-cell lymphomas,17-21 including Hodgkin lymphoma (HL).22 Contradictory to these inhibitory effects are recent reports linking high FOXO expression to poor prognosis and tumor-promoting activity in intestinal, neuronal, and hematological malignancies.23-29 The emerging concept of context-specific functions of FOXO proteins in cancer is substantially supported and extended in a breast cancer study in which FOXO activation needs optimal balance in order to promote cancer progression.30
In GC B-cell–derived non-HLs, nonsynonymous FOXO1 mutations, which disrupt the N-terminal AKT recognition motif around T24, are recurrent. Approximately 10% of diffuse large B-cell lymphoma patients carry heterozygous FOXO1 mutations, almost half of which (12 of 26) favor the nuclear localization of the transcription factor.31,32 In BL, similar incidences were described by Schmitz and colleagues analyzing primary tumors and cell lines7 and our mouse model of BL pathogenesis confirmed the acquisition of FOXO1 mutations (2 of 5 lymphomas) during tumorigenesis.6 Inferior treatment response rates were linked to mutant (mt) FOXO1 in diffuse large B-cell lymphoma and follicular lymphomas, demonstrating the clinical relevance of FOXO1 mutations.32,33
To clarify the impact of FOXO1 in B-cell tumorigenesis, we took advantage of newly generated and established mouse and human BL cell lines and genetically engineered the FOXO1 locus in these cells. Here, we report that nuclear FOXO1 and PI3K activity coexist in BL cells and that nuclear FOXO1 promotes tumor growth in murine and human lymphomas. Moreover, we provide evidence that FOXO1 mt BL cells are addicted in their proliferation and survival to the expression of nuclear FOXO1, thus highlighting FOXO1-dependent signaling pathways as attractive candidates for developing tumor-specific therapeutic approaches.
Methods
Cell lines
The lymphoma cell lines DG75, BL41, BL2, CA46, Namalwa, Ramos, Raji, L1236, KM-H2, and HDLM2 were obtained from the German Collection of Microorganisms and Cell Cultures (DSMZ) or ATCC. BL60 and LY47 were provided by G. M. Lenoir (International Agency for Research on Cancer, Lyon, France); Salina and Seraphine were provided by A. Rickinson (University of Birmingham, Birmingham, United Kingdom). Cell lines were authenticated by multiplex cell authentication (Multiplexion). Mouse cell lines were generated from BL-like tumors in Cγ1-cre,R26StopFLMYC,R26StopFLP110* animals.6
Immunofluorescence analysis
Cells were stained with α-FOXO1 antibody (Cell Signaling) and goat-anti rabbit immunoglobulin G–Alexa 568 (Invitrogen) as previously described.11 Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) and Leica-SP8 inverted confocal or KEYENCE microscopes were used for image acquisition.
Immunoprecipitation and western blot analysis
Cell lysate (500 µg) was immunoprecipitated using α-FOXO1 or α-acetylated lysine (both Cell Signaling) antibodies and protein A magnetic Dynabeads (Invitrogen) as previously described.34 For detection of posttranslational modifications, we treated the cells with inhibitors and extracted protein according to published protocols.35,36
For western blot analysis, radioimmunoprecipitation assay (RIPA) buffer extracts were fractionated on 10% sodium dodecyl sulfate polyacrylamide gels, electroblotted to polyvinylidene difluoride membranes, and reacted with α-phosphorylated AKT (pAKT; S473), α-AKT, α-phosphorylated FOXO1 (pFOXO1; T24 or S256 or S319), α-FOXO1, α-O-GlcNAc, α-Histone 3, α-14-3-3 (pan) (all Cell Signaling) and α-β-actin (Sigma-Aldrich) antibodies. Immunoreactivity was determined using ECL substrate buffer (Thermo Fisher). Nuclear and cytoplasmic extracts were prepared as previously published.32
Microarray analysis
Gene-expression profiling was performed on Affymetrix GeneChip Mouse Genome 430 2.0 arrays following the manufacturer’s recommendations (Affymetrix). Affymetrix GeneChip array data were preprocessed using Affymetrix Expression Console and normalized through the robust multiarray average (RMA) implementation in the Expression Console.
Results
Nuclear FOXO1 expression and PI3K activity coexist in BL cells
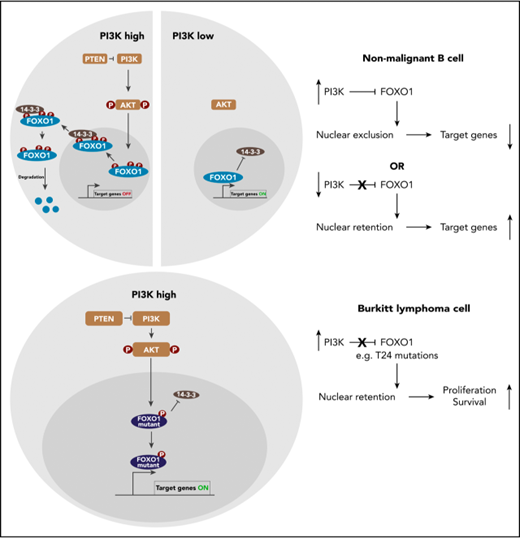
In BL, the reported PI3K activation is expected to ablate FOXO1 by AKT-mediated nuclear export and subsequent degradation of the transcription factor. However, in a panel of human BL cell lines, we determined high FOXO1 expression at the transcript and protein level in comparison with HL cell lines (supplemental Figure 1A-B, available on the Blood Web site). Strikingly, FOXO1 was located both in the cytoplasm and the nucleus of the cells (Figure 1A-B; supplemental Figure 1C-D). Reproducibly, FOXO1 appeared less abundant in cytoplasmic and nuclear extracts compared with total cell lysates in western blot analyses. Further experiments excluded FOXO1 loss during protein fractionation and sample loading, thus antibody detection might be hampered by epitope masking in the samples. In lymphomas arising from our BL mouse model and thus exhibiting constitutive PI3K pathway activation due to P110* transgene expression,6,37 FOXO1 localization defined 2 distinct lymphoma groups: malignancies where the transcription factor was predominantly nuclear and others defined by cytoplasmic FOXO1 (Figure 1C).
Nuclear FOXO1 and PI3K activity coexist in BL cells. (A) Immunofluorescence analysis of human BL cell lines using FOXO1 antibody (red). Cell nuclei were counterstained with DAPI (blue; scale bar, 5 μm). (B) Western blot analysis of FOXO1 expression in subcellular fractions of human BL cells. The purity of the cytoplasmic and nuclear fraction was determined by actin (ACTB) and histone H3 (H3) antibodies, respectively. Data are representative of at least 2 experiments. (C) Immunofluorescence analysis of FOXO1 in mouse BL cell lines as shown in panel A. (D) Western blot analysis of phospho-AKT (pAKT [S473]), AKT, phospho-FOXO1 (pFOXO1 [T24]), and FOXO1 expression in human BL cell lines. Cells were treated either with the PI3K inhibitor wortmannin (+) or dimethyl sulfoxide (DMSO) (−) 1 hour prior to protein extraction. ACTB served as loading control. Data are representative of 2 experiments. (E) Detection of pAKT(S473) in mouse BL-like cell lines by intracellular FACS analysis. After fixation and permeabilization, lymphoma cells were incubated with pAKT(S473) antibody (= antibody) or the antibody plus pAKT(S473) blocking peptide (= blocking peptide). Data are representative of 2 experiments. C, cytoplasmic fraction; KO, FOXO1 knockout cell; N, nuclear fraction; W, whole-cell lysate.
Nuclear FOXO1 and PI3K activity coexist in BL cells. (A) Immunofluorescence analysis of human BL cell lines using FOXO1 antibody (red). Cell nuclei were counterstained with DAPI (blue; scale bar, 5 μm). (B) Western blot analysis of FOXO1 expression in subcellular fractions of human BL cells. The purity of the cytoplasmic and nuclear fraction was determined by actin (ACTB) and histone H3 (H3) antibodies, respectively. Data are representative of at least 2 experiments. (C) Immunofluorescence analysis of FOXO1 in mouse BL cell lines as shown in panel A. (D) Western blot analysis of phospho-AKT (pAKT [S473]), AKT, phospho-FOXO1 (pFOXO1 [T24]), and FOXO1 expression in human BL cell lines. Cells were treated either with the PI3K inhibitor wortmannin (+) or dimethyl sulfoxide (DMSO) (−) 1 hour prior to protein extraction. ACTB served as loading control. Data are representative of 2 experiments. (E) Detection of pAKT(S473) in mouse BL-like cell lines by intracellular FACS analysis. After fixation and permeabilization, lymphoma cells were incubated with pAKT(S473) antibody (= antibody) or the antibody plus pAKT(S473) blocking peptide (= blocking peptide). Data are representative of 2 experiments. C, cytoplasmic fraction; KO, FOXO1 knockout cell; N, nuclear fraction; W, whole-cell lysate.
As PI3K pathway activity is a major determinant of FOXO1’s subcellular localization, we determined phosphorylation of AKT at S473 in human and mouse lymphoma cells. By this criterion, human BL cells exhibited PI3K activity, albeit at variable intensity, and pAKT levels positively correlated with FOXO1 phosphorylation at T24 (Figure 1D; supplemental Figure 1E). PI3K inhibition by wortmannin efficiently prevented AKT and FOXO1 phosphorylation in the lymphoma cells, confirming an intact PI3K-AKT-FOXO1 axis (Figure 1D). In the mouse BL-like tumors expressing P110*, PI3K was uniformly active as indicated by AKT S473 phosphorylation (Figure 1E).
Recurrent FOXO1 mutations lock the transcription factor in the nucleus
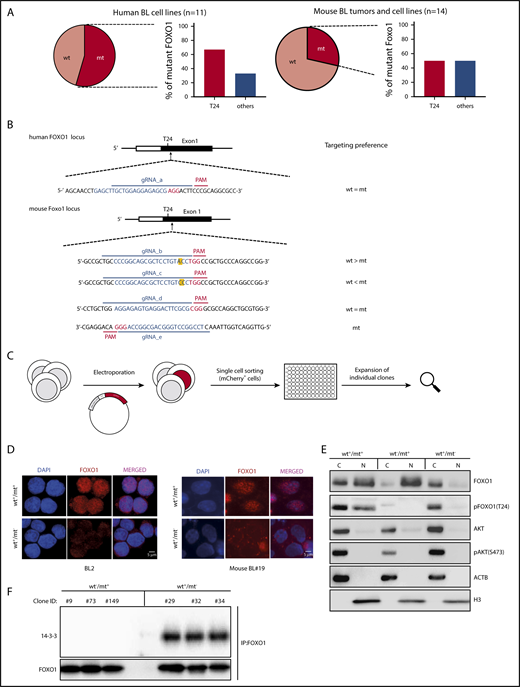
Sanger sequencing of genomic DNA isolated from 11 human BL cell lines and 14 murine BL-like tumors verified nonsynonymous FOXO1 mutations in both tumors (Figure 2A; supplemental Figure 2A). Six human cell lines carried nonsynonymous and heterozygous FOXO1 mutations. In 4 of these lines, the mutations targeted AKT-dependent phosphorylation at the N-terminus of the protein: the mt allele encoded aberrant FOXO1 in which either the AKT-recognition motif was disrupted (R21H and T24I) or deleted (M1V).32 Because of their functional redundancy, cell lines carrying these mutations will be classified as “mt T24” throughout the paper.
Mutations impairing T24 phosphorylation lock FOXO1 in the cell nucleus. (A) Pie charts indicating the incidence of nonsynonymous FOXO1 mutations in human BL cell lines (left) and mouse BL-like tumors (right). The bar diagrams show the percentage of FOXO1 mutations involving the AKT-dependent phosphorylation site at T24 (T24) or other positions (others) in human and mouse BL. (B) Design of FOXO1 gRNAs used for CRISPR/Cas9 experiments in human and mouse BL cells. CRISPR/Cas9 target sequences are given in blue (protospacer) and red (PAM). In the mouse FOXO1 locus the nucleotide affected by the T24 mutation is marked in yellow. (C) Experimental scheme for the generation of isogenic cell line clones analyzed in panels D-F. After electroporation and transient coexpression of gRNA, Cas9 and mCherry individual reporter-positive cells were FACS-based sorted and expanded for analysis. (D) Immunofluorescence analysis of FOXO1 (red) in CRISPR/Cas9-modified human and mouse BL cells. Parental cell lines (BL2 and mouse BL#19) carrying heterozygous FOXO1 T24 mutations (wt+/mt+) and isogenic cell line clones in which the mt alleles were ablated (wt+/mt−) are shown. Cell nuclei were counterstained with DAPI (blue; scale bar, 5 μm). (E) Western blot analysis of FOXO1 expression in subcellular fractions of mouse BL#19 cells (wt+/mt+) and isogenic cell line clones in which the wt (wt−/mt+) or the mt Foxo1 allele (wt+/mt−) was specifically ablated. The purity of the cytoplasmic and nuclear fraction was determined by actin (ACTB) and histone H3 (H3) antibodies, respectively. In addition, pFOXO1 (T24), pAKT (S473), and AKT expression were detected by the appropriate antibodies. Data are representative of 2 experiments. (F) Western blot analysis of 14-3-3 protein expression after immunoprecipitation with FOXO1 antibody in isogenic mouse BL cell lines (as described in panel D). Per genotype, 3 individual cell line clones were analyzed. The membrane was also incubated with anti-FOXO1 antibody to verify precipitation of the transcription factor. Data are representative of 2 experiments. C, cytoplasmic fraction; N, nuclear fraction.
Mutations impairing T24 phosphorylation lock FOXO1 in the cell nucleus. (A) Pie charts indicating the incidence of nonsynonymous FOXO1 mutations in human BL cell lines (left) and mouse BL-like tumors (right). The bar diagrams show the percentage of FOXO1 mutations involving the AKT-dependent phosphorylation site at T24 (T24) or other positions (others) in human and mouse BL. (B) Design of FOXO1 gRNAs used for CRISPR/Cas9 experiments in human and mouse BL cells. CRISPR/Cas9 target sequences are given in blue (protospacer) and red (PAM). In the mouse FOXO1 locus the nucleotide affected by the T24 mutation is marked in yellow. (C) Experimental scheme for the generation of isogenic cell line clones analyzed in panels D-F. After electroporation and transient coexpression of gRNA, Cas9 and mCherry individual reporter-positive cells were FACS-based sorted and expanded for analysis. (D) Immunofluorescence analysis of FOXO1 (red) in CRISPR/Cas9-modified human and mouse BL cells. Parental cell lines (BL2 and mouse BL#19) carrying heterozygous FOXO1 T24 mutations (wt+/mt+) and isogenic cell line clones in which the mt alleles were ablated (wt+/mt−) are shown. Cell nuclei were counterstained with DAPI (blue; scale bar, 5 μm). (E) Western blot analysis of FOXO1 expression in subcellular fractions of mouse BL#19 cells (wt+/mt+) and isogenic cell line clones in which the wt (wt−/mt+) or the mt Foxo1 allele (wt+/mt−) was specifically ablated. The purity of the cytoplasmic and nuclear fraction was determined by actin (ACTB) and histone H3 (H3) antibodies, respectively. In addition, pFOXO1 (T24), pAKT (S473), and AKT expression were detected by the appropriate antibodies. Data are representative of 2 experiments. (F) Western blot analysis of 14-3-3 protein expression after immunoprecipitation with FOXO1 antibody in isogenic mouse BL cell lines (as described in panel D). Per genotype, 3 individual cell line clones were analyzed. The membrane was also incubated with anti-FOXO1 antibody to verify precipitation of the transcription factor. Data are representative of 2 experiments. C, cytoplasmic fraction; N, nuclear fraction.
In the mouse lymphomas, we identified heterozygous Foxo1 mutations in altogether 4 of 14 cases, 2 of which affected the T24 AKT-phosphorylation site. The somatic origin of the mutations was confirmed by tail DNA analysis and, in both mouse and human tumor cells, we detected mt FOXO1 at the transcript level, confirming its expression (data not shown).
To evaluate the impact of FOXO1 in BL cells, we genetically modified human and mouse lymphoma cells through clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) mutagenesis. Various guide RNAs (gRNAs) targeting the FOXO1 locus were designed (Figure 2B): in human cells, the gRNA edited FOXO1 at its mt and wild-type (wt) allele (gRNA_a). In mouse tumor cells, multiple gRNAs (gRNA_b-d) with different targeting efficiencies were available (supplemental Figure 2B and data not shown). Only in mouse BL#19 cells did the replacement of threonine to proline at position 24 create a unique protospacer adjacent motif (PAM) sequence, allowing us to target this mt allele specifically (gRNA_e).
To prove a causative link between nuclear FOXO1 expression and T24 phosphorylation site disruption, FOXO1 mt lymphoma cells were transfected to transiently express FOXO1 gRNA and Cas9. Cas9 expression was coupled to an mCherry reporter via a self-cleaving peptide sequence (T2A) allowing fluorescence-activated cell sorter (FACS)-based isolation of Cas9-expressing cells before individual cell clone expansion. Immunofluorescence analysis revealed cytoplasmic FOXO1 staining in isogenic human and mouse cell line clones where the mt locus was selectively inactivated (wt+/mt−) (Figure 2D; supplemental Figure 2C). Western blot analysis of mouse cells, in which either mt (wt+/mt−) or wt (wt−/mt+) FOXO1 was ablated, confirmed the nuclear predominance of mt FOXO1 in the lymphoma cells (Figure 2E; supplemental Figure 2D). Impaired interaction between 14-3-3 and T24 mt FOXO1 could explain this finding (Figure 2F).
FOXO1 ablation impairs BL growth
The counterintuitive finding of abundant nuclear FOXO1 expression in BL compared with HL cells led us to evaluate the effects of FOXO1 ablation on lymphoma growth by transiently coexpressing Cas9, a gRNA targeting wt and mt FOXO1 alleles and the reporter mCherry in mouse BL-like cells. Two days after electroporation, successfully transfected cells were sorted and the bulk of mCherry-positive cells was analyzed at the indicated time points after sorting (Figure 3A).
Nuclear FOXO1 promotes lymphoma growth. (A) Experimental scheme of CRISPR/Cas9-mediated FOXO1 ablation in murine BL cells. After transient expression of Cas9 and the Foxo1 gRNA_d, which highly efficiently targets wt and mt Foxo1, reporter-positive cells were FACS-based sorted and the bulk of tumor cells analyzed in panels B-C. CRISPR/Cas9 target sequences are given in blue (protospacer) and red (PAM). (B) Foxo1 T7EI assay in CRISPR/Cas9-modified mouse BL cells at indicated time points after FACS-based sorting of mCherry-positive cells. Two cell lines characterized by a heterozygous T24 mutation (mouse BL#19 and #82) were analyzed as well as 2 cell lines exclusively expressing wt FOXO1 (mouse BL#81 and #88). Arrows indicate the expected positions of DNA bands cleaved by mismatch-sensitive T7EI. The calculated rate of indel mutagenesis after quantification of band intensities is shown in the graphs. (C) Foxo1 sequence analysis after CRISPR/Cas9 editing in mouse BL cell lines (mouse BL#19 and #82) at indicated time points after FACS-based sorting. Pie charts depict the distribution of CRISPR edited (targeted) vs untargeted sequences. The total number of sequences analyzed is given in the center of the chart. (D) Experimental setup for stable FOXO1 ablation in human BL cells. Cas9 coupled with an mCherry fluorescent reporter was introduced to the cells by lentiviral infection. Single mCherry-positive cells were FACS-based sorted, and stable expressing Cas9 clones were further expanded. In a second step, Cas9-positive clones were infected with a lentiviral construct encoding FOXO1 gRNA coupled to a BFP fluorescent reporter. mCherry- and BFP-coexpressing cells were sorted and Sanger sequencing was performed in genomic DNA extracted from the bulk of tumor cells at indicated time points. The protospacer (PAM) sequence is given in blue (red) as in panel A. (E) FOXO1 sequence analysis after CRISPR/Cas9 editing in human BL cell lines (Namalwa, BL2, and CA46) at indicated time points after FACS-based sorting. CRISPR-edited sequences were analyzed and the distribution of “out-of-frame” vs “in-frame“ sequences is shown in the pie charts. The total number of sequences analyzed is given in the center of the chart.
Nuclear FOXO1 promotes lymphoma growth. (A) Experimental scheme of CRISPR/Cas9-mediated FOXO1 ablation in murine BL cells. After transient expression of Cas9 and the Foxo1 gRNA_d, which highly efficiently targets wt and mt Foxo1, reporter-positive cells were FACS-based sorted and the bulk of tumor cells analyzed in panels B-C. CRISPR/Cas9 target sequences are given in blue (protospacer) and red (PAM). (B) Foxo1 T7EI assay in CRISPR/Cas9-modified mouse BL cells at indicated time points after FACS-based sorting of mCherry-positive cells. Two cell lines characterized by a heterozygous T24 mutation (mouse BL#19 and #82) were analyzed as well as 2 cell lines exclusively expressing wt FOXO1 (mouse BL#81 and #88). Arrows indicate the expected positions of DNA bands cleaved by mismatch-sensitive T7EI. The calculated rate of indel mutagenesis after quantification of band intensities is shown in the graphs. (C) Foxo1 sequence analysis after CRISPR/Cas9 editing in mouse BL cell lines (mouse BL#19 and #82) at indicated time points after FACS-based sorting. Pie charts depict the distribution of CRISPR edited (targeted) vs untargeted sequences. The total number of sequences analyzed is given in the center of the chart. (D) Experimental setup for stable FOXO1 ablation in human BL cells. Cas9 coupled with an mCherry fluorescent reporter was introduced to the cells by lentiviral infection. Single mCherry-positive cells were FACS-based sorted, and stable expressing Cas9 clones were further expanded. In a second step, Cas9-positive clones were infected with a lentiviral construct encoding FOXO1 gRNA coupled to a BFP fluorescent reporter. mCherry- and BFP-coexpressing cells were sorted and Sanger sequencing was performed in genomic DNA extracted from the bulk of tumor cells at indicated time points. The protospacer (PAM) sequence is given in blue (red) as in panel A. (E) FOXO1 sequence analysis after CRISPR/Cas9 editing in human BL cell lines (Namalwa, BL2, and CA46) at indicated time points after FACS-based sorting. CRISPR-edited sequences were analyzed and the distribution of “out-of-frame” vs “in-frame“ sequences is shown in the pie charts. The total number of sequences analyzed is given in the center of the chart.
Genome editing of the Foxo1 locus in mouse cells that express nuclear FOXO1 due to a T24 mutation (mouse BL#19 and #82) resulted in growth retardation (Figure 3B): in the T7EI assay, which is sensitive for CRISPR/Cas9-induced DNA mismatches, the proportion of Cas9-targeted cells decreased over time indicating that these cells were outcompeted by their counterparts with intact Foxo1 loci. In contrast, the detrimental effect of FOXO1 targeting was absent in mouse lymphoma cells exclusively expressing wt and thus cytoplasmic FOXO1 (mouse BL#81 and #88) (Figure 3B). In T24 mt mouse cells exposed to Foxo1 editing, Sanger sequencing of the Foxo1 locus in the bulk population of transfected cells confirmed this finding (Figure 3C; supplemental Figure 3A): although wt and mt Foxo1 alleles were initially modified by Cas9 with similar efficiency in all cell lines, targeted mt Foxo1 rapidly disappeared over time in the cultures, in contrast to the targeted wt allele.
For the human BL cells, where transient transfection was highly inefficient in most cell lines, we chose viral infection to stably apply CRISPR/Cas9 technology (Figure 3D). Cells were lentivirally infected to express Cas9 and mCherry reporter via a T2A sequence. After FACS-based selection of mCherry-positive cells, single clones were expanded and transduced by another lentivirus encoding the FOXO1 gRNA and a blue fluorescent protein (BFP) reporter gene. Cas9- and gRNA-positive cells by mCherry and BFP coexpression were bulk sorted prior to analysis.
Because human BL cells expressed nuclear FOXO1 regardless of its mutation status, we sequenced FOXO1 in T24 mt (Namalwa and BL2) and wt (CA46) lymphoma cells (Figure 3E; supplemental Figure 3B). In all cases, the proportion of cells carrying out-of-frame FOXO1 mutations after Cas9 editing dropped over time, indicating selection for FOXO1 protein–expressing cells. Moreover, T24 mt lymphoma cells predominantly preserved FOXO1 expression from the mt allele, as out-of-frame changes in the wt allele outnumbered the ones detected in the mt allele (Figure 3E). To analyze both FOXO1 alleles in individual tumor cells, we isolated single mCherry- and BFP-coexpressing cells and performed FOXO1 Sanger sequencing after clonal expansion. In the isolated clones, in-frame editing in at least 1 of the FOXO1 alleles was detectable (data not shown), demonstrating the dependency of the lymphoma cells on FOXO1 expression.
In contrast to the adverse effects of nuclear FOXO1 ablation in BL were our results in HL cells (supplemental Figure 3C): throughout the analysis, a large proportion of sequences was modified by out-of-frame mutations after CRISPR/Cas9 editing in 2 individual cell lines. Because of the constant or even increased ratio of out-of-frame/in-frame mutations over time, we concluded that HL cells expanded indefinitely after FOXO1 inactivation.
Nuclear FOXO1 induces proliferation and survival in BL
In Foxo1 knockout (KO) clones from T24 mt (mouse BL#82) or wt (mouse BL#81 and #88) BL cells, we studied lymphoma cell growth after FOXO1 ablation (Figure 4A). In accordance with our previous data, KO clones generated from the T24 mt cell line scarcely expanded over time, in contrast to control (CT) clones with intact Foxo1 alleles. This effect was specific for murine lymphoma cells expressing mt FOXO1 as the adverse phenotype of FOXO1 ablation was absent in clones arising from Foxo1 wt cell lines. Sanger sequencing and western blot analysis confirmed Foxo1 inactivation in all cases (supplemental Figure 4A-B). Cell death and cell cycle analyses revealed an increase of dead cells and a decrease of cells in the S phase of the cell cycle after Foxo1 deletion in T24 mt cells (Figure 4B-C).
Nuclear FOXO1 induces proliferation and survival in mouse BL cell lines. (A) Growth curves of mouse BL cell lines and their isogenic Foxo1 KO clones. Cells characterized by a heterozygous T24 mutation (mouse BL #82) or cell lines exclusively expressing wt FOXO1 (mouse BL#81 and #88) were analyzed. Individual clones of the parental cell lines (CT) and FOXO1-ablated clones (KO) were analyzed over time. The graph summarizes data of 3 experiments. Bars indicate the standard deviation. ****P < .0001 (Wilcoxon–Mann-Whitney test). (B) Representative FACS analysis of 5-bromo-2′-deoxyuridine (BrdU) and 7-aminoactinomycin D (7-AAD) staining in parental and Foxo1 KO cells. For each cell line, a single clone per condition (CT and KO) is shown. The gating strategy for dead (R1) and living (R2) cells is depicted. Numbers indicate the gate frequency (in percent). (C) Quantification of dead cells (R1 in panel B) and proliferating cells (G0/G1; S; G2/M in panel B) in cell line clones as analyzed in panel A. Bars indicate the standard deviation. *P < .05; **P < .01; ***P < .001; ****P < .0001 (Wilcoxon–Mann-Whitney test). ns, not significant.
Nuclear FOXO1 induces proliferation and survival in mouse BL cell lines. (A) Growth curves of mouse BL cell lines and their isogenic Foxo1 KO clones. Cells characterized by a heterozygous T24 mutation (mouse BL #82) or cell lines exclusively expressing wt FOXO1 (mouse BL#81 and #88) were analyzed. Individual clones of the parental cell lines (CT) and FOXO1-ablated clones (KO) were analyzed over time. The graph summarizes data of 3 experiments. Bars indicate the standard deviation. ****P < .0001 (Wilcoxon–Mann-Whitney test). (B) Representative FACS analysis of 5-bromo-2′-deoxyuridine (BrdU) and 7-aminoactinomycin D (7-AAD) staining in parental and Foxo1 KO cells. For each cell line, a single clone per condition (CT and KO) is shown. The gating strategy for dead (R1) and living (R2) cells is depicted. Numbers indicate the gate frequency (in percent). (C) Quantification of dead cells (R1 in panel B) and proliferating cells (G0/G1; S; G2/M in panel B) in cell line clones as analyzed in panel A. Bars indicate the standard deviation. *P < .05; **P < .01; ***P < .001; ****P < .0001 (Wilcoxon–Mann-Whitney test). ns, not significant.
To link the detrimental effects of FOXO1 ablation to its nuclear absence, we evaluated the growth characteristics of isogenic cell line clones of mouse BL#19 in which we individually targeted the mt or wt Foxo1 allele, helped by the mutation-specific PAM sequence in this mouse lymphoma. Immunofluorescence stainings for FOXO1 expression showed strong nuclear FOXO1 in clones where mt Foxo1 allele was intact (wt+/mt+ and wt−/mt+) (Figure 5A). In contrast, isogenic cells in which FOXO1 expression was restricted to the wt allele (wt+/mt−) displayed cytoplasmic FOXO1 expression. Ablation of nuclear FOXO1 by editing of the mt allele negatively impacted on tumor cell growth (Figure 5B): wt+/mt− and wt−/mt− cell line clones did not expand as their mt Foxo1-proficient counterparts (wt+/mt+ and wt−/mt+) over time. Consistently, loss of the mt allele increased the number of dead cells and resulted in impaired cell cycle progression, whereas loss of the cytoplasmic form had no detectable effect (Figure 5C).
Cytoplasmic FOXO1 is not required for survival and proliferation of T24 mt mouse cells. (A) Immunofluorescence analysis of mouse BL cells using FOXO1 antibody (red). Parental cells (mouse BL#19) characterized by a heterozygous T24 mutation (wt+/mt+) and isogenic cell line clones in which the wt (wt−/mt+) or the mt Foxo1 allele (wt+/mt−) was ablated are shown. Foxo1 KO cells (wt−/mt−) complete the analysis. Cell nuclei were counterstained with DAPI (blue; scale bar, 5 μm). (B) Growth curves of parental cell line clones (wt+/mt+) and isogenic cell line clones that express mt (wt−/mt+) or wt FOXO1 (wt+/mt−). Foxo1 KO cells (wt−/mt−) complete the analysis. The graph summarizes data of 3 experiments. Bars indicate the standard deviation. **P < .01; ***P < .001 (Wilcoxon–Mann-Whitney test). (C) Quantification of dead cells and proliferating cells in cell line clones as analyzed in panel B. FACS analysis of BrdU and 7-AAD staining was used to determine the cellular phenotype. Bars indicate the standard deviation. *P < .05; ***P < .001; ****P < .0001 (Wilcoxon–Mann-Whitney test).
Cytoplasmic FOXO1 is not required for survival and proliferation of T24 mt mouse cells. (A) Immunofluorescence analysis of mouse BL cells using FOXO1 antibody (red). Parental cells (mouse BL#19) characterized by a heterozygous T24 mutation (wt+/mt+) and isogenic cell line clones in which the wt (wt−/mt+) or the mt Foxo1 allele (wt+/mt−) was ablated are shown. Foxo1 KO cells (wt−/mt−) complete the analysis. Cell nuclei were counterstained with DAPI (blue; scale bar, 5 μm). (B) Growth curves of parental cell line clones (wt+/mt+) and isogenic cell line clones that express mt (wt−/mt+) or wt FOXO1 (wt+/mt−). Foxo1 KO cells (wt−/mt−) complete the analysis. The graph summarizes data of 3 experiments. Bars indicate the standard deviation. **P < .01; ***P < .001 (Wilcoxon–Mann-Whitney test). (C) Quantification of dead cells and proliferating cells in cell line clones as analyzed in panel B. FACS analysis of BrdU and 7-AAD staining was used to determine the cellular phenotype. Bars indicate the standard deviation. *P < .05; ***P < .001; ****P < .0001 (Wilcoxon–Mann-Whitney test).
To test whether the effects of nuclear FOXO1 on lymphoma growth were independent of PI3K activation, we applied genome editing in mouse BL#19 cells at the Rosa 26 locus where the P110* transgene was inserted in 1 of the alleles. In stably Cas9-expressing cells, we retrovirally expressed various gRNAs targeting P110*, MYC, and the wt Rosa 26 (R26) locus. In contrast to control cells in which P110* was not targeted (empty vector; gRNA_R26), cells expressing a P110* gRNA were as rapidly outcompeted by their noninfected counterparts as cells in which MYC was modified (supplemental Figure 5A). To confirm the requirement for PI3K activity in these cells, we transiently expressed Cas9 and gRNA_P110* in mouse BL#19 cells and sorted transfected cells either in bulk or as single cells. Sequencing analysis at an early and late time point after transfection showed strong selection for in-frame mutations over time (supplemental Figure 5B), indicating dependency of the mt FOXO1-expressing tumor cells on PI3K signaling.
Insights into the function of nuclear FOXO1 were gained from global gene expression profiles in cell line clones after Foxo1 locus editing. Principal component analysis (PCA) and analysis of variance–based variance analysis resolved the genotypes into distinct clusters (Figure 6A; supplemental Figure 5C): mt Foxo1-proficient samples (wt+/mt+ and wt−/mt+) grouped together, whereas wt Foxo1-proficient (wt+/mt−) and KO (wt−/mt−) samples formed independent nonoverlapping clusters.
Nuclear FOXO1 regulates gene expression. (A) Unsupervised PCA based on global transcript expression generated from individual cell line clones (n = 3 per genotype) as analyzed in Figure 5. (B) Hierarchical clustering of transcripts (n = 1500) differentially expressed in at least 1 of the groups (FDR q value < 0.05). (C) Summary of gene ontology enrichment analysis findings based on transcripts specifically deregulated by mt FOXO1 (n = 210 upregulated; n = 103 downregulated).
Nuclear FOXO1 regulates gene expression. (A) Unsupervised PCA based on global transcript expression generated from individual cell line clones (n = 3 per genotype) as analyzed in Figure 5. (B) Hierarchical clustering of transcripts (n = 1500) differentially expressed in at least 1 of the groups (FDR q value < 0.05). (C) Summary of gene ontology enrichment analysis findings based on transcripts specifically deregulated by mt FOXO1 (n = 210 upregulated; n = 103 downregulated).
Although Foxo1 KO samples were most different in comparison with the others, gene-expression changes in mt Foxo1-proficient samples (wt+/mt+ and wt−/mt+) were closely related, suggesting a dominant effect of mt FOXO1 on the transcriptome. To identify target genes of mt FOXO1 in BL, we compared the transcript-level changes in the following groups: FOXO1 KO (wt−/mt−), FOXO1 WT (wt+/mt−), and FOXO1 MT (wt−/mt+ and wt+/mt+). Of 20 000 transcripts that were present (absolute value > 100) in at least 1 of the groups, 1500 genes exhibited a false discovery rate q value < 0.05 and were defined as differentially expressed (Figure 6B). Hierarchical clustering of these genes showed not only a large set of genes being coregulated in FOXO1 MT and FOXO1 WT samples, but also genes exclusively regulated by the presence of the mt Foxo1 allele (Figure 6B). Gene-ontology enrichment analysis connected mt Foxo1-specific upregulated genes with processes related to cell cycle, cell division, and proliferation whereas downregulated genes contained regulators of apoptosis and metabolism (Figure 6C; supplemental Table 1). Although these results could be partially attributable to the different proliferative capacities of the samples (see Figure 5), tumor-promoting genes were significantly enriched in the mt FOXO1-specific gene set.
In GC B cells, the DZ transcriptional program is highly enriched in FOXO1-regulated genes.11 Thus, we determined the enrichment of FOXO1-controlled DZ genes in our BL cell line clones after FOXO1 editing. A list of potential FOXO1 target genes was generated by intercrossing genes differentially expressed in DZ/LZ cells with the ones from the comparison of FOXO1-proficient vs -deficient GC B cells (for details, see Sander et al11 ). Gene-set enrichment analysis revealed downregulation of FOXO1-induced DZ genes (Sander_DZ_up) in Foxo1 KO tumor samples (wt−/mt−; and upregulation of Sander_DZ_down genes), thereby suggesting shared FOXO1 target genes in nonmalignant and malignant GC B cells (supplemental Figure 5D).
Nuclear wt FOXO1 can substitute for mt FOXO1
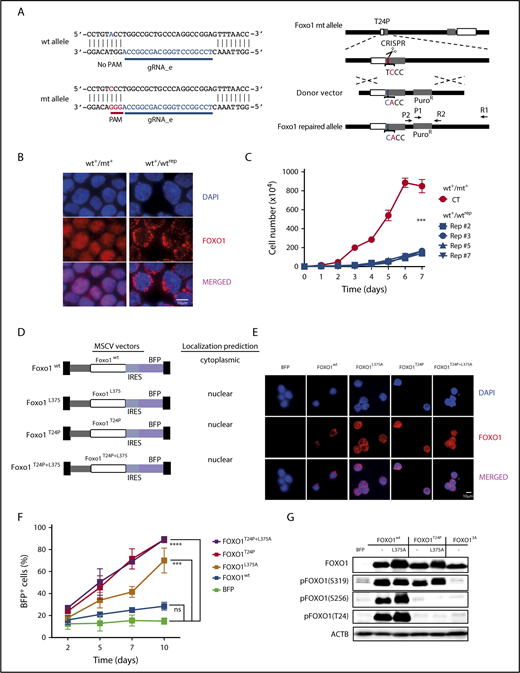
We confirmed that the single-nucleotide change in mt FOXO1 was responsible for its tumor-promoting effects when we repaired the endogenous Foxo1 locus in T24 mt mouse BL cells (Figure 7A): a vector encoding a gRNA specifically targeting the mt Foxo1 allele and Cas9 were electroporated in the cells, together with a donor plasmid encoding FOXO1 wt exon 1. To select for positively recombined cells, a puromycin-resistance gene was introduced into the donor plasmid as well as a silent mutation in the FOXO1 sequence to identify the repaired allele in sequencing analysis. The repair of the mt allele in individually recovered cell line clones was verified by Sanger sequencing. Western blot analysis demonstrated an increase in T24 phosphorylation of FOXO1, confirming the proper function of the corrected allele (supplemental Figure 6A-B). Indeed, the single-nucleotide exchange that corrected the T24 position and thus repaired the AKT-phosphorylation site of FOXO1 resulted in the nuclear exclusion of the transcription factor (Figure 7B), and cytoplasmic (wt) FOXO1 did not support lymphoma cell growth (Figure 7C).
T24 mutation or NES deficiency of FOXO1 act redundantly on mouse BL growth. (A) Schematic depiction of CRISPR/Cas9 targeting at the endogenous mouse Foxo1 locus to repair the T24 mutation. The Foxo1 gRNA_e was selected to specifically target the mt allele because of the lack of the PAM sequence at the wt allele. (B) Immunofluorescence analysis of mouse BL cells using FOXO1 antibody (red). Parental cells (mouse BL#19) carrying a heterozygous Foxo1 T24 mutation (wt+/mt+) and an isogenic cell line clone in which the mt allele was repaired to express exclusively wt FOXO1 (wt+/wtrep) are shown. Cell nuclei were counterstained with DAPI (blue; scale bar, 10 μm). (C) Growth curves of parental cells (wt+/mt+) and isogenic cell line clones (wt+/wtrep) in which the mt allele was repaired by CRISPR/Cas9 genome editing. The graph summarizes data of 3 experiments. Bars indicate the standard deviation. ***P < .001 (Wilcoxon-Mann-Whitney test). (D) Murine stem cell virus–based vector constructs for overexpression of wt and mt FOXO1. Transgene expression is coupled to BFP via an internal ribosome entry site sequence. (E) Immunofluorescence analysis of infected mouse BL cells using FOXO1 antibody (red). Foxo1 KO cells were either transduced with an empty vector (BFP) or the constructs described in panel D. Cell nuclei were counterstained with DAPI (blue; scale bar, 10 μm). (F) Percentage of BFP expressing cells over time. After infection of Foxo1 KO cells with the constructs described in panel D, the proportion of transgene-expressing cells was monitored by FACS at indicated time points. The graph summarizes the results of 2 infections. Bars indicate the standard deviation. ***P < .001; ****P < .0001 (Wilcoxon-Mann-Whitney test). (G) Western blot analysis of AKT-dependent FOXO1 phosphorylation in mouse BL cells infected with the constructs depicted in panel D. Cells expressing mt FOXO1 AAA (FOXO13A), which is impaired in T24, S256, and S319 phosphorylation, were included as controls. ACTB served as loading control. Data are representative of 2 experiments.
T24 mutation or NES deficiency of FOXO1 act redundantly on mouse BL growth. (A) Schematic depiction of CRISPR/Cas9 targeting at the endogenous mouse Foxo1 locus to repair the T24 mutation. The Foxo1 gRNA_e was selected to specifically target the mt allele because of the lack of the PAM sequence at the wt allele. (B) Immunofluorescence analysis of mouse BL cells using FOXO1 antibody (red). Parental cells (mouse BL#19) carrying a heterozygous Foxo1 T24 mutation (wt+/mt+) and an isogenic cell line clone in which the mt allele was repaired to express exclusively wt FOXO1 (wt+/wtrep) are shown. Cell nuclei were counterstained with DAPI (blue; scale bar, 10 μm). (C) Growth curves of parental cells (wt+/mt+) and isogenic cell line clones (wt+/wtrep) in which the mt allele was repaired by CRISPR/Cas9 genome editing. The graph summarizes data of 3 experiments. Bars indicate the standard deviation. ***P < .001 (Wilcoxon-Mann-Whitney test). (D) Murine stem cell virus–based vector constructs for overexpression of wt and mt FOXO1. Transgene expression is coupled to BFP via an internal ribosome entry site sequence. (E) Immunofluorescence analysis of infected mouse BL cells using FOXO1 antibody (red). Foxo1 KO cells were either transduced with an empty vector (BFP) or the constructs described in panel D. Cell nuclei were counterstained with DAPI (blue; scale bar, 10 μm). (F) Percentage of BFP expressing cells over time. After infection of Foxo1 KO cells with the constructs described in panel D, the proportion of transgene-expressing cells was monitored by FACS at indicated time points. The graph summarizes the results of 2 infections. Bars indicate the standard deviation. ***P < .001; ****P < .0001 (Wilcoxon-Mann-Whitney test). (G) Western blot analysis of AKT-dependent FOXO1 phosphorylation in mouse BL cells infected with the constructs depicted in panel D. Cells expressing mt FOXO1 AAA (FOXO13A), which is impaired in T24, S256, and S319 phosphorylation, were included as controls. ACTB served as loading control. Data are representative of 2 experiments.
Because human BL cells are characterized by nuclear FOXO1 regardless of its mutation status, we intended to mimic the existence of nuclear wt FOXO1 in the mouse cells. To study tumor cell expansion under this condition, we enforced expression of T24 wt FOXO1 lacking an intact nuclear export signal (NES)38 and appropriate controls (Figure 7D) in a FOXO1 KO clone of a FOXO1 mt lymphoma cell line (mouse BL#19). As expected, expression of wt FOXO1 resulted in its constitutive phosphorylation at T24 (due to the intrinsic PI3K pathway activation in the cells) and the cytoplasmic localization of the transcription factor (FOXO1wt) (Figure 7E; supplemental Figure 7A). Lack of T24 phosphorylation and nuclear FOXO1 was detected in cells expressing T24 mt FOXO1, in the presence (FOXO1T24P) or absence of an intact NES (FOXO1T24P+L375A). Nuclear localization of T24 wt FOXO1 whose NES was ablated (FOXO1L375A) coincided with variable levels of T24 phosphorylation (supplemental Figure 7A; Figure 7G). In contrast to cytoplasmic T24 wt FOXO1, the nuclear T24 mt was able to strongly enhance lymphoma growth in the FOXO1-deficient tumor cells (Figure 7F; supplemental Figure 7B).
Besides their impact on the subcellular localization, posttranslational modifications control the transcriptional activity of FOXO proteins and thereby their function.9,35,39,40 Thus, we studied selected FOXO1 modifications after overexpression of the transcription factor in mouse FOXO1 KO cells. Phosphorylation at all known AKT-phosphorylation sites (T24, S256, and S319) was strong in T24 wt FOXO1, irrespective of the sequence integrity of the NES and the subcellular localization of the transcription factor (Figure 7G). In constrast, FOXO1T24P selectively lacked T24 and S256 phosphorylation in mouse BL cells. The functional significance of this effect remains unclear, however, because FOXO1L375A significantly rescued the growth of FOXO1-deficient BL cells (Figure 7F). FOXO1 AAA (FOXO13A), a mutant with alanine substitutions at all AKT-phosphorylation sites, was included in our analysis to demonstrate specificity of the antibodies used. We also analyzed lysine acetylation and O-glycosylation in T24 wt and mt FOXO1, but failed to detect significant differences (supplemental Figure 7C-D).
Discussion
The GC reaction is fundamental for effective pathogen defense, but prone to tumorigenesis. The transcription factor FOXO1 is one of the prominent determinants of the GC reaction required for GC compartmentalization and DZ maintenance.
Abundant FOXO1 expression in proliferating DZ cells is counterintuitive given its antiproliferative function in nonmalignant cells outside of the GC reaction. However, the recurrent FOXO1 mutations in BL, the tumor-promoting activity of nuclear FOXO1 in mouse and human BL, and overlapping gene sets controlled by FOXO1 in DZ and BL cells suggest shared FOXO1 functions in nonmalignant and malignant GC B cells. Although Foxo1 deletion in early (or pre-) GC B cells does not interfere with the proliferative capacity of the cells,11 its absence at a later stage of GC B-cell differentiation negatively affects cell expansion.41 Similarly, in GC B-cell–derived HL cells, FOXO1 acts as a bona fide tumor-suppressor gene: its downregulation is essential for the expansion of the malignant cells22 and correlates with plasma cell differentiation blockade.42 In contrast, BL cells with their characteristic MYC translocation abuse FOXO1 to promote their growth. Interestingly, this is not a feature of MYC-driven lymphomagenesis in general, as FOXO1 has been shown to have tumor-suppressor activity in Eµ-MYC transgenic mice.43 In this latter model, B-cell transformation is biased toward early stages of B-cell development44 where FOXO1 is associated with cell cycle blockade.45
Transcriptional control of key target genes by FOXO1 is most attractive to explain its tumor-promoting function. A prerequisite for its transcription factor function is FOXO1’s localization in the nucleus. PI3K-mediated AKT activation antagonizes its nuclear retention whereas mutations preventing AKT-mediated phosphorylation foster the coexistence of active PI3K and nuclear FOXO1 in the tumor cells, a scenario not shared by nonmalignant B cells throughout development and maturation. As in the presence of MYC overexpression, PI3K signaling promotes malignant transformation of GC B cells6 and nuclear FOXO1 has tumor-promoting activity in the resulting lymphomas (present study); concomitant PI3K and FOXO1 activity in BL seem well suited to give the tumor cells a selective advantage. Indeed, both factors act nonredundantly in lymphoma growth as mouse BL cells expressing nuclear FOXO1 will stop expanding after either mt FOXO1 ablation or inactivation of PI3K signaling.
One mechanism of nuclear retention of FOXO1, regardless of PI3K activity in mouse BL cells, is disturbed FOXO1 binding to the scaffold protein 14-3-3 by mutating the T24-phosphorylation site. In mouse lymphomas, T24 mt and therefore nuclear FOXO1 was responsible for proliferation and survival. This effect could be partially mimicked by wt FOXO1 lacking the NES. Although mutations around the T24 position can explain the nuclear localization of FOXO1 in the presence of constitutive PI3K pathway activation in the mouse model and also heterozygous mt human BL cells (Figure 2D), the molecular mechanisms allowing FOXO1 to persist in the nucleus of human BL cells lacking FOXO1 mutations remain elusive. Possible explanations relate to the synergy in human BL of PI3K-pathway activation and BCR signaling,7 leading perhaps to intermittent rather than continuous PI3K activity; or redox-state–dependent protein interactions between FOXO1 and nuclear transporters46 ; or posttranslational modifications other than AKT-dependent phosphorylation that might promote the nuclear translocation of wt FOXO1 as has been described for other FOXO members and cellular contexts.23,47
Posttranslational modifications can also influence the transcription factor activity of FOXO proteins and may differ between T24 mt and wt FOXO1. Although the present data do not support a role of AKT-dependent phosphorylation in this respect and also demonstrated lysine acetylation and O-glycosylation in both T24 wt and mt nuclear FOXO1, a more detailed analysis of posttranslational FOXO1 modifications will be needed to settle this point.
Our gene-expression data provide evidence that mt FOXO1 positively controls a defined gene set that is significantly enriched for proproliferative and antiapoptotic genes. However, the substantial overlap of coregulated genes in the presence of mt (nuclear) and wt (predominantly cytoplasmic) FOXO1 was surprising. Persistence of wt FOXO1 in the cell nuclei, albeit very inefficient in comparison with mt FOXO1 (see Figure 2E), might be responsible for this effect.
Although prominent in the transcriptome analyses, the tumor-promoting function of nuclear FOXO1 is unlikely to be restricted to its effects on cell viability and cell cycle progression. Control of redox homeostasis, metabolism, and growth factor signaling40 may well contribute to its oncogenic activity in BL.
Taken together, we provide evidence that FOXO1 is abundantly expressed and recurrently mutated in both human and mouse BL tumors. Human BL cells are characterized by the predominantly nuclear localization of FOXO1, and ablation of nuclear FOXO1 leads to proliferation and survival defects in both human and mouse lymphoma cells. We conclude that nuclear FOXO1 has oncogenic function in BL (and potentially other GC-derived B-cell lymphomas), tightly associated with its nuclear localization.
The complete microarray data reported in this article have been deposited in the Gene Expression Omnibus database (accession number GSE119437).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank A. Mann, C. Grosse, J. Pempe, and K. Petsch for technical assistance; R. Lauhkonen-Seitz for administrative assistance; and the Sander and Rajewsky laboratory members for critical comments and suggestions. The authors thank M. Janz and the group of M. Sieweke for sharing samples and resources. The authors are grateful to R. Dalla-Favera and D. Dominguez-Sola for discussions and the communication of unpublished results.
The work was supported by the Else Kröner-Fresenius-Stiftung (2014-A191 [S.S. and K.R.]), the European Research Council (ERC; Advanced Grant ERC-AG-LS6 [K.R.]), and the Deutsche Krebshilfe (S.S. is a junior group leader in the Max Eder Program).
Authorship
Contribution: K.R. and S.S. designed experiments and supervised all aspects of the project; E.K., V.T.C., T.S., and E.T. performed experiments; E.K., V.T.C., T.S., K.B., T.U., L.B., and J.L.S. analyzed data; T.Z. contributed vital reagents; E.K., K.R., and S.S. wrote the manuscript; and all authors discussed results and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Sandrine Sander, Adaptive Immunity and Lymphoma, German Cancer Research Center (DKFZ)/National Center for Tumor Diseases Heidelberg (NCT), Im Neuenheimer Feld 280, 69120 Heidelberg, Germany; e-mail: sandrine.sander@nct-heidelberg.de; or Klaus Rajewsky, Immune Regulation and Cancer, Max-Delbrück-Center for Molecular Medicine in the Helmholtz Alliance, Robert-Rössle-Str 10, 13125 Berlin, Germany; e-mail: klaus.rajewsky@mdc-berlin.de.
REFERENCES
Author notes
E.K. and V.T.C. contributed equally.

![Figure 1. Nuclear FOXO1 and PI3K activity coexist in BL cells. (A) Immunofluorescence analysis of human BL cell lines using FOXO1 antibody (red). Cell nuclei were counterstained with DAPI (blue; scale bar, 5 μm). (B) Western blot analysis of FOXO1 expression in subcellular fractions of human BL cells. The purity of the cytoplasmic and nuclear fraction was determined by actin (ACTB) and histone H3 (H3) antibodies, respectively. Data are representative of at least 2 experiments. (C) Immunofluorescence analysis of FOXO1 in mouse BL cell lines as shown in panel A. (D) Western blot analysis of phospho-AKT (pAKT [S473]), AKT, phospho-FOXO1 (pFOXO1 [T24]), and FOXO1 expression in human BL cell lines. Cells were treated either with the PI3K inhibitor wortmannin (+) or dimethyl sulfoxide (DMSO) (−) 1 hour prior to protein extraction. ACTB served as loading control. Data are representative of 2 experiments. (E) Detection of pAKT(S473) in mouse BL-like cell lines by intracellular FACS analysis. After fixation and permeabilization, lymphoma cells were incubated with pAKT(S473) antibody (= antibody) or the antibody plus pAKT(S473) blocking peptide (= blocking peptide). Data are representative of 2 experiments. C, cytoplasmic fraction; KO, FOXO1 knockout cell; N, nuclear fraction; W, whole-cell lysate.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/132/25/10.1182_blood-2018-06-856203/5/m_blood856203f1.png?Expires=1764958494&Signature=olACxoC6d9wuRBm6WN3kDpZ4vTm~clTYDYWTCUHQCxnGBlQEd1M6ldT7wijO2tNFVG0~1j-pb~0~tffxFmjbid-6cIRM8bric3hoZ7IHFfyyJLkai3w5jK-E87oTgRAJCgZjyZdd0MignVqqNk~JJxG1nVjvKj3oRIGRLFTjzOZtWY09smMfId~aflYmCq82iZ98pGkheNRCJcWr3ZiT0IODHHT2pGrDy-eQOJ5MnPJ6J3D0B5mP6dxujNNG0esbIni7ebyOENhCU5NvIxkOQK13r37QF0DEGRH8IyxQTw1InOBQiO8WZal7mYrgAfLn6cf7S2PTCIzbZ~VAu90vtw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal