TO THE EDITOR:
Familial bone marrow failure (BMF) syndromes present typically in children and younger adults.1-3 A number of germline (GL) mutations in genes such as DDX41,4 RUNX1,5 ETV6,6 GATA2,7 and ANKRD268 have been implicated in the pathogenesis of familial myelodysplastic syndromes (MDSs) and define a disease class of myeloid neoplasms with GL predisposition.9 GL SAMD9 mutations arise in MIRAGE (myelodysplasia, infection, restriction of growth, adrenal hypoplasia, genital phenotypes, and enteropathy) syndrome patients,10,11 whereas GL SAMD9L mutations occur in pediatric MDS and BMF patients.7,12,13 SAMD9 and SAMD9L are proximal on 7q21.2. GL variants in these genes enhance their physiologic growth-inhibitory function and are thus gains of function (GOF).12,14,15 Supporting this notion, somatically acquired −7/del(7q), or loss-of-function (LOF) missense and truncating mutations affecting the same SAMD9/SAMD9L mutant allele, revert their GL mutation to escape its inhibitory effects.10-12,15-17 We report a 9-month-old infant with familial thrombocytopenia with a SAMD9L variant, marrow normocellularity, and the absence of megakaryocytes, which resolved following 4 months of transfusion support (supplemental Figure 1; available on the Blood Web site). Whole exome sequencing (WES) identified a novel heterozygous GL variant in a conserved amino acid region of SAMD9L (Trp517Arg) with likely deleterious consequences predicted by in silico analysis (supplemental Table 1). Sequencing of the patients’ families confirmed that the GL variant was of paternal origin. Although SAMD9 and SAMD9L GL mutations were proposed to be GOF hits in pediatric MDS,7,10,12,14 less consequential alterations could have a longer latency and also be present in sporadic adult MDS. We thus examined our sequencing data on 799 adults with presumed acquired MDS, BMF, and other diseases and searched for variants found in these genes in public WES data (n = 349 myeloid neoplasms). We used WES or targeted capture sequencing in which the entire coding regions were sequenced (supplemental Methods, supplemental Figure 2, and supplemental Tables 2 and 3). Defining rare variants as those present in <0.1% of ethnically matched healthy controls (supplemental Figure 3), we found 26 rare SAMD9/SAMD9L GL variants in 24 patients (3%; Figure 1A and supplemental Table 4). Among them, 1/12 in SAMD9 and 3/14 in SAMD9L were not previously reported (Figure 1B). All but 2 variants were heterozygous, and all but 3 were missense. In total, 4% of MDS patients (n = 21/575) and 3% of BMF patients (n = 3/105) carried variants in these genes. We made conservative algorithms to predict functional effects of missense variants by using SAMD9 GL variants reported in previous papers10,11 or public databases of registered healthy donors (supplemental Table 5). Receiver operating characteristic curves of 8 different algorithms were created (supplemental Figure 4 and supplemental Table 6); variants with ≥3 positive scores out of the top 5 algorithms were defined as “pathogenic” (sensitivity 30%, specificity 88%, positive predictive value 78%, and negative predictive value 47%; supplemental Figure 5). Eleven missense variants (48%, n = 11/23) were predicted to be pathogenic (supplemental Table 7). These results were similar to those obtained using the same inclusion criteria in public WES databases of myeloid neoplasms (2.0%, n = 7/349, pathogenic missense variants 57%, n = 4/7; supplemental Table 8).18-22
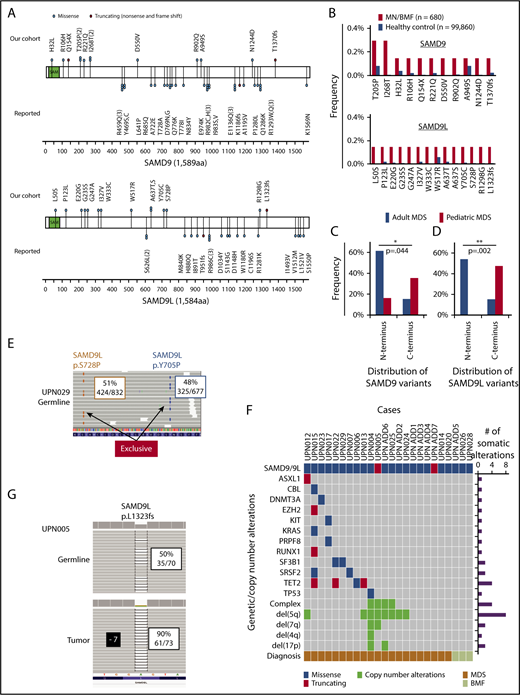
Characteristic of GL SAMD9/SAMD9L variants in adult MDS. (A) Positions and types of GL SAMD9 and SAMD9L variants detected in adult patients with MDS and BMF (above the 2 genes). Reported SAMD9 and SAMD9L variants in familial pediatric MDS and BMFs are also depicted (below the 2 genes). Functional sterile α motif (SAM) is shown in green. (B) Frequencies of specific SAMD9 and SAMD9L GL variants in our cohort and ethnically matched healthy controls. Distribution of GL SAMD9 (C) and SAMD9L (D) variants between adult and pediatric MDS. Three genetic regions (N terminus, enter, and C terminus) were defined with equal number of base pairs and compared with each other. P values were calculated by Fisher’s exact test. (E) Biallelic GL variants of SAMD9L. Representative view of 2 independent GL SAMD9L variants (Tyr705Pro, Ser728Pro) in a patient (UPN029). Variant allele frequencies and counts of sequence reads in GL samples (CD3 positive lymphocyte) are shown in boxes. (F) Landscape of somatic mutations and CNAs in adult MDS patients with GL SAMD9/SAMD9L variants. Type of mutations, cytogenetics, and diagnosis, together with the number of somatic mutations/CNAs, are color coded as indicated. (G) A patient (UPN005) with a GL frameshift SAMD9L variant (Leu1323fs) and −7 simultaneously. Variant allele frequencies and counts of sequence reads in GL (CD3 positive lymphocyte) and tumor (bone marrow) are shown in boxes.
Characteristic of GL SAMD9/SAMD9L variants in adult MDS. (A) Positions and types of GL SAMD9 and SAMD9L variants detected in adult patients with MDS and BMF (above the 2 genes). Reported SAMD9 and SAMD9L variants in familial pediatric MDS and BMFs are also depicted (below the 2 genes). Functional sterile α motif (SAM) is shown in green. (B) Frequencies of specific SAMD9 and SAMD9L GL variants in our cohort and ethnically matched healthy controls. Distribution of GL SAMD9 (C) and SAMD9L (D) variants between adult and pediatric MDS. Three genetic regions (N terminus, enter, and C terminus) were defined with equal number of base pairs and compared with each other. P values were calculated by Fisher’s exact test. (E) Biallelic GL variants of SAMD9L. Representative view of 2 independent GL SAMD9L variants (Tyr705Pro, Ser728Pro) in a patient (UPN029). Variant allele frequencies and counts of sequence reads in GL samples (CD3 positive lymphocyte) are shown in boxes. (F) Landscape of somatic mutations and CNAs in adult MDS patients with GL SAMD9/SAMD9L variants. Type of mutations, cytogenetics, and diagnosis, together with the number of somatic mutations/CNAs, are color coded as indicated. (G) A patient (UPN005) with a GL frameshift SAMD9L variant (Leu1323fs) and −7 simultaneously. Variant allele frequencies and counts of sequence reads in GL (CD3 positive lymphocyte) and tumor (bone marrow) are shown in boxes.
When we checked for differences in the position of mutations in SAMD9 and SAMD9L between adult MDS and pediatric MDS, GL SAMD9/SAMD9L variants in sporadic adult MDS did not overlap with those reported in familial pediatric MDS. Within 3 gene regions (N-terminal, center, and C-terminal) with an equal number of base pairs in each (supplemental Table 9), more SAMD9/SAMD9L GL variants were located in the N terminus in adult MDS than in pediatric MDS (Figure 1C-D; SAMD9: 8/13 vs 5/31, P = .044; SAMD9L: 7/13 vs 0/19, P = .002). Analysis of historical data yielded similar results (N terminus 71%, n = 5/7; supplemental Table 8).
Different distributions of alterations in adult vs pediatric MDS suggest distinct mechanistic consequences, as described for other genes. For instance, opposite functional consequences have been found for RHOA mutations in angioimmunoblastic T-cell lymphoma vs those in T-cell leukemia/lymphoma, leading to a dominant-negative LOF effect.23 Similarly, GOF EZH2 mutations in diffuse large B-cell lymphoma are in contrast to LOF EZH2 defects in myeloid neoplasms.24 Two independent SAMD9L variants were simultaneously present in 2 patients. One (UPN029) had biallelic alterations (Tyr705Pro and Ser728Pro in trans) as evidenced by exclusivity of independent reads (Figure 1E), while the 2 remaining variants (SAMD9 His32Leu, SAMD9L Leu50Ser) in adult MDS were located in the sterile α motif domain. Three-dimensional structure mapping suggests that these variants likely caused major structural perturbations (supplemental Figure 6).
To better elucidate the pathogenic role of GL SAMD9/SAMD9L variants, we analyzed copy number alterations (CNAs) and somatic mutations known to be associated with myeloid malignancies. In 24 patients with rare SAMD9/SAMD9L GL variants, 63% (15/24) had at least 1 somatic mutation or CNA (Figure 1F). Recurrent lesions included del(5q) (8/24 in adults vs 1/24 in children; P = .022), complex karyotype, TET2 mutations, and −7/del(7q) (2/24 in adults vs 22/32 in children; P = .0037) (supplemental Table 10).7,10,11,13 Notably, in 1 patient with −7 (UPN005), the clonal burden for SAMD9L Leu1323fs variant was higher in MDS cells than in T lymphocytes (Figure 1G), suggesting that the wild-type allele was deleted and the retained allele was a truncating variant (biallelic LOF). This finding is in contrast to the genetic reversion either through loss of −7/del(7q), 7q uniparental disomy, or by additional in cis somatic mutations in the same allele harboring GOF SAMD9/SAMD9L mutations described in children.10-12,25
SAMD9/SAMD9L negatively regulate proliferation.10,11,15 To assess the effects of rare variants identified in adults with MDS and BMFs, we tested the growth of HEK293 cells with inducible expression of SAMD9 or SAMD9L proteins. We evaluated all 11 unique pathogenic missense variants identified in 14 patients (our cohort 10, public database 4), 1 missense variant (Trp517Arg) identified in our familial case,1 frameshift variant with −7, and 1 reported pediatric alteration. Induction of FLAG-SAMD9 or TagRFP-SAMD9L proteins (Figure 2A) resulted in a mild growth restriction compared with controls, as previously reported (Figures 2B-C, and supplemental Figure 7).10-12 Compared with WT-SAMD9, Thr205Pro-SAMD9, Ile247Thr-SAMD9, and Leu574Pro-SAMD9 showed more proliferation, whereas Ile268Thr-SAMD9 and Asp550Val-SAMD9L showed no significant differences vs WT (Figure 2B). For SAMD9L, 6 variants (Glu220Gly, Leu1323fs, Cys228Tyr, Trp517Arg, Gly235Ser, and Trp333Cys) had higher proliferation rates than WT-SAMD9L (Figure 2C). Thus, 9/13 variants (69%) enhanced cell growth vs WT, indicating that inherent physiologic antiproliferative effects were affected (LOF; supplemental Table 11). In contrast, pediatric MDS variants showed less cell growth consistent with a “GOF”10,11 effect. The Trp517Arg alteration from our familial case (albeit with an atypical presentation) showed LOF, suggesting that not all GL alterations are GOF and should be functionally tested.
Functional impact of SAMD9/SAMD9L mutants. (A) Doxycycline-induced expression of rTagRFP-SAMD9L protein in HEK293 cells. (B-C) Cell proliferation curves of HEK293 cells with inducible expression (wild-type [WT] or mutant) of SAMD9/SAMD9L proteins; SAMD9 (B), SAMD9L (C). SAMD9/SAMD9L WT protein (black circles) had slower cell growth than noninduced HEK293 cells (white circles). GOF SAMD9L variant His880Glu was used as a positive control of slower cell growth than WT. Growth curve data are representative of 3 independent assays performed in triplicate. Error bars indicate standard errors. (D) Model of SAMD9/SAMD9L GL variant’s physiology in MDS. GOF SAMD9/SAMD9L GL variants suppress proliferation of human hematopoietic stem/progenitor cells (HSPCs), could be removed by genetic reversions that are rapidly selected for in pediatric MDS without additional secondary hits (upper panel). In contrast, LOF SAMD9/SAMD9L GL variants found in adult MDS cases increase cell proliferation of HSPCs, were not subject to somatic reversion, and were accompanied by additional secondary hits. The greater number of hits needed explains the longer latency in adult MDS (lower panel).
Functional impact of SAMD9/SAMD9L mutants. (A) Doxycycline-induced expression of rTagRFP-SAMD9L protein in HEK293 cells. (B-C) Cell proliferation curves of HEK293 cells with inducible expression (wild-type [WT] or mutant) of SAMD9/SAMD9L proteins; SAMD9 (B), SAMD9L (C). SAMD9/SAMD9L WT protein (black circles) had slower cell growth than noninduced HEK293 cells (white circles). GOF SAMD9L variant His880Glu was used as a positive control of slower cell growth than WT. Growth curve data are representative of 3 independent assays performed in triplicate. Error bars indicate standard errors. (D) Model of SAMD9/SAMD9L GL variant’s physiology in MDS. GOF SAMD9/SAMD9L GL variants suppress proliferation of human hematopoietic stem/progenitor cells (HSPCs), could be removed by genetic reversions that are rapidly selected for in pediatric MDS without additional secondary hits (upper panel). In contrast, LOF SAMD9/SAMD9L GL variants found in adult MDS cases increase cell proliferation of HSPCs, were not subject to somatic reversion, and were accompanied by additional secondary hits. The greater number of hits needed explains the longer latency in adult MDS (lower panel).
GL GOF SAMD9/SAMD9L variants were followed by additional somatic inactivating hits in the same allele (cis configuration) to recover cell proliferation.11,12 An analogous mechanism is imparted by −7/del(7q) leading to deletion of the mutant allele (supplemental Table 12). These patients can present in infancy and progress early10,11 (Figure 2D, upper). In an opposite scenario, GL LOF SAMD9/SAMD9L variants increased cell proliferation and were not subject to somatic reversion. Instead, MDS likely evolved by protracted acquisition of secondary hits (Figure 2D, lower). A parallel can be drawn between SAMD9/SAMD9L and DDX41 mutations, which can lead to later onset MDS.4 Similar to these mutations, functionally dichotomous SAMD9/SAMD9L variants could account for their distinct consequences.
To summarize, SAMD9 and SAMD9L LOF GL variants exist in 3% of adult MDS and are located more in the N terminus relative to pediatric GOF GL variants, which exist more in the C terminus. Genetic reversions via −7/del(7q) or additional in cis somatic mutations10-12 are rare in adult MDS. SAMD9/SAMD9L variants encountered in adult MDS tend to be LOF hits and thus convey different pathophysiologic effects.
The online version of this article contains a data supplement.
Acknowledgments
This work was supported by grants from the National Heart, Lung, and Blood Institute, National Institutes of Health (R01HL-118281, R01HL123904, R01HL128425, and R01HL132071) (J.P.M.); the Aplastic Anemia (AA) and Myelodysplastic Syndromes (MDS) Foundation (J.P.M.); the Edward P. Evans Foundation (J.P.M.); the Cancerfonden and Barncancerfonden (J.C.); the Instituto de Salud Carlos III, Ministerio de Economia y Competividad, Spain (PI/14/00013) (F.S.); 2014 SGR225 (GRE) Generalitat de Catalunya by José Carreras Leukämie-Stiftung (AR 14/34) (F.S.); Centres de Recerca de Catalunya Programme/Generalitat de Catalunya (F.S.); Fundació Internacional Josep Carreras (F.S.); the “la Caixa” Foundation (F.S.); and the Daiichi Sankyo Foundation of Life Science (Y.N.).
Authorship
Contribution: Y.N., B.P.P., C.M.H, H.M., V.P., M.A., B.K.J., T.R., V.A., and K.Y. performed experiments of molecular study and data analysis; S.L. and T.L. were committed to bioinformatics analysis of sequencing data; T.K., F.S., R.H., S.O., M.A.S., and M.W.W. collected specimens and were involved in planning the project; S.N., Y.G., and H.S. performed functional analysis; Y.N., J.C., and J.P.M. generated figures and tables and wrote the manuscript; Y.N. and J.P.M. led the entire project; and all authors participated in discussions and interpretation of the data and results.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Yasunobu Nagata, Lerner Research Institute NE6, Cleveland Clinic, 9500 Euclid Ave, Cleveland, OH, 44195; e-mail: ysnagata-tky@umin.ac.jp; and Jaroslaw P. Maciejewski, Lerner Research Institute NE6, Cleveland Clinic, 9500 Euclid Ave, Cleveland, OH; e-mail: maciejj@ccf.org.
REFERENCES
Author notes
Y.N. and S.N. contributed equally to this study.

![Figure 2. Functional impact of SAMD9/SAMD9L mutants. (A) Doxycycline-induced expression of rTagRFP-SAMD9L protein in HEK293 cells. (B-C) Cell proliferation curves of HEK293 cells with inducible expression (wild-type [WT] or mutant) of SAMD9/SAMD9L proteins; SAMD9 (B), SAMD9L (C). SAMD9/SAMD9L WT protein (black circles) had slower cell growth than noninduced HEK293 cells (white circles). GOF SAMD9L variant His880Glu was used as a positive control of slower cell growth than WT. Growth curve data are representative of 3 independent assays performed in triplicate. Error bars indicate standard errors. (D) Model of SAMD9/SAMD9L GL variant’s physiology in MDS. GOF SAMD9/SAMD9L GL variants suppress proliferation of human hematopoietic stem/progenitor cells (HSPCs), could be removed by genetic reversions that are rapidly selected for in pediatric MDS without additional secondary hits (upper panel). In contrast, LOF SAMD9/SAMD9L GL variants found in adult MDS cases increase cell proliferation of HSPCs, were not subject to somatic reversion, and were accompanied by additional secondary hits. The greater number of hits needed explains the longer latency in adult MDS (lower panel).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/132/21/10.1182_blood-2017-05-787390/5/m_blood787390f2.png?Expires=1765892209&Signature=oS0rxeRHQHAlCsPkoNSmb8A9lOqGARRljUqslRZSawTN9fir9KVD8yqNvLF3-pkCzg2Xei8XNbymUv2HLClhhhoc-MCTUqJJ0q4KPRPVB3Y8Y27jrwqHen0odd50s4jdBDqULKJ5Iqz5cjPFxBd-zNzikiCxwBiabs22xjSvQWBqUcHGQxi~K0TrTHYkQzzn8BQYz8RM8k05T4OkhRn1irVWT7R7iUG6B3nE0IRUWBNTm3H74J81BFYW-qVvueVB7JI0rWOPkEz65VVaqBrQDkrCB3KPvm2HbU~CAiC2e~pG32LcqU1norOqaJT1HPJyAsKJCxlJNCFRkec-8XNWnA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal