Key Points
First-in-class MIR17PTi enables 1-shot downregulation of miR-17-92 in vitro and in vivo, with favorable pharmacokinetic profile.
MIR17PTi affects homeostatic MYC/miR-17-92 FFLs in MM cells, resulting in strong anti-MM activity.
Abstract
The microRNA (miRNA) cluster miR-17-92 is oncogenic and represents a valuable therapeutic target in c-MYC (MYC)–driven malignancies. Here, we developed novel LNA gapmeR antisense oligonucleotides (ASOs) to induce ribonuclease H–mediated degradation of MIR17HG primary transcripts and consequently prevent biogenesis of miR-17-92 miRNAs (miR-17-92s). The leading LNA ASO, MIR17PTi, impaired proliferation of several cancer cell lines (n = 48) established from both solid and hematologic tumors by on-target antisense activity, more effectively as compared with miR-17-92 inhibitors. By focusing on multiple myeloma (MM), we found that MIR17PTi triggers apoptosis via impairment of homeostatic MYC/miR-17-92 feed-forward loops (FFLs) in patient-derived MM cells and induces MYC-dependent synthetic lethality. We show that alteration of a BIM-centered FFL is instrumental for MIR17PTi to induce cytotoxicity in MM cells. MIR17PTi exerts strong in vivo antitumor activity in nonobese diabetic severe combined immunodeficient mice bearing clinically relevant models of MM, with advantageous safety and pharmacokinetic profiles in nonhuman primates. Altogether, MIR17PTi is a novel pharmacological tool to be tested in early-phase clinical trials against MM and other MYC-driven malignancies.
Introduction
miR-17-92 is an oncogenic cluster of microRNAs (miRNAs) encoded by MIR17HG at 13q31.3.1,2 According to ENSEMBL genome browser (GRCh38.p10), 3 long noncoding RNAs (lncRNAs) are produced by the alternative splicing of an MIR17HG primary transcript (MIR17HG-PT), with only lncRNAs MIR17HG-202 and MIR17HG-203 (hereafter referred to as pri-mir-17-92) generating the 6 miR-17-92 mature miRNAs (miR-17-92s), including miR-17/-18a/-19a/-20a/-19b-1/-92a-1.1,2 This cluster is steadily upregulated in solid and hematologic malignancies, mainly because of amplification of the genomic locus or by transcriptional mechanisms.1,2 On the basis of their seed regions, miR-17-92s are grouped into 4 different families (miR-17, miR-18, miR-19, and miR-92), which share most target genes.1,2 These latter include several tumor suppressors, such as the proapoptotic factor BIM (also known as BCL2L11), thus explaining the tumor-promoting functions of miR-17-92s.1,2
A key transcriptional regulator of MIR17HG is the transcription factor c-MYC (MYC).3 This oncogene drives the onset and progression of several human malignancies, including multiple myeloma (MM),4 B-cell lymphomas (BCLs),5 and triple-negative breast cancer,6 which carry the trait of hard-to-treat tumors.7 miR-17-92 ensures cellular homeostasis during MYC-driven tumorigenesis by counteracting the MYC apoptotic signal. This effect is achieved by the cooperative activity of all 6 miR-17-92s, which coordinately fine tune MYC expression and suppress the MYC-related apoptotic program.3,8 The close interplay between MYC and miR-17-92, along with the lack of therapeutic tools for the direct targeting of MYC,9 underscores the relevance of miR-17-92 as an attractive druggable target in MYC-driven cancer. In this scenario, the targeting of miR-17-92 is likely to frame as an MYC-dependent synthetic lethal (SL) approach, whereby it specifically produces cytotoxicity in the presence of hyperactive/deregulated MYC.10
On these bases, we hypothesized that a powerful antitumor effect would be achieved by inducing simultaneous downregulation of all miR-17-92s, and we developed a novel strategy that allows their 1-shot inhibition via the targeting of miR-17-92 primary transcript by ribonuclease (RNase) H–activating antisense oligonucleotides (ie, gapmeRs). Our efforts led to the preclinical development as an anticancer agent of first-in-class MIR17PTi (MIR17HG primary transcript inhibitor). We demonstrate its therapeutic activity in the context of MM, where MIR17PTi promotes a newly identified MYC-dependent SL via the alteration of BIM-centered MYC/miR-17-92 feed-forward loops (FFLs).
To our knowledge, this is the first report of miRNA primary transcript (pri-miRNA) therapeutics with translational value in human cancer.
Material and methods
Cells
Human cell lines and primary cells were grown at 37°C, 5% CO2. Detailed information is included in supplemental Methods (available on the Blood Web site).
Antisense oligonucleotides, miRNA mimics/inhibitors, and short hairpin RNAs
The long noncoding LNA gapmerRs listed in Table 1 were custom designed and purchased from Exiqon (Vedbaek, Denmark). Detailed information on other oligonucleotides used in our work is reported in supplemental Methods.
IDs, sequences, and lengths of 7 miR-17-92 LNA gapmeRs and 3 controls
| ID . | Sequence 5′-3′ . | Length, mer . |
|---|---|---|
| LNA gapmeR_02 | ACATCGACACAATAA | 15 |
| LNA gapmeR_05 | TCAGTAACAGGACAGT | 16 |
| LNA gapmeR_06 (MIR17PTi) | TACTTGCTTGGCTT | 14 |
| LNA gapmeR_10 | ATGCAAAACTAACAGA | 16 |
| LNA gapmeR_12 | GAAGGAAAATAGCAGGC | 16 |
| LNA gapmeR_15 | AGCACTCAACATCAGC | 16 |
| LNA gapmeR_16 | CGACAGGCCGAAGCT | 15 |
| Scr-NC (also known as negative control A) | GCTCCCTTCAATCCAA | 15 |
| Mix-MIR17PTi | TACTTGCTTGGCTT | 14 |
| ID . | Sequence 5′-3′ . | Length, mer . |
|---|---|---|
| LNA gapmeR_02 | ACATCGACACAATAA | 15 |
| LNA gapmeR_05 | TCAGTAACAGGACAGT | 16 |
| LNA gapmeR_06 (MIR17PTi) | TACTTGCTTGGCTT | 14 |
| LNA gapmeR_10 | ATGCAAAACTAACAGA | 16 |
| LNA gapmeR_12 | GAAGGAAAATAGCAGGC | 16 |
| LNA gapmeR_15 | AGCACTCAACATCAGC | 16 |
| LNA gapmeR_16 | CGACAGGCCGAAGCT | 15 |
| Scr-NC (also known as negative control A) | GCTCCCTTCAATCCAA | 15 |
| Mix-MIR17PTi | TACTTGCTTGGCTT | 14 |
Gymnosis
Cells were seeded at low plating density to reach confluence on the final day of the experiments (day 6). Cell numbers at plating ranged from 0.5 × 103 to 2.5 × 103 in 96-well plates, from 2.5 × 104 to 10 × 104 in 12-well plates, and from 1 × 105 to 3 × 105 in 6-well plates.
Transient and stable transfection of cells
Detailed information can be found in supplemental Methods.
CRISPR/CAS9-mediated genome editing
Detailed information can be found in supplemental Methods.
Survival assay/detection of apoptosis/cell-cycle analysis
Detailed information can be found in supplemental Methods.
Synergism quantification
Detailed information can be found in supplemental Methods section.
Reverse transcription and qRT-PCR
RNA extraction, reverse transcription, and quantitative real-time polymerase chain reaction amplification (qRT-PCR) were performed as previously described.11 Detailed information is included in supplemental Methods.
Western blot analysis
Protein extraction and western blot analysis were performed as previously described.11 Detailed information is included in supplemental Methods.
Confocal microscopy
Detailed information can be found in supplemental Methods.
Gene expression profiling
Detailed information can be found in supplemental Methods.
NOD SCID mice and in vivo models of human MM
Male CB-17 nonobese diabetic (NOD) severe combined immunodeficient (SCID) mice (6 to 8 weeks old; Harlan Laboratories, Inc., Indianapolis, IN) were housed and monitored in our animal research facility. All experimental procedures and protocols were approved by the institutional ethical committee (Magna Graecia University) and conducted according to protocols approved by the National Directorate of Veterinary Services (Italy). Detailed information can be found in supplemental Methods.
Immunohistochemistry and in situ hybridization
These experiments were performed as recently described by our group.12
Nonhuman primates study
These experiments were performed as recently described by our group.12
Statistical analyses
All in vitro experiments were repeated at least 3 times and performed in triplicate; a representative experiment is shown in figures. Statistical significance of differences was determined using Student t test, with minimal level of significance specified as P < .05. Statistical significance of the in vivo growth inhibition was determined using Student t test. The minimal level of significance was specified as P < .05. All statistical analyses were performed using GraphPad software. Graphs were obtained using GraphPad software.
Results
MIR17PTi prevents biogenesis of miR-17-92s
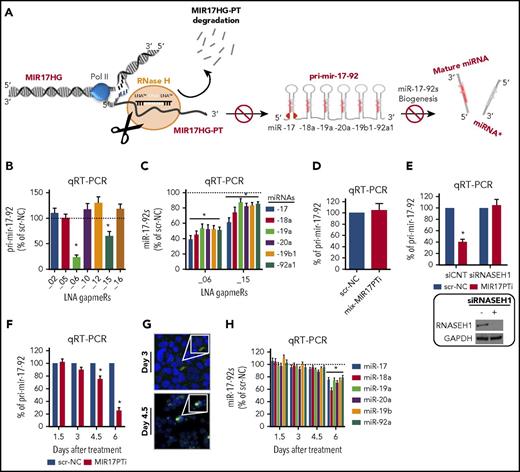
MIR17HG-PT, as well as pri-mir-17-92, is located within the cell nucleus.13 This environment is selectively enriched with RNase H, an enzyme that catalyzes nonspecific cleavage of RNA strands within DNA/RNA heteroduplexes.14 Therefore, we attempted to induce degradation of nascent MIR17HG-PT and pri-mir-17-92 by LNA gapmeRs, a highly efficient and recently developed class of RNase H–activating antisense oligonucleotides (ASOs; Figure 1A).14 We first screened LNA-modified gapmeRs for their ability to inhibit miR-17-92 cluster in transfected 293T cells; of 7 original molecules, only gapmeR_06 and gapmeR_15 efficiently downregulated pri-mir-17-92 (Figure 1B) and all 6 miR-17-92s (Figure 1C; supplemental Figure 1A). Because gapmeR_06 showed higher inhibitory efficiency, it was selected for further development (hereafter named MIR17PTi). Importantly, MIR17PTi downregulated miR-17-92 cluster by activating RNase H; in fact, chemical modifications of MIR17PTi within the RNase H–recruiting domain (the resulting oligo was named mix-MIR17PTi) abrogated its inhibitory activity (Figure 1D); consistently, MIR17PTi failed to downregulate pri-mir-17-92 after RNA interference (RNAi)–mediated silencing of RNase H1 (Figure 1E). MIR17PTi also produced the downregulation of lncRNA MIR17HG-201 (supplemental Figure 1B), a shorter transcript produced by the alternative splicing of MIR17HG-PT. This isoform does not contain the MIR17PTi-targeted sequence, confirming that MIR17HG-PT is primarily targeted by MIR17PTi.
Development of MIR17PTi. (A) Illustration summarizing the activity of LNA gapmeRs with regard to the miR-17-92 cluster. (B) qRT-PCR analysis of pri-mir-17-92 expression in 293T 2 days after transfection with miR-17-92 LNA gapmeRs or scr-NC (25 nM). (C) qRT-PCR analysis of miR-17-92s in 293T 2 days after transfection with mir-17-92 LNA gapmeRs or scr-NC (25 nM). (D) qRT-PCR analysis of pri-mir-17-92 expression in 293T 2 days after transfection with mix-MIR17PTi or scr-NC (25 nM). (E) qRT-PCR analysis of pri-mir-17-92 (left) and western blotting of RNase H1 (right) in 293T cotransfected with either small interfering RNAs (siRNAs) targeting RNase H1 (siRNASE H1; 25 nM) or scrambled siRNAs (siCNT; 25 nM) and MIR17PTi or scr-NC (25 nM; 3-day time point). GAPDH was used as protein loading control. (F) qRT-PCR analysis of pri-mir-17-92 in 293T during a time-course exposure (days 1.5, 3, 4.5, and 6, every 36 hours) to MIR17PTi or scr-NC (10 µM). (G) Confocal microscopy analysis of 293T after 3 or 4.5 days of exposure to FAM-labeled MIR17PTi (10 µM). Cell nuclei are evidenced by 4′,6-diamidino-2-phenylindole staining (original magnification ×40). (H) qRT-PCR analysis of miR-17-92s in 293T during a time-course exposure (days 1.5, 3, 4.5, and 6, every 36 hours) to MIR17PTi or scr-NC (10 µM). The qRT-PCR results are average expression levels after normalization with GAPDH (for pri-mir-17-92) or RNU44 (for miR-17-92s) and ΔΔCt calculations. Data from 1 of 3 independent experiments are shown in each panel. *P < .05.
Development of MIR17PTi. (A) Illustration summarizing the activity of LNA gapmeRs with regard to the miR-17-92 cluster. (B) qRT-PCR analysis of pri-mir-17-92 expression in 293T 2 days after transfection with miR-17-92 LNA gapmeRs or scr-NC (25 nM). (C) qRT-PCR analysis of miR-17-92s in 293T 2 days after transfection with mir-17-92 LNA gapmeRs or scr-NC (25 nM). (D) qRT-PCR analysis of pri-mir-17-92 expression in 293T 2 days after transfection with mix-MIR17PTi or scr-NC (25 nM). (E) qRT-PCR analysis of pri-mir-17-92 (left) and western blotting of RNase H1 (right) in 293T cotransfected with either small interfering RNAs (siRNAs) targeting RNase H1 (siRNASE H1; 25 nM) or scrambled siRNAs (siCNT; 25 nM) and MIR17PTi or scr-NC (25 nM; 3-day time point). GAPDH was used as protein loading control. (F) qRT-PCR analysis of pri-mir-17-92 in 293T during a time-course exposure (days 1.5, 3, 4.5, and 6, every 36 hours) to MIR17PTi or scr-NC (10 µM). (G) Confocal microscopy analysis of 293T after 3 or 4.5 days of exposure to FAM-labeled MIR17PTi (10 µM). Cell nuclei are evidenced by 4′,6-diamidino-2-phenylindole staining (original magnification ×40). (H) qRT-PCR analysis of miR-17-92s in 293T during a time-course exposure (days 1.5, 3, 4.5, and 6, every 36 hours) to MIR17PTi or scr-NC (10 µM). The qRT-PCR results are average expression levels after normalization with GAPDH (for pri-mir-17-92) or RNU44 (for miR-17-92s) and ΔΔCt calculations. Data from 1 of 3 independent experiments are shown in each panel. *P < .05.
In transfection-free or gymnotic conditions,14 the timing of pri-mir-17-92 downregulation (Figure 1F) was concomitant with nucleus entrance of a FAM-labeled MIR17PTi (Figure 1G). We did not observe any further increase of nuclear uptake at day 6 as compared with day 4.5 (data not shown), suggesting that the strongest downregulation of pri-mir-17-92 observed at day 6 comes from prolonged activity of MIR17PTi in the nucleus. Importantly, downregulation of pri-mir-17-92 was followed by decrease of miR-17-92s (Figure 1H); both effects (inhibition of pri-mir-17-92 and miR-17-92s) occurred in a dose-dependent fashion (supplemental Figure 1C-D).
These findings demonstrate that targeting of MIR17HG-PT by LNA gapmeRs prevents the biogenesis of all miR-17-92s.
MIR17PTi affects proliferation of several CCLs
We analyzed MIR17PTi activity by gymnotic exposure in cancer cells and found that it significantly affected the in vitro growth of several cancer cell lines (CCLs; n = 48) established from either solid or hematologic tumors (Figure 2A). Intriguingly, the latter showed the strongest sensitivity to MIR17PTi as well as the highest pri-mir-17-92 expression within the Cancer Cell Line Encyclopedia (supplemental Figure 2A). MIR17PTi did not affect the proliferation of 4 of 5 NM-CLs (Figure 2A); indeed, proliferation of THLE-2 (liver) and HK-2 (kidney) NM-CLs was not significantly affected even at concentrations up to fivefold higher (50 μM) than those used in this screening phase (10 μM; supplemental Figure 3A). By qRT-PCR analysis, we found that MIR17PTi efficiently downregulated pri-mir-17-92 and miR-17-92s both in MIR17PTi-sensitive and in MIR17PTi-resistant cells, indicating that MIR17PTi resistance did not depend on inefficient uptake of the oligo (supplemental Figure 3B-C). Moreover, we further confirmed this pattern of sensitivity/resistance to MIR17PTi by transfecting a subset of both CCLs and NM-CLs (supplemental Figure 3D). In CCLs, mix-MIR17PTi did not affect proliferation (supplemental Figure 3E), whereas LNA gapmeR-15, which downregulates both pri-mir-17-92 and miR-17-92s, performed similarly to MIR17PTi (supplemental Figure 3F-G).
In vitro anticancer activity of MIR17PTi. (A) CCK-8 assay of CCLs (n = 48) and nonmalignant cell lines (NM-CLs; n = 5) exposed for 6 days to MIR17PTi or scr-NC (10 µM). Data are represented as percentage of MIR17PTi-treated live cells (absorbance), as compared with scr-NC. Dashed lines indicate 20% (upper line) and 80% (bottom line) growth inhibition. (B) CCK-8 proliferation assay of 8 CCLs (P3HR1 [diffuse large B cell lymphoma (DLBCL)], SULTAN [Burkitt lymphoma (BL)], JeKo-1, Maver [mantle cell lymphoma (MCL)], AMO1, KMS-12-BM, NCI-H929, RPMI-8226 [MM]) transfected with indicated miRNA inhibitors or MIR17PTi (25 nM). Data are represented as compared (percentage) with live cells after scr-NC transfection. (C) CCK-8 proliferation assay of RPMI-8226 and AMO1 after transfection with miR-NC-inhibitors (150 nM) or pooled miR-17-92 inhibitors (25 nM each) or scr-NC (25 nM) or MIR17PTi (25 nM). Data are represented as compared (percentage) with live cells after mock (RNase-free water) transfection. (D) CCK-8 proliferation assay of: AMO1 2 days after cotransfection with scr-NC (5 or 25 nM; showed as single point because no difference was detected in percentage of live cells) or MIR17PTi (5 or 25 nM) and miR-NC-mimics (60 nM) or pooled miR-17-92 mimics (10 nM each) (left) and AMO1 transduced with an empty lentiviral vector or an miR-17-92 lentiviral vector and then exposed for 6 days to scr-NC or MIR17PTi (0.5 µM) (right). Data from 1 of 3 independent experiments are shown in each panel. *P < .05. AML, acute myeloid leukemia; BC, breast cancer; MPM, malignant pleural mesothelioma; PC, pancreatic cancer; TCL, T-cell leukemia.
In vitro anticancer activity of MIR17PTi. (A) CCK-8 assay of CCLs (n = 48) and nonmalignant cell lines (NM-CLs; n = 5) exposed for 6 days to MIR17PTi or scr-NC (10 µM). Data are represented as percentage of MIR17PTi-treated live cells (absorbance), as compared with scr-NC. Dashed lines indicate 20% (upper line) and 80% (bottom line) growth inhibition. (B) CCK-8 proliferation assay of 8 CCLs (P3HR1 [diffuse large B cell lymphoma (DLBCL)], SULTAN [Burkitt lymphoma (BL)], JeKo-1, Maver [mantle cell lymphoma (MCL)], AMO1, KMS-12-BM, NCI-H929, RPMI-8226 [MM]) transfected with indicated miRNA inhibitors or MIR17PTi (25 nM). Data are represented as compared (percentage) with live cells after scr-NC transfection. (C) CCK-8 proliferation assay of RPMI-8226 and AMO1 after transfection with miR-NC-inhibitors (150 nM) or pooled miR-17-92 inhibitors (25 nM each) or scr-NC (25 nM) or MIR17PTi (25 nM). Data are represented as compared (percentage) with live cells after mock (RNase-free water) transfection. (D) CCK-8 proliferation assay of: AMO1 2 days after cotransfection with scr-NC (5 or 25 nM; showed as single point because no difference was detected in percentage of live cells) or MIR17PTi (5 or 25 nM) and miR-NC-mimics (60 nM) or pooled miR-17-92 mimics (10 nM each) (left) and AMO1 transduced with an empty lentiviral vector or an miR-17-92 lentiviral vector and then exposed for 6 days to scr-NC or MIR17PTi (0.5 µM) (right). Data from 1 of 3 independent experiments are shown in each panel. *P < .05. AML, acute myeloid leukemia; BC, breast cancer; MPM, malignant pleural mesothelioma; PC, pancreatic cancer; TCL, T-cell leukemia.
We next evaluated the anticancer activity of MIR17PTi as compared with the individual targeting of miR-17-92s. At this end, we transfected different CCLs from hematologic malignancies (n = 8) with either MIR17PTi or synthetic inhibitors for each miR-17-92 and found, as shown in Figure 2B, that MIR17PTi affected proliferation of all CCLs significantly more efficiently than miR-17-92 inhibitors. Importantly, the antiproliferative activity of MIR17PTi was closely reproduced by cotransfecting all miR-17-92 inhibitors (pooled miR-17-92 inhibitors; Figure 2C), and the activity of MIR17PTi was significantly rescued by concomitant overexpression of all miR-17-92s by either synthetic mimics or lentivectors (Figure 2D).
These findings indicate that cancer cells from different tissues, especially those of hematologic origin, are sensitive to MIR17PTi. Moreover, they strengthen the rationale for targeting the entire miR-17-92 cluster via MIR17HG-PT degradation instead of inhibiting individual mature miRNA components.
MIR17PTi is highly active in pMM cells and HMCLs
Further investigation to establish proof of concept for the potential therapeutic value of MIR17PTi was carried out using MM as the working platform. In patients with MM, by querying a proprietary microarray data set, we found that expression of pri-mir-17-92 and miR-17-92s is significantly upregulated at the most advanced stage (namely, secondary plasma-cell leukemia; supplemental Figure 4A). RNA sequencing analysis of 320 patients with newly diagnosed MM from the IFM/DFCI 2009 clinical study (registered at www.clinicaltrials.gov as #NCT0191060)15 indicated that high pri-mir-17-92 expression correlates with worse clinical outcome (overall and progression-free survival; supplemental Figure 4B), even if at this stage it is not significantly upregulated as compared with healthy donors (n = 16; data not shown). These data suggested that in patients with MM, higher miR-17-92 expression is associated with a more aggressive phenotype, such as end-stage disease (secondary plasma-cell leukemia) and/or poor-prognosis subgroups yet to be defined.
In human myeloma cell lines (HMCLs), we found that MIR17PTI affected proliferation in a dose-dependent fashion (Figure 3A), and the timing of this effect overlapped with downregulation of miR-17-92s (Figure 3B). Growth inhibition was ascribed to G0/G1 cell-cycle arrest (Figure 3C; supplemental Figure 5), apoptosis (Figure 3C; supplemental Figures 6 and 7), and senescence (Figure 3D) and was even more evident in methylcellulose-based long-term cultures, where the growth of colonies was abrogated at nanomolar concentrations (Figure 3E). Importantly, MIR17PTi was still active against HMCLs made resistant to conventional anti-MM agents, such as dexamethasone (MM.1R), melphalan (LR7), bortezomib (ABZB) or carfilzomib (ACFZ; Figure 3F), and survival signals from cocultured human BM stromal cells did not rescue MIR17PTi activity (Figure 3G; supplemental Figure 8A). Moreover, combination of MIR17PTi with dexamethasone, melphalan, or bortezomib produced in most cases synergistic effects (Figure 3H; supplemental Figure 8B).
In vitro activity of MIR17PTi in HMCLs and primary MM (pMM) cells. (A) Dose-dependent activity of MIR17PTi (6-day time point) in HMCLs (n = 10) as assessed by CCK-8 proliferation assay. Live cells are represented as compared (percentage) with untreated controls. (B) Time-dependent activity of MIR17PTi (1 µM) in AMO1, as assessed by qRT-PCR analysis of pri-mir-17-92 (left) and miR-17-92s (middle). The results shown are average pri-mir-17-92 or miRNA expression levels after normalization with glyceraldehyde-3-phosphate dehydrogenase or RNU44 and ΔΔCt calculation (expressed as percentage). Time-dependent activity of MIR17PTi (1 µM) in AMO1, as assessed by CCK-8 assay (right pane). (C) Cell-cycle analysis of AMO1 exposed for 6 days to MIR17PTi (1 µM) or sc-NC (1 µM). (D) Flow cytometric analysis of senescent AMO1 cells (β-galactosidase positive) exposed for 6 days to MIR17PTi (1 µM) o scr-NC (1 µM). (E) Methylcellulose clonogenic assay of AMO1 exposed to MIR17PTi (1 µM) o scr-NC (1 µM; 2-week time point). (F) CCK-8 proliferation assay of HMCLs sensitive (MM.1S, U266, AMO1) or resistant (MM.1R, LR7, ABZB, ACFZ) to conventional anti-MM agents (dexamethasone, melphalan, bortezomib, carfilzomib) exposed to different concentrations of MIR17PTi for 6 days. (G) CCK-8 assay of human bone marrow (BM) stromal cells (hBMSCs), AMO1, or AMO1 cocultured with hBMSCs after 6 days of exposure to indicated concentrations of MIR17PTi. (H) Table showing combination indexes resulting from combinatorial treatments of AMO1 with MIR17PTi and dexamethasone or melphalan or bortezomib (6-day time point). (I) Flow cytometric analysis of 7-AAD–stained pMM cells (n = 13) exposed for 8 days to MIR17PTi or scr-NC (2.5 µM). pMM cells were cultured adherent to GFP+ HS-5 stromal cells (patients 1-3; patients with intramedullary disease) or physically separated from HS-5 by membranes (patients 4-11; patients with intramedullary disease) or alone (patients 12-13; patients with extramedullary disease at relapse [secondary plasma-cell leukemia (sPCL)]). Lower panel (within the frame) shows median activity of MIR17PTi in sensitive pMM cells (n = 10) from the left panel. *P < .05.
In vitro activity of MIR17PTi in HMCLs and primary MM (pMM) cells. (A) Dose-dependent activity of MIR17PTi (6-day time point) in HMCLs (n = 10) as assessed by CCK-8 proliferation assay. Live cells are represented as compared (percentage) with untreated controls. (B) Time-dependent activity of MIR17PTi (1 µM) in AMO1, as assessed by qRT-PCR analysis of pri-mir-17-92 (left) and miR-17-92s (middle). The results shown are average pri-mir-17-92 or miRNA expression levels after normalization with glyceraldehyde-3-phosphate dehydrogenase or RNU44 and ΔΔCt calculation (expressed as percentage). Time-dependent activity of MIR17PTi (1 µM) in AMO1, as assessed by CCK-8 assay (right pane). (C) Cell-cycle analysis of AMO1 exposed for 6 days to MIR17PTi (1 µM) or sc-NC (1 µM). (D) Flow cytometric analysis of senescent AMO1 cells (β-galactosidase positive) exposed for 6 days to MIR17PTi (1 µM) o scr-NC (1 µM). (E) Methylcellulose clonogenic assay of AMO1 exposed to MIR17PTi (1 µM) o scr-NC (1 µM; 2-week time point). (F) CCK-8 proliferation assay of HMCLs sensitive (MM.1S, U266, AMO1) or resistant (MM.1R, LR7, ABZB, ACFZ) to conventional anti-MM agents (dexamethasone, melphalan, bortezomib, carfilzomib) exposed to different concentrations of MIR17PTi for 6 days. (G) CCK-8 assay of human bone marrow (BM) stromal cells (hBMSCs), AMO1, or AMO1 cocultured with hBMSCs after 6 days of exposure to indicated concentrations of MIR17PTi. (H) Table showing combination indexes resulting from combinatorial treatments of AMO1 with MIR17PTi and dexamethasone or melphalan or bortezomib (6-day time point). (I) Flow cytometric analysis of 7-AAD–stained pMM cells (n = 13) exposed for 8 days to MIR17PTi or scr-NC (2.5 µM). pMM cells were cultured adherent to GFP+ HS-5 stromal cells (patients 1-3; patients with intramedullary disease) or physically separated from HS-5 by membranes (patients 4-11; patients with intramedullary disease) or alone (patients 12-13; patients with extramedullary disease at relapse [secondary plasma-cell leukemia (sPCL)]). Lower panel (within the frame) shows median activity of MIR17PTi in sensitive pMM cells (n = 10) from the left panel. *P < .05.
In ex vivo viability assays, MIR17PTi was cytotoxic to pMM cells derived from patients at either BM microenvironment–dependent (n = 11) or BM microenvironment–independent (n = 2) stage of disease (Figure 3I; detailed information on pMM cells are reported in supplemental Figure 9A). Consistently with tumor-selective killing activity, MIR17PTi did not affect viability of either CD138+ cells from patients with the premalignant condition of monoclonal gammopathy of undetermined significance (n = 3; supplemental Figure 9B) or peripheral blood mononuclear cells from healthy donors (n = 2; supplemental Figure 9C).
These data highlight the strong therapeutic potential of MIR17PTi against MM.
MIR17PTi affects MYC transcriptional network in pMM cells
By microarray-based gene expression profiling (GEP), we explored the molecular perturbations produced by MIR17PTi in pMM cells (n = 4; patients 5, 6, 7, and 9) cocultured with HS-5 stromal cells. Unsupervised hierarchical clustering segregated samples based on treatment, suggesting a coherent transcriptome modulation (Figure 4A). Ingenuity pathway analysis of GEP data highlighted MYC as the most significantly affected upstream regulator (P = 3.17E−06; data not shown). Consistently, gene set enrichment analysis16,17 identified MYC_TARGET_V1 as the most significantly enriched hallmark collection in the positive phenotype (ie, genes upregulated by MIR17PTi; Figure 4B), and in-depth analysis of the MYC_TARGET_V1 founder gene sets (n = 404) showed the specific enrichment of transcriptional signatures of MYC-upregulated target genes (Figure 4C). MIR17PTi induced similar transcriptional alterations in the highly sensitive AMO1 cells (supplemental Figure 10B-C). Conversely, treatment of MIR17PTi-resistant pMM cells (n = 2; patients 10 and 11) did not alter the MYC transcriptional signaling, as assessed by GEP analysis, even if pri-mir-17-92 was significantly downregulated (data not shown).
Molecular perturbation induced by MIR17PTi in pMM cells. (A) Hierarchical clustering of pMM cells (n = 4; patients 5, 6, 7, and 9) exposed for 6 days to MIR17PTi (2.5 µM) or equimolar scr-NC. (B) Table of gene sets from the Hallmark collection enriched with genes upregulated by MIR17PTi (positive phenotype) in sensitive pMM cells (patients 5, 6, 7, and 9). Number of genes in each set (size), the normalized enrichment score (NES), and test of statistical significance (false discovery rate [FDR] q value) are highlighted. (C) Enrichment plots of 3 representative transcriptional signatures of MYC-upregulated target genes in the positive phenotype of sensitive pMM cells exposed to MIR17PTi (2.5 µM) for 6 days. (D) Venn diagram–intersecting genes upregulated by MIR17PTi with genes predicted to be miR-17-92 targets (by miRcode) and genes validated as miR-17-92 interactors in CLIP sequencing experiments (starBase v2.0). (E) Representation of MYC/miR-17-92 FFLs. In red are reported genes with a fold change (FC) increase >1.5. (F) qRT-PCR analysis of pri-mir-17-92 and BIM, BZW2, DUSP2, NAP1L1, or VDAC1 messenger RNAs (mRNAs) in pMM cells (n = 3) exposed to MIR17PTi (2.5 µM) for 6 days. The results shown are average pri-mir-17-92 or mRNA expression levels after normalization with glyceraldehyde-3-phosphate dehydrogenase and ΔΔCt calculations. *P < .05.
Molecular perturbation induced by MIR17PTi in pMM cells. (A) Hierarchical clustering of pMM cells (n = 4; patients 5, 6, 7, and 9) exposed for 6 days to MIR17PTi (2.5 µM) or equimolar scr-NC. (B) Table of gene sets from the Hallmark collection enriched with genes upregulated by MIR17PTi (positive phenotype) in sensitive pMM cells (patients 5, 6, 7, and 9). Number of genes in each set (size), the normalized enrichment score (NES), and test of statistical significance (false discovery rate [FDR] q value) are highlighted. (C) Enrichment plots of 3 representative transcriptional signatures of MYC-upregulated target genes in the positive phenotype of sensitive pMM cells exposed to MIR17PTi (2.5 µM) for 6 days. (D) Venn diagram–intersecting genes upregulated by MIR17PTi with genes predicted to be miR-17-92 targets (by miRcode) and genes validated as miR-17-92 interactors in CLIP sequencing experiments (starBase v2.0). (E) Representation of MYC/miR-17-92 FFLs. In red are reported genes with a fold change (FC) increase >1.5. (F) qRT-PCR analysis of pri-mir-17-92 and BIM, BZW2, DUSP2, NAP1L1, or VDAC1 messenger RNAs (mRNAs) in pMM cells (n = 3) exposed to MIR17PTi (2.5 µM) for 6 days. The results shown are average pri-mir-17-92 or mRNA expression levels after normalization with glyceraldehyde-3-phosphate dehydrogenase and ΔΔCt calculations. *P < .05.
To address the transcriptional changes directly produced in pMM cells after miR-17-92 downregulation, we searched for miR-17-92 targets among the genes upregulated by MIR17PTi. At this aim, GEP data from MIR17PTi-sensitive pMM cells were matched with computationally predicted and CLIP sequencing–validated direct interactors of miR-17-92s (Figure 4D). This approach disclosed several upregulated genes (n = 65), which are bona fide targets of miR-17-92s in pMM cells (Figure 4D; supplemental Table 1), and consistently with a direct regulation by miR-17-92s, transduction of AMO1 or U266 with an miR-17-92 lentiviral vector produced the downregulation of some of these genes (supplemental Figure 11A-D). Importantly, approximately half of these miR-17-92 direct targets present MYC binding loci within the promoter region (28 of 65), and some have been already validated as transcriptional targets of MYC (15 of 65), even in B cells (13 of 6518-22 ; Figure 4E; supplemental Table 1). GEP data were validated in pMM cells (patients 4, 5, and 6) by qRT-PCR analysis of BIM, BZW2, DUSP2, NAP1L1, and VDAC1 (Figure 4F) and in AMO1 either by qRT-PCR or western blotting (supplemental Figure 11E-F).
These findings suggest that MIR17PTi upregulates a subset of genes that are transcriptionally regulated by MYC, pointing to the existence of an MYC/miR-17-92 coregulated network that might exert homeostatic functions in MM cells and led us to hypothesize that MIR17PTi exerts its anti-MM activity via the redirection of an MYC-related transcriptional program toward apoptotic cell death.
MIR17PTi triggers MYC-dependent SL
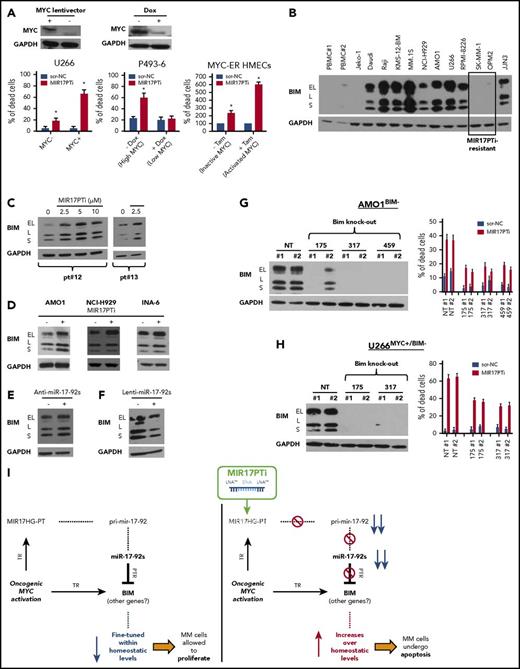
We investigated whether an active MYC transcriptional program sensitizes MM cells to MIR17PTi; to this aim, we transduced MYC-defective U26611 with a lentiviral vector carrying the MYC open reading frame (U266MYC+) or with an empty vector (U266MYC−). Both pri-mir-17-92 and miR-17-92s were increased in U266MYC+ (supplemental Figure 12A), which is consistent with an MYC upstream regulation on MIR17HG in MM cells as well. This assumption was further corroborated by the in silico analysis of a proprietary microarray data set (GSE70323) that showed positive correlation between MYC and MIR17HG in patients with MM (supplemental Figure 12B). More importantly, MIR17PTi was considerably more cytotoxic to U266MYC+ as compared with U266MYC− (Figure 5A), supporting the idea that MIR17PTi triggers an MYC-dependent SL in MM cells.
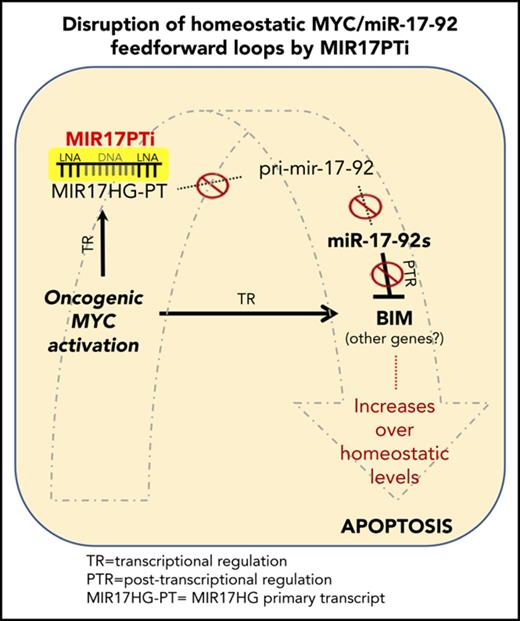
Molecular mechanism underlying MIR17PTi proapoptotic activity. (A) 7-AAD flow cytometric assay of U266MYC−/U266MYC+ after 6 days of treatment with MIR17PTi (2.5 µM) or scr-NC (2.5 µM); western blotting of MYC protein in U266MYC− and U266MYC+ is reported. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as protein loading control (left). 7-AAD flow cytometric assay of P493-6 after 6 days of treatment with MIR17PTi (2.5 µM) or scr-NC (2.5 µM), in presence or absence of doxycycline (dox); western blotting of MYC protein in P493-6 cultured for 2 days with or without doxycycline is reported. GAPDH was used as protein loading control (middle). Trypan blue exclusion staining in MYC-ER human mammary epithelial cells (HMECs) 2 days after transfection with MIR17PTi (50 nM) or scr-NC (50 nM) and cultured with or without tamoxifen (tam) (right). (B) Western blotting of BIM in lysates from healthy donor peripheral blood mononuclear cells (PBMCs; n = 2), Jeko-1 (mantle cell lymphoma; BIM null), Daudi, Raji (Burkitt lymphoma), and indicated HMCLs. GAPDH was used as protein loading control. (C) Western blot analysis of BIM in lysates from pMM cells (patients 12 and 13; extramedullary MM) exposed to MIR17PTi for 6 days at indicated concentrations. GAPDH was used as protein loading control. (D) Western blotting of BIM in AMO1, NCI-H929, or INA-6 exposed for 6 days to MIR17PTi (AMO1, 1 µM; NCI-H929 and INA-6, 2.5 µM) or equimolar scr-NC. GAPDH was used as protein loading control. (E) Western blot analysis of BIM in lysates from AMO1 transfected with miR-NC inhibitor (150 nM) or pooled miR-17-92 inhibitors (25 nM each; 2-day time point). GAPDH was used as protein loading control. (F) Western blot analysis of BIM in lysates from AMO1 transduced with an empty lentiviral vector or an miR-17-92 lentiviral vector. GAPDH was used as protein loading control. (G) Western blot analysis of BIM (upper panel) and flow cytometric analysis of 7-AAD–stained cells (lower panel) in CRISPR/CAS9 genome-edited AMO1BIM− cells. GAPDH was used as protein loading control. Flow cytometry was performed after 6 days of exposure to MIR17PTi (1 µM) or scr-NC (1 µM). (H) Western blot analysis of BIM (upper panel) and flow cytometric analysis of 7-AAD–stained cells (lower panel) in CRISPR/CAS9 genome-edited U266MYC+/BIM− cells. GAPDH was used as protein loading control. Flow cytometry was performed after 6 days of exposure to MIR17PTi (2.5 µM) or scr-NC (2.5 µM). (I) Proposed model of MYC/miR-17-92 FFLs and MIR17PTi mechanism of action in MM cells. Data from 1 of 3 independent experiments are shown in each panel. *P < .05. NT, not targeting; PTR, posttranscriptional regulation; TR, transcriptional regulation.
Molecular mechanism underlying MIR17PTi proapoptotic activity. (A) 7-AAD flow cytometric assay of U266MYC−/U266MYC+ after 6 days of treatment with MIR17PTi (2.5 µM) or scr-NC (2.5 µM); western blotting of MYC protein in U266MYC− and U266MYC+ is reported. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as protein loading control (left). 7-AAD flow cytometric assay of P493-6 after 6 days of treatment with MIR17PTi (2.5 µM) or scr-NC (2.5 µM), in presence or absence of doxycycline (dox); western blotting of MYC protein in P493-6 cultured for 2 days with or without doxycycline is reported. GAPDH was used as protein loading control (middle). Trypan blue exclusion staining in MYC-ER human mammary epithelial cells (HMECs) 2 days after transfection with MIR17PTi (50 nM) or scr-NC (50 nM) and cultured with or without tamoxifen (tam) (right). (B) Western blotting of BIM in lysates from healthy donor peripheral blood mononuclear cells (PBMCs; n = 2), Jeko-1 (mantle cell lymphoma; BIM null), Daudi, Raji (Burkitt lymphoma), and indicated HMCLs. GAPDH was used as protein loading control. (C) Western blot analysis of BIM in lysates from pMM cells (patients 12 and 13; extramedullary MM) exposed to MIR17PTi for 6 days at indicated concentrations. GAPDH was used as protein loading control. (D) Western blotting of BIM in AMO1, NCI-H929, or INA-6 exposed for 6 days to MIR17PTi (AMO1, 1 µM; NCI-H929 and INA-6, 2.5 µM) or equimolar scr-NC. GAPDH was used as protein loading control. (E) Western blot analysis of BIM in lysates from AMO1 transfected with miR-NC inhibitor (150 nM) or pooled miR-17-92 inhibitors (25 nM each; 2-day time point). GAPDH was used as protein loading control. (F) Western blot analysis of BIM in lysates from AMO1 transduced with an empty lentiviral vector or an miR-17-92 lentiviral vector. GAPDH was used as protein loading control. (G) Western blot analysis of BIM (upper panel) and flow cytometric analysis of 7-AAD–stained cells (lower panel) in CRISPR/CAS9 genome-edited AMO1BIM− cells. GAPDH was used as protein loading control. Flow cytometry was performed after 6 days of exposure to MIR17PTi (1 µM) or scr-NC (1 µM). (H) Western blot analysis of BIM (upper panel) and flow cytometric analysis of 7-AAD–stained cells (lower panel) in CRISPR/CAS9 genome-edited U266MYC+/BIM− cells. GAPDH was used as protein loading control. Flow cytometry was performed after 6 days of exposure to MIR17PTi (2.5 µM) or scr-NC (2.5 µM). (I) Proposed model of MYC/miR-17-92 FFLs and MIR17PTi mechanism of action in MM cells. Data from 1 of 3 independent experiments are shown in each panel. *P < .05. NT, not targeting; PTR, posttranscriptional regulation; TR, transcriptional regulation.
We next explored MIR17PTi activity in P493-6 and MYC-ER HMECs, where MYC expression can be conditionally modulated. Specifically, P493-6 is an immortalized B-cell line carrying a tetracycline-regulated MYC,23 and MYC-ER is an immortalized HMEC line engineered with an inducible MYC and estrogen receptor fusion protein (MYC-ER).24 These cells represent valuable experimental models of BCLs and triple-negative breast cancers, respectively, which are prototypical MYC-driven diseases5,6 where miR-17-92 is commonly upregulated.25,26 Both pri-mir-17-92 and miR-17-92s increased in P493-6 and MYC-ER HMECs when MYC expression or activity, respectively, was turned on (supplemental Figure 12A). Again, treatment with MIR17PTi effectively induced cell death only at MYC-on cells, without any significant cytotoxicity when MYC was silenced or inactive (Figure 5A).
These data provide the formal proof that treatment with MIR17PTi is a new MYC-dependent SL approach in MM and other cancers.
Impairment of a BIM-centered MYC/miR-17-92 FFL mediates MIR17PTi anti-MM activity
To shed light on the molecular mechanisms underlying MYC-dependent SL by MIR17PTi, we next focused on MYC/miR-17-92 coregulated targets in MM cells, and of these, the tumor suppressor and proapoptotic factor BIM, a BH3-only BCL2 family member, seemed of particular interest. In fact, MYC positively regulates BIM transcription27 while different miR-17-92s repress its expression at the posttranscriptional level,1-3 thus forming a prototypical miRNA-mediated FFL.28 Moreover, BIM is the primary effector of the MYC-induced apoptotic program in MYC-overexpressing cells,27 with haploinsufficient tumor suppressor activity in B cells.18,27 Interestingly, OPM-2 and SK-MM-1, which were the only 2 MIR17PTi-resistant HMCLs, did not express detectable levels of BIM protein (Figure 5B) or mRNA (data not shown), likely because of epigenetic repression of its transcription (in fact, neither OPM-2 nor SK-MM-1 bring about deletion of BCL2L11/BIM; data not shown). In healthy donor peripheral blood mononuclear cells, which express very low levels of BIM protein as compared with HMCLs (Figure 5B; supplemental Figure 13), MIR17PTi did not increase BIM expression (supplemental Figure 13). In contrast, MIR17PTi strongly upregulated BIM protein in pMM cells (Figure 5C) and different HMCLs (Figure 5D); upregulation of BIM was also produced by cotransfection of pooled miR-17-92 inhibitors (Figure 5E), whereas its protein expression was significantly reduced in AMO1 transduced with an miR-17-92 lentiral vector (Figure 5F). More importantly, CRISPR/Cas9-mediated knockout of BIM strongly antagonized MIR17PTi-mediated killing in both AMO1 (AMO1BIM−)- and U266MYC+/BIM−-generated clones (Figure 5G-H).
These results demonstrate that MIR17PTi triggers MYC-dependent SL in MM cells, largely via the impairment of a BIM-centered MYC/miR-17-92 FFL, which results in the upregulation of BIM up to proapoptotic level (Figure 5I). Of course, upregulation of other miR-17-92 targets likely contributes to its activity; indeed, MIR17PTi also increased the expression of relevant tumor suppressors such as CDKN1A (also known as p21), TP53, E2F1, and PTEN (supplemental Figure 13), which are validated targets of different miR-17-92s.2
In vivo anti-MM activity of MIR17PTi in NOD SCID mice
We investigated the in vivo activity of MIR17PTi in NOD SCID mice bearing subcutaneous MM xenografts (NCI-H929, AMO1luc+, and bortezomib-refractory ABZBluc+). Importantly, systemic administration (IV) of naked MIR17PTi significantly inhibited tumor growth (Figure 6A; supporting BLI images are reported in supplemental Figure 14A-B) and prolonged animal survival (Figure 6B), with a well-defined therapeutic window (hematoxylin and eosin staining of accumulation in organs is reported in supplemental Figure 15A). qRT-PCR analysis of pri-mir-17-92 confirmed productive antisense activity of MIR17PTi in retrieved tumors (Figure 6C), which also resulted in BIM upregulation at both mRNA and protein levels (Figure 6D). By in situ hybridization analysis, using a specific LNA probe for MIR17PTi, we showed a higher uptake in the kidney and liver, followed by BM, heart, and lung (supplemental Figure 15B), which is consistent with canonical LNA-modified ASO pharmacokinetics (PKs).29 By this approach, MIR17PTi was also detected in retrieved tumors (supplemental Figure 15B). Next, we investigated the anti-MM activity of MIR17PTi in the clinically relevant SCID-hu model,30 which allows the in vivo growth of BM-dependent MM cells within a recapitulated human BM milieu; in this model, MIR17PTi elicited strong anti-MM activity, as assessed by measurement of soluble interleukin-6 receptor levels in the plasma of treated vs untreated mice (Figure 6E).
In vivo anti-MM activity of MIR17PTi. (A) In vivo growth inhibition of NCI-H929, AMO1luc+, or ABZBluc+ subcutaneous xenografts by MIR17PTi (2 mg/kg). Treatments were performed on days 1, 4, 8, 15, and 22 (NCI-H929) or 1, 4, 8, and 11 (AMO1luc+ and ABZBluc+). Mice were also treated with bortezomib (BZB) as positive control (1 mg/kg on days 1, 4, 8, and 11). Numbers of xenografted mice: NCI-H929, n = 17 (no treatment [NT], n = 7; MIR17PTi, n =10); AMO1luc+, n = 13 (NT, n = 5; MIR17PTi, n = 5; BZB, n = 3); ABZBluc+, n = 12 (NT, n = 4; MIR17PTi, n = 5; BZB, n = 3). Treatments were performed systemically (IV). Tumor volumes at day 33 (NCI-H929) or day 12 (AMO1luc+ and ABZBluc+) after the first treatment are reported. Supporting images and BLI measurements are shown in supplemental Figure 14. (B) Survival curves (Kaplan-Meier) relative to mice reported in panel A (log-rank test P < .05). Survival was evaluated from the first day of treatment until death or euthanasia. Percentage of mice alive is shown. (C) qRT-PCR analysis of pri-mir-17-92 in retrieved NCI-H929 (n = 3) or AMO1luc+ (n = 1) subcutaneous xenografts 2 days after the last dose of the treatment period with MIR17PTi (on days 1, 4, 8, and 11), as compared with tumor retrieved from untreated animals (NCI-H929, n = 1; AMOluc+, n = 1). Animals used for this analysis were not considered for tumor growth and survival evaluations. The results shown are average pri-mir-17-92 expression levels after normalization with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and ΔΔCt calculations. (D) qRT-PCR analysis (left) and western blotting (right) of BIM in retrieved tumors (AMO1) from MIR17PTi-treated (n = 1) or -untreated (n = 1) mice. The qRT-PCR results shown are average BIM expression levels after normalization with GAPDH and ΔΔCt calculations. GAPDH was used as protein loading control. (E) Schematic representation of SCID-hu model (left) and INA-6 growth within human fetal bone chips subcutaneously implanted in NOD SCID mice (right). Plasmatic levels of soluble interleukin-6 receptor (IL-6R) were used to evaluate tumor growth. Systemic treatments with MIR17PTi (2 mg/kg; days 1, 4, 8, 15, and 22) started at day 22. Effects of MIR17PTi were evaluated at the end of treatment (day 44). A total of 8 mice were treated as follows: NT (n = 4), MIR17PTi (n = 4). *P < .05. BMM, BM microenvironment.
In vivo anti-MM activity of MIR17PTi. (A) In vivo growth inhibition of NCI-H929, AMO1luc+, or ABZBluc+ subcutaneous xenografts by MIR17PTi (2 mg/kg). Treatments were performed on days 1, 4, 8, 15, and 22 (NCI-H929) or 1, 4, 8, and 11 (AMO1luc+ and ABZBluc+). Mice were also treated with bortezomib (BZB) as positive control (1 mg/kg on days 1, 4, 8, and 11). Numbers of xenografted mice: NCI-H929, n = 17 (no treatment [NT], n = 7; MIR17PTi, n =10); AMO1luc+, n = 13 (NT, n = 5; MIR17PTi, n = 5; BZB, n = 3); ABZBluc+, n = 12 (NT, n = 4; MIR17PTi, n = 5; BZB, n = 3). Treatments were performed systemically (IV). Tumor volumes at day 33 (NCI-H929) or day 12 (AMO1luc+ and ABZBluc+) after the first treatment are reported. Supporting images and BLI measurements are shown in supplemental Figure 14. (B) Survival curves (Kaplan-Meier) relative to mice reported in panel A (log-rank test P < .05). Survival was evaluated from the first day of treatment until death or euthanasia. Percentage of mice alive is shown. (C) qRT-PCR analysis of pri-mir-17-92 in retrieved NCI-H929 (n = 3) or AMO1luc+ (n = 1) subcutaneous xenografts 2 days after the last dose of the treatment period with MIR17PTi (on days 1, 4, 8, and 11), as compared with tumor retrieved from untreated animals (NCI-H929, n = 1; AMOluc+, n = 1). Animals used for this analysis were not considered for tumor growth and survival evaluations. The results shown are average pri-mir-17-92 expression levels after normalization with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and ΔΔCt calculations. (D) qRT-PCR analysis (left) and western blotting (right) of BIM in retrieved tumors (AMO1) from MIR17PTi-treated (n = 1) or -untreated (n = 1) mice. The qRT-PCR results shown are average BIM expression levels after normalization with GAPDH and ΔΔCt calculations. GAPDH was used as protein loading control. (E) Schematic representation of SCID-hu model (left) and INA-6 growth within human fetal bone chips subcutaneously implanted in NOD SCID mice (right). Plasmatic levels of soluble interleukin-6 receptor (IL-6R) were used to evaluate tumor growth. Systemic treatments with MIR17PTi (2 mg/kg; days 1, 4, 8, 15, and 22) started at day 22. Effects of MIR17PTi were evaluated at the end of treatment (day 44). A total of 8 mice were treated as follows: NT (n = 4), MIR17PTi (n = 4). *P < .05. BMM, BM microenvironment.
These data demonstrate relevant in vivo anti-MM activity of MIR17PTi as a single agent, capable of overcoming the protective effects of the BM milieu.
Safety and PK profiles of MIR17PTi in nonhuman primates
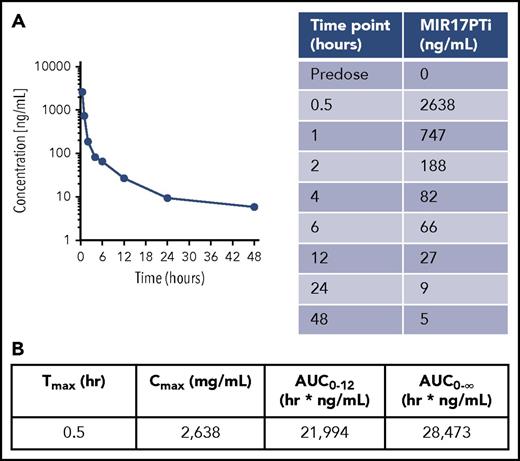
We investigated the safety and plasmatic PKs of MIR17PTi in nonhuman primates (nonnaïve cynomolgus monkeys). The amount per injection was equivalent to the amount administered to NOD SCID mice, after interspecies dose conversion.31 To assess the safety profile, MIR17PTi was injected on days 1, 4, 8, 15, and 22 (same schedule used in mice), and as a control, an additional animal was injected IV with saline solution. At the end of the treatment period, no signs of toxicity were recorded (supplemental Figures 16 and 17). MIR17PTi was then injected IV as a single dose in a third animal to explore the plasmatic PK profile. Plasma was collected at the following time points: predose and at 0.5, 1, 2, 4, 6, 12, 24,and 48 hours. Plasmatic MIR17PTi, as detected by PNA-HPCL assay, peaked at a Tmax of 0.5 hour, with a Cmax of 2638 ng/mL (Figure 7A). Compared with the area under the concentration-time curve value from 0 hours to infinity of 28 473 hr × ng/mL, the area under the concentration-time curve value from 0 to 12 hours of 21 994 hr × ng/mL indicated that >75% of MIR17PTi had left the plasma within 12 hours (Figure 7B). These findings were consistent with the favorable PK profile of LNA-modified ASOs, which assumes rapid plasmatic clearance via tissue uptake.12,29
PK profile of MIR17PTi in Cynomolgus monkeys. (A) Plasmatic PK profile of MIR17PTi after injection of a Cynomolgus monkey with a single dose (0.504 mg/kg) (left). Sampling was performed at the time points indicated in the table (right). (B) Plasmatic PK parameters of MIR17PTi in Cynomolgus monkeys. AUC, area under the concentration-time curve.
PK profile of MIR17PTi in Cynomolgus monkeys. (A) Plasmatic PK profile of MIR17PTi after injection of a Cynomolgus monkey with a single dose (0.504 mg/kg) (left). Sampling was performed at the time points indicated in the table (right). (B) Plasmatic PK parameters of MIR17PTi in Cynomolgus monkeys. AUC, area under the concentration-time curve.
Discussion
We developed and here describe a new strategy for the therapeutic targeting of miRNA network in human cancer. This approach exploits RNase H–activating LNA gapmeRs to induce the degradation of pri-miRNA and consequently prevent the biogenesis of related mature miRNAs. Recent evidence has demonstrated that these third-generation ASOs predominantly act on nascent primary transcripts (nascent pri-miRNAs in this study) before being processed by the splicing machinery.32,33 As compared with canonical miRNA inhibitors, LNA gapmeRs better adapt to the targeting of miRNA clusters; in this case, indeed, degradation of pri-miRNAs leads to the concomitant downregulation of all miRNA members of the cluster. On these bases, we identified the miR-17-92 cluster as a target with high translational value in the context of human cancers,3 and from among different originally designed LNA gapmeRs, we selected MIR17PTi for its antisense activity. Consistent with our initial hypothesis, MIR17PTi efficiently downregulated pri-mir-17-92 and all miR-17-92s in an RNase H–dependent manner; by contrast, we failed to induce these effects via RNAi, possibly because of the absence or alternative functions of RNAi machinery within the cell nucleus.34,35 Our study spanned from conceptual drug design to preclinical investigation in nonhuman primates.
Previous reports have suggested that the oncogenicity of miR-17-92 may be confined to 1 or few miRNAs in specific cellular contexts,36 thus supporting the rational design of miR-17-92–based therapeutic strategies. Our data indeed confirm that CCLs may be differently addicted to miR-17-92 members; however, they instead support the potential advantage of concomitant downregulation of the entire cluster at least in hematologic cancers. In fact, MIR17PTi antagonized the proliferation of hematologic CCLs more efficiently than synthetic inhibitors of miR-17-92s. Moreover, hematologic CCLs from the same malignancy such as mantle cell lymphoma or MM were differently sensitive to the inhibition of each miR-17-92, making very likely the failure of an miRNA-based therapeutic approach in the context of the intra-/interclonal heterogeneity that features human cancer.37,38 Actually, our results can be explained by taking into account the fact that miR-17-92s have redundant functions because of the sharing of most of the mRNA targets; it is reasonable that, upon individual antagonism of each cluster member, the remaining miR-17-92s could act by compensatory mechanisms. Moreover, it must be noted that MIR17PTi also induces the downregulation of lncRNA MIR17HG-201/-202/-203, pre-mir-17-92 (pre-mir-17/-18a/-19a/-20a/-19b/-92a), and miR-17-92 complementary strands (miR-17-3p/miR-18a-3p/miR-19a-5p/miR-20a-3p/miR-19b-5p/miR-92a-5p) that are potentially endowed with their own functions and can contribute to MIR17PTi anticancer activity. This possibility is also suggested by the incomplete rescue obtained by enforcing the expression of all miR-17-92s. However, this point has not been addressed in our work and requires further investigation.
We used MM, an MYC-driven malignancy of terminally differentiated B cells (ie, plasma cells),39 as the main investigational platform for the preclinical development of MIR17PTi. In a proprietary RNA sequencing data set of 320 patients with newly diagnosed MM, we disclosed the association of high pri-mir-17-92 levels with worse clinical outcome. In vitro, MIR17PTi efficiently killed HMCLs and pMM cells, even cocultured with human BM stromal cells and/or HS-5. At the molecular level, we found that the induction of an MYC-related and proapoptotic transcriptional network underlies the anti-MM activity of MIR17PTi. We explained this outcome by hypothesizing that MIR17PTi impairs a functional interplay between MYC and miR-17-92, which follows the general scheme of homeostatic miRNA-mediated FFLs,28,40 and we formally proved our hypothesis by demonstrating that active MYC downstream signaling, achieved by enforcing MYC expression (U266MYC+ or P493-6DOX−) or activity (MYC-ER HMECTAM+), strongly sensitizes to MIR17PTi killing. We explored the molecular mechanisms underlying this synthetic lethal effect in MM cells by focusing on MYC/miR-17-92 coregulated targets. Specifically, the tumor suppressor BIM gained our attention, given its established role as the primary effector of the MYC apoptotic program.18,27 The finding that MIR17PTi-resistant MM cells (ie, OPM2 and SK-MM-1) did not express detectable BIM protein/mRNA prompted us to investigate MIR17PTi activity in HMCLs after BIM knockout (ie, AMO1BIM− and U266MYC+/BIM−), and in these experimental conditions, the killing activity of MIR17PTI was strongly antagonized. On these bases, we speculate that MYC/miR-17-92 FFLs are required to maintain the expression of BIM, and likely other coregulated genes, at homeostatic levels that allow MM cells to proliferate, and we conclude that MIR17PTi triggers MYC-dependent SL by affecting these homeostatic FFLs, resulting in the upregulation of BIM (and likely other genes) over a homeostatic threshold at levels prompting MM cells toward apoptosis. Overall, this mechanism is likely to occur specifically in cancer cells, where oncogenic MYC controls the transcription of apoptotic genes; it is known, in fact, that this transcriptional activity is not relevant when MYC is expressed within a physiologic range.41
MIR17PTi antagonized in vivo growth of human MM cells as a single agent. This was demonstrated in 4 different and clinically relevant murine models, including those refractory to conventional anti-MM agents (ABZB) and those in which MM cells grow within a recapitulated human BM microenvironment (ie, SCID-hu).30 Moreover, the potential therapeutic benefit of treatment with MIR17PTi was further corroborated by its synergistic anti-MM activity in combination with conventional and clinically active agents, such as dexamethasone, melphalan, or bortezomib. These data indeed provide the rational framework for the design of novel MIR17PTi-based drug combinations for the treatment of patients with MM.
To our knowledge, this is the first report of pri-miRNA therapeutics with translational value for cancer therapy. We believe that this study opens a new avenue for targeting the miRNA network in human cancer and provides in MIR17PTi a powerful weapon against MM and other hard-to-treat MYC-driven tumors.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dirk Eick (Molecular Epigenetics, Helmholtz-Zentrum München), Stephen J. Elledge (Division of Genetics, Harvard Medical School, Brigham and Women’s Hospital), and Christoph Driessen (Cantonal Hospital St. Gallen, St. Gallen, Switzerland) for providing relevant cellular models; Emilia Dora Giovannone and Valter Agosti for help in the generation of genome-edited cell lines (ie, cell sorting and single-cell plating); and Ivana Criniti (Magna Graecia University, Catanzaro, Italy) for editorial assistance.
This work was mainly supported by the Italian Association for Cancer Research (AIRC) through its Special Program for Molecular Clinical Oncology–5 per mille 2010/15 and Extension Program No. 9980 2016/18 (principal investigator P. Tassone) and also by the Innovative Immunotherapeutic Treatments of Human Cancer Multi Unit Regional Program No. 16695 (cofinanced by AIRC and the CARICAL foundation). This work was also partially supported by National Institutes of Health grants P50-100707 (N.C.M.) and P01-155258/07 (N.C.M. and M.F.) and by AIRC grant IG1 6722 (A.N.).
Authorship
Contribution: E.M., L.B., and C.F. conducted most in vitro and in vivo experiments; E.M. and A.G. performed experiments with the SCID-hu animal model; M.T.D.M., L.B., and F.S. performed GEP (E.M. analyzed GEP data); M.E.G.C. performed drug combination studies and analyzed data with CalcuSyn; M.M. conducted bioinformatic analysis of gene and miRNA expression in patients with MM; M.K.S. analyzed RNA sequencing data; M.A.S. performed the methylcellulose assay; M.R.P. performed confocal microscopy analysis; S.S. performed electron microscopy analysis; M.F. established U266MYC+ cells and contributed to interpretation of data; N.M.F. suggested the use of LNA gapmeRs to target the miR-17-92 cluster and contributed to the design and interpretation of key experiments; N.A. and M.R. contributed to the design and interpretation of key experiments; A.N. and N.C.M. provided reagents and contributed to interpretation of data; all authors read and edited the manuscript; and E.M., P. Tagliaferri, and P. Tassone designed research studies and wrote the manuscript.
Conflict-of-interest disclosure: N.M.F. is an employee of Exiqon. The remaining authors declare no competing financial interests.
Correspondence: Pierfrancesco Tassone, Magna Graecia University, Viale Europa, 88100 Catanzaro, Italy; e-mail: tassone@unicz.it.

![Figure 2. In vitro anticancer activity of MIR17PTi. (A) CCK-8 assay of CCLs (n = 48) and nonmalignant cell lines (NM-CLs; n = 5) exposed for 6 days to MIR17PTi or scr-NC (10 µM). Data are represented as percentage of MIR17PTi-treated live cells (absorbance), as compared with scr-NC. Dashed lines indicate 20% (upper line) and 80% (bottom line) growth inhibition. (B) CCK-8 proliferation assay of 8 CCLs (P3HR1 [diffuse large B cell lymphoma (DLBCL)], SULTAN [Burkitt lymphoma (BL)], JeKo-1, Maver [mantle cell lymphoma (MCL)], AMO1, KMS-12-BM, NCI-H929, RPMI-8226 [MM]) transfected with indicated miRNA inhibitors or MIR17PTi (25 nM). Data are represented as compared (percentage) with live cells after scr-NC transfection. (C) CCK-8 proliferation assay of RPMI-8226 and AMO1 after transfection with miR-NC-inhibitors (150 nM) or pooled miR-17-92 inhibitors (25 nM each) or scr-NC (25 nM) or MIR17PTi (25 nM). Data are represented as compared (percentage) with live cells after mock (RNase-free water) transfection. (D) CCK-8 proliferation assay of: AMO1 2 days after cotransfection with scr-NC (5 or 25 nM; showed as single point because no difference was detected in percentage of live cells) or MIR17PTi (5 or 25 nM) and miR-NC-mimics (60 nM) or pooled miR-17-92 mimics (10 nM each) (left) and AMO1 transduced with an empty lentiviral vector or an miR-17-92 lentiviral vector and then exposed for 6 days to scr-NC or MIR17PTi (0.5 µM) (right). Data from 1 of 3 independent experiments are shown in each panel. *P < .05. AML, acute myeloid leukemia; BC, breast cancer; MPM, malignant pleural mesothelioma; PC, pancreatic cancer; TCL, T-cell leukemia.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/132/10/10.1182_blood-2018-03-836601/4/m_blood836601f2.jpeg?Expires=1767713208&Signature=a4dwHrzYt2~j0g2XLZ1zfgoyssTjacaiZM~1IKcTJEst-q6-1AB454Th2BKbJmvOiAfaDAxOzfqNz9DKV0cqmYhEGTzsNtJf5JFME-W793A~Q~INHyJUFeHfe1~1ypnd7Df0ERuYrn1A~LT3DJ6LsPO6jkIFu7xuufrfdRElbqmdV1g0J6RX803SVYYqm67Uekl9GX~9iAeeq0CR92t0tTeeQTWiHQ6pa4vquLO45RsNLH0X0GVH47cW5eZK5fw5~Fv8ou3bGG442n-bT~i~AgZQcJImMxG-~mh3bkMeWl1klmerLxuYzgu8vPECyAmTotLA-MqxoPyfkvX4Wq5NBw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 3. In vitro activity of MIR17PTi in HMCLs and primary MM (pMM) cells. (A) Dose-dependent activity of MIR17PTi (6-day time point) in HMCLs (n = 10) as assessed by CCK-8 proliferation assay. Live cells are represented as compared (percentage) with untreated controls. (B) Time-dependent activity of MIR17PTi (1 µM) in AMO1, as assessed by qRT-PCR analysis of pri-mir-17-92 (left) and miR-17-92s (middle). The results shown are average pri-mir-17-92 or miRNA expression levels after normalization with glyceraldehyde-3-phosphate dehydrogenase or RNU44 and ΔΔCt calculation (expressed as percentage). Time-dependent activity of MIR17PTi (1 µM) in AMO1, as assessed by CCK-8 assay (right pane). (C) Cell-cycle analysis of AMO1 exposed for 6 days to MIR17PTi (1 µM) or sc-NC (1 µM). (D) Flow cytometric analysis of senescent AMO1 cells (β-galactosidase positive) exposed for 6 days to MIR17PTi (1 µM) o scr-NC (1 µM). (E) Methylcellulose clonogenic assay of AMO1 exposed to MIR17PTi (1 µM) o scr-NC (1 µM; 2-week time point). (F) CCK-8 proliferation assay of HMCLs sensitive (MM.1S, U266, AMO1) or resistant (MM.1R, LR7, ABZB, ACFZ) to conventional anti-MM agents (dexamethasone, melphalan, bortezomib, carfilzomib) exposed to different concentrations of MIR17PTi for 6 days. (G) CCK-8 assay of human bone marrow (BM) stromal cells (hBMSCs), AMO1, or AMO1 cocultured with hBMSCs after 6 days of exposure to indicated concentrations of MIR17PTi. (H) Table showing combination indexes resulting from combinatorial treatments of AMO1 with MIR17PTi and dexamethasone or melphalan or bortezomib (6-day time point). (I) Flow cytometric analysis of 7-AAD–stained pMM cells (n = 13) exposed for 8 days to MIR17PTi or scr-NC (2.5 µM). pMM cells were cultured adherent to GFP+ HS-5 stromal cells (patients 1-3; patients with intramedullary disease) or physically separated from HS-5 by membranes (patients 4-11; patients with intramedullary disease) or alone (patients 12-13; patients with extramedullary disease at relapse [secondary plasma-cell leukemia (sPCL)]). Lower panel (within the frame) shows median activity of MIR17PTi in sensitive pMM cells (n = 10) from the left panel. *P < .05.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/132/10/10.1182_blood-2018-03-836601/4/m_blood836601f3.jpeg?Expires=1767713208&Signature=3QDdeQWdAm3-ati7VsoNSD8nrDuzJoK3GCNPytHARSPBjxAN-4B6t7p0I9J2nnEMpsrmpuhjGNeo2TZqsZsrO6JVHM9F9PfhUVRgcsh~9~aL5b1z5TN7a-25WpmPUMMx1wYi0CXN1-ubPl9EmbLLFFJkXrnnKGtRHvX3FqR12eiSIgy5dKltIY6xJ98y3BTAo3qEcOYLmpF9NUnNzbr~-feHB2Fbp9RakEkc~uxDJ23BzmMQcazdYRFS1G8UmjPbjdKfHuOU2qmDFWHZL0sgDYBawrjWv~Lr3GxUPv64T-6xqBL-N9SF3umRzKE5lnKCprWQjoZz0rgSTZ5i7bZPEA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 4. Molecular perturbation induced by MIR17PTi in pMM cells. (A) Hierarchical clustering of pMM cells (n = 4; patients 5, 6, 7, and 9) exposed for 6 days to MIR17PTi (2.5 µM) or equimolar scr-NC. (B) Table of gene sets from the Hallmark collection enriched with genes upregulated by MIR17PTi (positive phenotype) in sensitive pMM cells (patients 5, 6, 7, and 9). Number of genes in each set (size), the normalized enrichment score (NES), and test of statistical significance (false discovery rate [FDR] q value) are highlighted. (C) Enrichment plots of 3 representative transcriptional signatures of MYC-upregulated target genes in the positive phenotype of sensitive pMM cells exposed to MIR17PTi (2.5 µM) for 6 days. (D) Venn diagram–intersecting genes upregulated by MIR17PTi with genes predicted to be miR-17-92 targets (by miRcode) and genes validated as miR-17-92 interactors in CLIP sequencing experiments (starBase v2.0). (E) Representation of MYC/miR-17-92 FFLs. In red are reported genes with a fold change (FC) increase >1.5. (F) qRT-PCR analysis of pri-mir-17-92 and BIM, BZW2, DUSP2, NAP1L1, or VDAC1 messenger RNAs (mRNAs) in pMM cells (n = 3) exposed to MIR17PTi (2.5 µM) for 6 days. The results shown are average pri-mir-17-92 or mRNA expression levels after normalization with glyceraldehyde-3-phosphate dehydrogenase and ΔΔCt calculations. *P < .05.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/132/10/10.1182_blood-2018-03-836601/4/m_blood836601f4.jpeg?Expires=1767713208&Signature=aLIaFBWnwJ6TR8cOFQ~ZU0C0dnUkoAftqNQBPJkFYl1A41p9RpVz8moQ8TK1Ul1~DeVeXkSTX2-N0GvR-PxXoiG4evqJwTNV6~xvQovFTL-GHELsI80JnMfqf8a4KnPI-UzTtaoPt4sVNsDYXBpd9BV6dR7c1l~0OPU7mTMS45tr~pzqi0f6ur2clG0g76Uxq743wwocOeMAAeE~YUNq70NwB4zAlsREIgDi6OiOqcikKkNedLm6PJjYwdrgofaU4-tOFPNFZk2jjKU9rLug8idYh-5F~pHfWnwZxsDUI-w273FOEz13XgXsw-BwX~Ju33C9nV-bdgdNwMltFvW4rg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 6. In vivo anti-MM activity of MIR17PTi. (A) In vivo growth inhibition of NCI-H929, AMO1luc+, or ABZBluc+ subcutaneous xenografts by MIR17PTi (2 mg/kg). Treatments were performed on days 1, 4, 8, 15, and 22 (NCI-H929) or 1, 4, 8, and 11 (AMO1luc+ and ABZBluc+). Mice were also treated with bortezomib (BZB) as positive control (1 mg/kg on days 1, 4, 8, and 11). Numbers of xenografted mice: NCI-H929, n = 17 (no treatment [NT], n = 7; MIR17PTi, n =10); AMO1luc+, n = 13 (NT, n = 5; MIR17PTi, n = 5; BZB, n = 3); ABZBluc+, n = 12 (NT, n = 4; MIR17PTi, n = 5; BZB, n = 3). Treatments were performed systemically (IV). Tumor volumes at day 33 (NCI-H929) or day 12 (AMO1luc+ and ABZBluc+) after the first treatment are reported. Supporting images and BLI measurements are shown in supplemental Figure 14. (B) Survival curves (Kaplan-Meier) relative to mice reported in panel A (log-rank test P < .05). Survival was evaluated from the first day of treatment until death or euthanasia. Percentage of mice alive is shown. (C) qRT-PCR analysis of pri-mir-17-92 in retrieved NCI-H929 (n = 3) or AMO1luc+ (n = 1) subcutaneous xenografts 2 days after the last dose of the treatment period with MIR17PTi (on days 1, 4, 8, and 11), as compared with tumor retrieved from untreated animals (NCI-H929, n = 1; AMOluc+, n = 1). Animals used for this analysis were not considered for tumor growth and survival evaluations. The results shown are average pri-mir-17-92 expression levels after normalization with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and ΔΔCt calculations. (D) qRT-PCR analysis (left) and western blotting (right) of BIM in retrieved tumors (AMO1) from MIR17PTi-treated (n = 1) or -untreated (n = 1) mice. The qRT-PCR results shown are average BIM expression levels after normalization with GAPDH and ΔΔCt calculations. GAPDH was used as protein loading control. (E) Schematic representation of SCID-hu model (left) and INA-6 growth within human fetal bone chips subcutaneously implanted in NOD SCID mice (right). Plasmatic levels of soluble interleukin-6 receptor (IL-6R) were used to evaluate tumor growth. Systemic treatments with MIR17PTi (2 mg/kg; days 1, 4, 8, 15, and 22) started at day 22. Effects of MIR17PTi were evaluated at the end of treatment (day 44). A total of 8 mice were treated as follows: NT (n = 4), MIR17PTi (n = 4). *P < .05. BMM, BM microenvironment.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/132/10/10.1182_blood-2018-03-836601/4/m_blood836601f6.jpeg?Expires=1767713208&Signature=d4Ubybpe4cZxABZvXbinGbIq4H-ZRelohMRMsPcCVK7VudodVt8brck0wMlpvn--~TJuNgNS0YlxdFdSTKwhCpwfcJVuYNmoWZdo0bMf0lVw7uTzftREDAVsYyi4-ziTCgyDosqBblgkndfG4erahBBZkIdHJorgXRLNrvzono~zK-krMEsJzDpxlfZWbMYjh7qEcbxBIObP6jql8~AUAka1507bMaaCkA045oXRVtYgp0qWMqWyOj4fxWibdwZ6pqOqkVgp2YNq0P7nsacGuFY2O2AxCM7dKoUuZZ3c70nOIUCunUiDqzgzyXRIhL0T4TNJlCTKnP-RLiCfTSESDA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal