Key Points
Analysis of mutations causing nonclassical FD defined the hepcidin-binding site in the central cavity of Fpn.
Hepcidin inhibits iron export through Fpn not only by causing Fpn endocytosis, but also by occluding the transporter.
Abstract
Nonclassical ferroportin disease (FD) is a form of hereditary hemochromatosis caused by mutations in the iron transporter ferroportin (Fpn), resulting in parenchymal iron overload. Fpn is regulated by the hormone hepcidin, which induces Fpn endocytosis and cellular iron retention. We characterized 11 clinically relevant and 5 nonclinical Fpn mutations using stably transfected, inducible isogenic cell lines. All clinical mutants were functionally resistant to hepcidin as a consequence of either impaired hepcidin binding or impaired hepcidin-dependent ubiquitination despite intact hepcidin binding. Mapping the residues onto 2 computational models of the human Fpn structure indicated that (1) mutations that caused ubiquitination-resistance were positioned at helix-helix interfaces, likely preventing the hepcidin-induced conformational change, (2) hepcidin binding occurred within the central cavity of Fpn, (3) hepcidin interacted with up to 4 helices, and (4) hepcidin binding should occlude Fpn and interfere with iron export independently of endocytosis. We experimentally confirmed hepcidin-mediated occlusion of Fpn in the absence of endocytosis in multiple cellular systems: HEK293 cells expressing an endocytosis-defective Fpn mutant (K8R), Xenopus oocytes expressing wild-type or K8R Fpn, and mature human red blood cells. We conclude that nonclassical FD is caused by Fpn mutations that decrease hepcidin binding or hinder conformational changes required for ubiquitination and endocytosis of Fpn. The newly documented ability of hepcidin and its agonists to occlude iron transport may facilitate the development of broadly effective treatments for hereditary iron overload disorders.
Introduction
Iron homeostasis is maintained by the hepcidin-ferroportin (Fpn) axis, which controls intestinal absorption of iron, as well as internal iron recycling and systemic distribution. Fpn is the only known cellular iron exporter in vertebrates and is the conduit through which iron is delivered into plasma (reviewed in Drakesmith et al1 ). The main tissues expressing Fpn include duodenal enterocytes absorbing dietary iron, hepatic and splenic iron-recycling macrophages, and hepatocytes, which export stored iron when demand is high. Hepcidin is a systemically acting iron-regulatory peptide hormone and the only known natural Fpn ligand. Hepcidin binding to Fpn causes the ubiquitination, endocytosis, and degradation of the ligand-receptor complex, thereby decreasing iron supply to plasma.2-4 Decreased Fpn activity, usually caused by high hepcidin levels, is manifested as an iron-restriction syndrome. The condition is exemplified by iron-refractory iron deficiency anemia (IRIDA) or anemia of inflammation,5,6 in which iron accumulates in recycling macrophages and enterocytes, but may become insufficient in other tissues. The other extreme of the spectrum of iron disorders is exemplified by hereditary hemochromatosis or β-thalassemia intermedia.7,8 In these disorders, Fpn is hyperactive, usually as a result of hepcidin deficiency, and leads to excessive iron absorption and toxic iron deposition in hepatocytes and other parenchymal cells, but relative iron depletion of macrophages. Understanding the mechanism of hepcidin-ferroportin interaction would allow improved targeting of this regulatory axis and development of novel treatments for iron disorders.
To study structural determinants of hepcidin-ferroportin interaction, we focused on Fpn mutations that cause ferroportin disease (FD), a rare, autosomal-dominant iron disorder. Specifically, we focused on nonclassical FD, which is characterized by hyperferritinemia with high transferrin saturation and iron overload primarily in hepatocytes, thought to result from Fpn gain-of-function mutations.9,10 In contrast, classical FD is characterized by hyperferritinemia with normal-to-low transferrin saturation that arises from Fpn mutations that cause decreased iron export (loss-of-function), and iron accumulates primarily in macrophages, which normally export very high amounts of iron after erythrophagocytosis.
We examined the structural and functional characteristics of clinical gain-of-function Fpn mutants associated with both hyperferritinemia and transferrin saturation >60% (Table 111-24 ). Additional nonclinical mutants were generated to probe the hepcidin-binding region in greater detail.
Nonclassical FD mutations
| Mutant . | Serum ferritin, µg/L . | Transferrin saturation, % . | Examples of pathological findings . | References . | Adult patients, n . |
|---|---|---|---|---|---|
| WT | 15-200 F | 15-45 both | 11 | ||
| 40-300 M | M and F | ||||
| C326Y* | 481-552F | 94-80 F | 11 | 2 F, 2 M | |
| 4232-4326 M | 96-100 M | ||||
| C326S* | Elevated | Elevated | Cirrhosis and arthritis | 12 | 1 M† |
| Y501C* | 43-199 F | 35-64 F | Asthenia, arthralgia, and hepatomegaly | 13 | 2 F, 2 M |
| 568-642 M | 94-104 M | ||||
| D504N* | 1514-1605 M | 89-102 M | Hepatomegaly | 14 | 2 M |
| N144D* | 7500 F | 99 M | Iron loading in hepatocytes and BDECs, cirrhosis | 15 | 1 F, 1 M |
| 10 510 M | |||||
| V72F* | 27-174 F | 31-35 F | Iron loading primarily in hepatocytes, but also in Kupffer cells, BDECs, and PTMs, fibrosis, and hepatic steatosis | 16 | 2 F, 3 M |
| 195-1091 M | 53-81 M | ||||
| Y64N‡ | 176-513 F | 47-86 F | Iron loading in Kupffer cells and hepatocytes, fatigue and tremors | 17 | 3 F, 5 M |
| 647-2,812 M | 91-98 M | ||||
| H507R‡ | 233 F | 46 F | Iron loading in hepatocytes, mild steatosis, and siderosis | 18, 19 | 1 F, 3 M |
| 785-3751 M | 93-100 M | ||||
| S71F‡ | 5500 | 70 | Uncharacterized | 20 | 1§ |
| G204S‡ | 4-5236 F | 2-100 F | Mixed iron loading, but mainly hepatocytes heterogeneous phenotype | 20, 21 | 3 F, 5 M |
| 508-3755M | 27-91 M | ||||
| D270V‡ | <1000 F | 78 F | Fatigue, fibrosis, iron loading in hepatocytes and Kupffer cells, hepatitis C | 22, 23 | 1 F, 1 M |
| 353-559 M|| | 37-41 M|| | ||||
| S338R‡ | 1990 M | 90 M | Iron loading in hepatocytes, Kupffer cells, and PTMs, mild steatosis | 24 | 1 M |
| Mutant . | Serum ferritin, µg/L . | Transferrin saturation, % . | Examples of pathological findings . | References . | Adult patients, n . |
|---|---|---|---|---|---|
| WT | 15-200 F | 15-45 both | 11 | ||
| 40-300 M | M and F | ||||
| C326Y* | 481-552F | 94-80 F | 11 | 2 F, 2 M | |
| 4232-4326 M | 96-100 M | ||||
| C326S* | Elevated | Elevated | Cirrhosis and arthritis | 12 | 1 M† |
| Y501C* | 43-199 F | 35-64 F | Asthenia, arthralgia, and hepatomegaly | 13 | 2 F, 2 M |
| 568-642 M | 94-104 M | ||||
| D504N* | 1514-1605 M | 89-102 M | Hepatomegaly | 14 | 2 M |
| N144D* | 7500 F | 99 M | Iron loading in hepatocytes and BDECs, cirrhosis | 15 | 1 F, 1 M |
| 10 510 M | |||||
| V72F* | 27-174 F | 31-35 F | Iron loading primarily in hepatocytes, but also in Kupffer cells, BDECs, and PTMs, fibrosis, and hepatic steatosis | 16 | 2 F, 3 M |
| 195-1091 M | 53-81 M | ||||
| Y64N‡ | 176-513 F | 47-86 F | Iron loading in Kupffer cells and hepatocytes, fatigue and tremors | 17 | 3 F, 5 M |
| 647-2,812 M | 91-98 M | ||||
| H507R‡ | 233 F | 46 F | Iron loading in hepatocytes, mild steatosis, and siderosis | 18, 19 | 1 F, 3 M |
| 785-3751 M | 93-100 M | ||||
| S71F‡ | 5500 | 70 | Uncharacterized | 20 | 1§ |
| G204S‡ | 4-5236 F | 2-100 F | Mixed iron loading, but mainly hepatocytes heterogeneous phenotype | 20, 21 | 3 F, 5 M |
| 508-3755M | 27-91 M | ||||
| D270V‡ | <1000 F | 78 F | Fatigue, fibrosis, iron loading in hepatocytes and Kupffer cells, hepatitis C | 22, 23 | 1 F, 1 M |
| 353-559 M|| | 37-41 M|| | ||||
| S338R‡ | 1990 M | 90 M | Iron loading in hepatocytes, Kupffer cells, and PTMs, mild steatosis | 24 | 1 M |
Based on the literature, the 12 Fpn mutations listed in the table are associated with elevated serum ferritin and transferrin saturation, although not every subject carrying the mutation had abnormal biochemical parameters or related pathology. Adult = ≥ 18 years.
BDEC, bile duct epithelial cell; F, female; M, male PTM, portal tract macrophage.
Mutants in which hepcidin binding is impaired based on the results of our study; of these, C326Y, C326S, Y501C, D504N, and N144D were more severely impaired.
Six descendants aged 8 to 16 had transferrin saturation 84-97%.
Mutants in which hepcidin binding is normal, but hepcidin-mediated ubiquitination is impaired based on the results of our study; of these, Y64N and H507R were most severely impaired, followed by S71F and G204S.
Source did not cite sex.
The patient’s parameters were recorded at 2 different times.
Our study builds on 2 important technical advances: improved cellular modeling of Fpn mutants and updated structural Fpn data. To avoid the limitations of transiently transfected cells with large variations of Fpn expression between individual cells and mutants,10,13,14,18,24-29 we established stable, inducible mammalian cell lines expressing wild-type (WT) or mutant Fpn from a defined genomic location, a Flp-In recombination locus. This allowed for a more accurate comparison and quantification of the mutation effects on iron export, Fpn stability, hepcidin binding, and hepcidin-induced Fpn ubiquitination and degradation, steps that were not individually assessed in earlier studies of hepcidin resistance.10,13,14,18,24-29
The lack of information about Fpn structure has also hampered understanding of hepcidin-Fpn interaction.28,30-32 Recently, Taniguchi et al identified the prokaryotic protein, BdFPN, from the bacterium Bdellovibrio bacteriovorus as an ortholog of Fpn and obtained its structure by X-ray crystallography.33 However, hepcidin itself and the hepcidin-binding site within Fpn are found only in vertebrates,34 so in this context, the BdFPN structure can only serve as a modeling constraint. We therefore used the available structural data from related members of the major facilitator superfamily, including eventually BdFPN, and incorporated the experimental results from the Fpn mutant analysis to develop a new model of hepcidin-Fpn interaction, which segregates the Fpn residues important for hepcidin-induced conformational change from those critical for hepcidin binding. As predicted by the model, we also experimentally demonstrated that hepcidin can function by occluding Fpn and preventing iron export even in the absence of endocytosis.
Materials and methods
Site-directed mutagenesis
Human Fpn-GFP in the pGFP-N3 vector27 was transferred into the pcDNA5/FRT/TO vector (Invitrogen). Single mutations were introduced using the QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent Technologies) and confirmed by sequencing. Mutagenesis primers are listed in supplemental Table 1 (available on the Blood Web site).
Stably transfected inducible cell lines
All mutant human Fpn-GFP pcDNA5/FRT/TO vector constructs were verified by sequencing. Using the Flp-In T-Rex system (Invitrogen K5600-01), HEK293T cells were transfected with the pcDNA5/FRT/TO vector encoding WT or mutant Fpn and pOG44 vector. Stable cell lines were established according to the manufacturer’s protocol. Doxycycline (dox) was used to induce expression of Fpn-GFP.
The characterization of these cell lines, including Fpn microscopy, Fpn cell surface localization, iron export as assessed by ferritin assay or radiolabeled iron export, hepcidin binding to Fpn, hepcidin-dependent ubiquitination of Fpn, Fpn stability, and hepcidin-dependent degradation of Fpn by western blotting, is described in the supplemental Methods.
Computational modeling
Structural models of human Fpn (hFpn) were generated using I-TASSER Iterative Threading ASSEmbly Refinement).35,36 This server predicted the protein structure and function of hFpn by threading hFpn onto program-selected crystal structures of other members of the major facilitator superfamily (Protein Data Bank entries: 4IKV, 3WDO, 4AV3, and 4M64), either not including (Figure 2) or including (supplemental Figure 7) the BdFPN crystal structure (Protein Data Bank entry: 5AYM) as a template. The models without and with the BdFPN template were generated respectively on 18 June 2015 and 20 January 2016.
Functional expression of hFpn in Xenopus oocytes
The pOX(+) oocyte expression plasmid vectors containing human WT or K8R Fpn-GFP were used to synthesize RNA in vitro. RNA-injected Xenopus oocytes were incubated for 4 to 5 days before being used in functional assays. Oocytes were treated for 0 to 4 hours with 10 μM hepcidin before assaying iron efflux. Oocytes were injected with 50 nL of 5 μM 55Fe, added as FeCl3 (Perkin-Elmer Life Science Products). The first-order rate constants (k) describing iron efflux >30 minutes was determined,37 and live-cell imaging of Fpn-GFP performed as described.37
Treatment of human red blood cells with hepcidin and hepcidin agonists
Human packed red blood cells (PRBCs) were obtained from the University of California, Los Angeles Department of Pathology and Laboratory Medicine. PRBC aliquots from 4 different donors were incubated in their citrate-phosphate-dextrose-adenine (CPDA-1) storage medium at 37°C for 24 hours in the presence of hepcidin (0.4 and 3.6 µM), PR73 (0.4 and 3.6 µM), or solvent. Samples were collected after 24 hours by centrifuging the PRBC at 5000 g for 8 minutes to obtain cell pellets. The supernatant was centrifuged again, and non–transferrin-bound iron (NTBI) was measured using enhanced labile plasma iron assay, as previously described.38 For protein isolation, cell pellets were lysed, and membranes were pelleted and lysed in RIPA buffer. After electrophoresis and transfer, western blotting was performed using anti-ferroportin antibody (human anti-human ferroportin 38C8, Amgen) and developed with SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific).
Mouse treatment with hepcidin agonists
C57BL/6 mice were injected intraperitoneally with PR73 (2 60-nmol doses, 12 hours apart) or solvent, and analyzed 4 hours after the second injection. Th3 mice were injected subcutaneously with 50 nmol minihepcidin M00939 and were analyzed over a time course of 0 to 36 hours after injection. We measured serum iron using a colorimetric assay, duodenal and splenic Fpn levels by western blotting, and serum hepcidin by enzyme-linked immunosorbent assay.
Additional details and methods can be found in the supplemental Methods and supplemental Figures 1-14.
Results
Fpn-GFP mutants localize to cell membrane and export iron
We first examined whether the gain-of-function Fpn mutants (Table 1) were expressed on the cell membrane and exported iron. Mutants were visualized by microscopy to assess membrane localization. Green fluorescence was visible on the cell membrane for all mutants (supplemental Figure 1). To quantify the amount of Fpn present on the cell surface compared with the total Fpn amount, we performed cell-surface, thiol-specific biotinylation targeting the Fpn C326 residue. All mutants were expressed on the cell surface similarly to WT (quantitation shown in Figure 1A, representative western blots in supplemental Figure 2). As expected, C326S mutant showed essentially no biotinylation, confirming the specificity of the method. We next assessed the iron-exporting function of the mutants by measuring intracellular ferritin as an index of remaining cellular iron stores. All mutants exported iron as indicated by decreased intracellular ferritin concentration after induction of Fpn-GFP expression with dox, compared with cells not induced with dox (Figure 1B).
Fpn mutants have either impaired hepcidin binding or intact hepcidin binding but impaired hepcidin-dependent ubiquitination, leading to varying hepcidin resistance. (A) HEK293T cells stably transfected with hFpn-GFP mutants or WT were induced with dox (+D) to express Fpn, surface-thiol biotinylated for 30 minutes, immunoprecipitated with anti-GFP antibody (Ab), and immunoblotted with streptavidin-HRP or anti-GFP Ab. Band intensity was normalized to total GFP and then further normalized to WT Fpn on each blot. Because mutant data within each western blot were normalized to the WT sample on the same blot, the WT value is always 1 and is without error bars. WT is included in the graph only for visual reference. (B) hFpn-GFP mutants were induced overnight with dox and then incubated for 24 hours in the presence of 25 μM ferric ammonium citrate (FAC). The intracellular ferritin concentration was normalized to the total protein concentration. Ferritin was normalized to the uninduced sample (−D) for each cell line (uninduced = 1). Data shown are means ± standard errors of the means of 3 to 5 independent experiments. For statistical analysis, the 2-tailed 1-sample Student t test (normally distributed data) or the 2-tailed 1-sample signed rank test (data with nonnormal distribution) was used with 1 as the comparison. The false discovery rate (FDR) procedure was used to determine significance (*significant). (C) Expression of WT and mutant hFpn-GFP was induced overnight or not, and cells were then incubated for 24 hours with 25 μM FAC ± 1 µg/ml (0.4 µM) hepcidin. The intracellular ferritin concentration was normalized to the total protein concentration and expressed relative to uninduced cells (ie, maximal ferritin levels for each mutant, −dox). (D) HEK293T cells expressing WT and mutant hFpn-GFP were treated with N-terminally biotinylated hepcidin for 30 minutes, immunoprecipitated with anti-GFP Ab, run under nonreducing conditions, and immunoblotted with streptavidin-HRP or anti-GFP Abs. The streptavidin signal was first normalized to total GFP, and then mutant hepcidin binding values were expressed as a fraction of hepcidin binding to WT Fpn. Because mutant data within each western blot were normalized to the WT sample on the same blot, the WT value is always 1 and is without error bars. WT is included in the graph only for visual reference. (E) Fpn-GFP WT and mutants were treated with hepcidin for 30 minutes, immunoprecipitated with anti-GFP Ab, and immunoblotted with anti–poly-/monoubiquitin (FK2) Ab or anti-GFP Ab. The ubiquitination signal was first normalized to the GFP signal, and then the mutant ubiquitination was expressed as a fraction of WT Fpn ubiquitination. As in panel B, WT is included only for visual reference. For statistical analysis in panels B and C, the 2-tailed 1-sample Student t test was used wtih 1 as the comparison, and for panel A, the 2-tailed Student t test (normally distributed data) or the Mann-Whitney rank sum test (data with nonnormal distribution) was used with WT as the comparison. After the P values were obtained, the FDR procedure was used to determine significance (*significant). Data shown are means ± standard errors of the means of 3 to 6 independent experiments. Hepcidin-binding mutants are denoted by red shades and ubiquitination mutants by green shades. Severely impaired mutants are denoted by bolder colors. Severe impairment was defined as ≤25% of WT values based on assessing the values from panels C-E for mutants with impaired hepcidin binding and panels C and E for impaired hepcidin-induced conformational change mutants. Also, for panels A-E, red and dark green indicate severe mutations, medium green indicates mild mutation, and pink and light green indicates borderline mutations.
Fpn mutants have either impaired hepcidin binding or intact hepcidin binding but impaired hepcidin-dependent ubiquitination, leading to varying hepcidin resistance. (A) HEK293T cells stably transfected with hFpn-GFP mutants or WT were induced with dox (+D) to express Fpn, surface-thiol biotinylated for 30 minutes, immunoprecipitated with anti-GFP antibody (Ab), and immunoblotted with streptavidin-HRP or anti-GFP Ab. Band intensity was normalized to total GFP and then further normalized to WT Fpn on each blot. Because mutant data within each western blot were normalized to the WT sample on the same blot, the WT value is always 1 and is without error bars. WT is included in the graph only for visual reference. (B) hFpn-GFP mutants were induced overnight with dox and then incubated for 24 hours in the presence of 25 μM ferric ammonium citrate (FAC). The intracellular ferritin concentration was normalized to the total protein concentration. Ferritin was normalized to the uninduced sample (−D) for each cell line (uninduced = 1). Data shown are means ± standard errors of the means of 3 to 5 independent experiments. For statistical analysis, the 2-tailed 1-sample Student t test (normally distributed data) or the 2-tailed 1-sample signed rank test (data with nonnormal distribution) was used with 1 as the comparison. The false discovery rate (FDR) procedure was used to determine significance (*significant). (C) Expression of WT and mutant hFpn-GFP was induced overnight or not, and cells were then incubated for 24 hours with 25 μM FAC ± 1 µg/ml (0.4 µM) hepcidin. The intracellular ferritin concentration was normalized to the total protein concentration and expressed relative to uninduced cells (ie, maximal ferritin levels for each mutant, −dox). (D) HEK293T cells expressing WT and mutant hFpn-GFP were treated with N-terminally biotinylated hepcidin for 30 minutes, immunoprecipitated with anti-GFP Ab, run under nonreducing conditions, and immunoblotted with streptavidin-HRP or anti-GFP Abs. The streptavidin signal was first normalized to total GFP, and then mutant hepcidin binding values were expressed as a fraction of hepcidin binding to WT Fpn. Because mutant data within each western blot were normalized to the WT sample on the same blot, the WT value is always 1 and is without error bars. WT is included in the graph only for visual reference. (E) Fpn-GFP WT and mutants were treated with hepcidin for 30 minutes, immunoprecipitated with anti-GFP Ab, and immunoblotted with anti–poly-/monoubiquitin (FK2) Ab or anti-GFP Ab. The ubiquitination signal was first normalized to the GFP signal, and then the mutant ubiquitination was expressed as a fraction of WT Fpn ubiquitination. As in panel B, WT is included only for visual reference. For statistical analysis in panels B and C, the 2-tailed 1-sample Student t test was used wtih 1 as the comparison, and for panel A, the 2-tailed Student t test (normally distributed data) or the Mann-Whitney rank sum test (data with nonnormal distribution) was used with WT as the comparison. After the P values were obtained, the FDR procedure was used to determine significance (*significant). Data shown are means ± standard errors of the means of 3 to 6 independent experiments. Hepcidin-binding mutants are denoted by red shades and ubiquitination mutants by green shades. Severely impaired mutants are denoted by bolder colors. Severe impairment was defined as ≤25% of WT values based on assessing the values from panels C-E for mutants with impaired hepcidin binding and panels C and E for impaired hepcidin-induced conformational change mutants. Also, for panels A-E, red and dark green indicate severe mutations, medium green indicates mild mutation, and pink and light green indicates borderline mutations.
Gain-of-function Fpn-GFP mutants display hepcidin resistance because of either impaired hepcidin binding or altered hepcidin-induced conformational change of Fpn with consequently decreased hepcidin-induced ubiquitination
Resistance of Fpn mutants to downregulation by hepcidin is thought be an essential component of nonclassical FD pathogenesis and manifests as continued cellular iron export despite exposure to hepcidin concentrations that cause iron retention in cells expressing WT Fpn. We therefore measured intracellular ferritin as an indicator of iron retention in cells treated with or without hepcidin for 24 hours and observed by microscopy the localization of Fpn after 24 hours. Finally, we quantified the stability of WT and mutant Fpn over 24 hours by western blotting, as well as Fpn degradation after treatment with hepcidin for 24 hours. All clinical mutants, with the exception of V72F, D270V, and S338R, were functionally hepcidin resistant compared with WT as demonstrated by lower ferritin retention after hepcidin treatment (Figure 1C), and less Fpn-GFP degradation as assessed by both microscopy (supplemental Figure 1) and western blotting compared with WT (supplemental Figure 3A, representative blots and supplemental Figure 3B, quantitation). To detect milder resistance, we exposed V72F and D270V mutants to a lower hepcidin concentration for a shorter time and measured iron export using radioactive iron release for ≤10 hours. V72F and D270V indeed displayed mild hepcidin resistance compared with WT Fpn (supplemental Figure 4; S71F and C326S mutants were added as controls manifesting strong resistance). We also examined S338R using the radioactive iron export assay, but with higher hepcidin concentrations (supplemental Figure 4), and also observed mild, but significant resistance to hepcidin (with N144D and C326S serving as controls for medium-to-strong resistance).
Of note, several mutants (Y64N, H507R, D270V and S338R) had decreased stability at 24 hours compared with WT (supplemental Figure 3A,C), which could partially counteract hepcidin resistance in vivo, and modulate the patient phenotype.
Two potential mechanisms can explain the hepcidin-resistant phenotype of the clinical Fpn mutants as shown in Figure 1C: (1) decreased hepcidin binding to Fpn, or (2) decreased hepcidin-induced endocytosis of Fpn despite intact hepcidin binding. To determine if the Fpn-GFP mutants bind hepcidin, we treated mutant cell lines with biotinylated hepcidin for 30 minutes, immunoprecipitated Fpn with anti-GFP antibody, and detected bound hepcidin by immunoblotting with streptavidin-horseradish peroxidase (HRP), taking advantage of the stability of the hepcidin-ferroportin complex in nonreducing sodium dodecyl sulfate-polyacrylamide gel electrophoresis.2,40 We identified 5 mutants that showed impaired hepcidin binding compared with WT (quantitation shown in Figure 1D and representative western blots in supplemental Figure 5): C326S, Y501C, D504N, N144D and V72F.
After hepcidin binding to Fpn, ubiquitination of several cytoplasmic lysines is required for endocytosis of Fpn.3 To assess hepcidin-induced ubiquitination, we treated Fpn-GFP mutants with hepcidin, immunoprecipitated with anti-GFP antibody, and probed for ubiquitin with an antibody that recognizes both poly-and monoubiquitinated proteins. We identified 6 mutants that had normal hepcidin binding, but variably deficient hepcidin-induced ubiquitination: Y64N and H507R showed severe impairment in ubiquitination, and S71F, G204S, D270V, and S338R were less impaired (quantitation shown in Figure 1E and representative western blots in Figure S6).
The results shown in Figure 1D-E (hepcidin binding and Fpn ubiquitination, respectively) were used as the basis for color assignment in additional figures in this manuscript (Figure 2, supplemental Figures 3, 7, and 10): red/pink indicates mutants with impaired hepcidin binding, and light/dark green indicates mutants that bind hepcidin normally but have variably impaired ubiquitination. The intensity of the color denotes the severity of the impairment, where bold colors indicate severe impairment, lighter colors indicate mild impairment, and the lightest colors (V72F, D270V, and S338R) indicate borderline impairment.
Fpn-GFP mutants map out a hepcidin-binding site in the central cavity and reveal peripheral hindrance of conformation-dependent ubiquitination
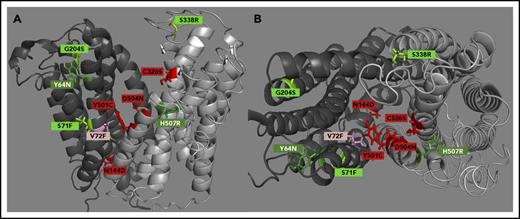
We next mapped the mutations causing impaired hepcidin binding (C326S, Y501C, D504N, N144D, and V72F) or impaired hepcidin-dependent ubiquitination (Y64N, H507R, S71F, G204S, D270V, and S338R) onto a computational structural model of hFpn (Figure 2). All the mutations that decreased hepcidin binding localized within the main cavity of Fpn, implicating 4 helices in the hepcidin-binding site (helices 2, 4, 7, and 11). All of the mutations causing impaired Fpn ubiquitination localized to the periphery of Fpn at the helix-helix interfaces, suggesting that these substitutions interfere with appropriate conformational change after hepcidin binding. A second computational model (supplemental Figure 7A-D), which included the crystal structure of BdFPN in the modeling, led to the same conclusion.
hFpn structure depicting clinically relevant mutant residues. (A) A side view of hFpn in its outward-facing state, with the N-terminus on the left and C-terminus on the right. (B) A top-down view of hFpn in its outward-facing state. D270V is not modeled because it is located in the unstructured intracellular loop of Fpn. The red/pink color denotes mutants with impaired hepcidin binding, and the green color denotes mutants with intact hepcidin binding, but variably impaired hepcidin-dependent ubiquitination. For simplicity, the mild and borderline mutants are labeled with the same light color.
hFpn structure depicting clinically relevant mutant residues. (A) A side view of hFpn in its outward-facing state, with the N-terminus on the left and C-terminus on the right. (B) A top-down view of hFpn in its outward-facing state. D270V is not modeled because it is located in the unstructured intracellular loop of Fpn. The red/pink color denotes mutants with impaired hepcidin binding, and the green color denotes mutants with intact hepcidin binding, but variably impaired hepcidin-dependent ubiquitination. For simplicity, the mild and borderline mutants are labeled with the same light color.
To refine the model of hepcidin binding to Fpn, we generated additional nonclinical mutations within helices 7 and 11 (supplemental Figure 7E) because our data suggest that these helices contribute most strongly to hepcidin binding. We tested F324A, F324Y, Y333A, Y333F, and F508Y with the same assays as clinical mutants, which showed that all were displayed on the membrane and exported iron (Figure 3A-B). Only F508Y exhibited impaired hepcidin binding, whereas the other mutants had impaired ubiquitination compared with the WT (Figure 3C-D). Ferritin retention as a marker of functional resistance to hepcidin showed that F508Y was strongly resistant despite its conservative amino acid substitution (Figure 3E; supplemental Figure 8), whereas for other residues, nonconservative substitutions (F324A and Y333A) were required for significant resistance (Figure 3E). Interestingly, even though F508Y bound hepcidin very poorly and failed to retain intracellular ferritin or radiolabeled iron, it was mostly degraded by 24 hours (supplemental Figure 9A-B), suggesting delayed internalization and degradation. The only unstable nonclinical mutant was F324A (supplemental Figure 9C), a ubiquitination-impaired mutant. Mapping the 5 nonclinical Fpn-GFP mutants onto our computational hFpn model (supplemental Figures 7 and 1) indicates that the deduced hepcidin-binding site may include F508. The model also highlights the role of peripheral Fpn mutations in impeding ubiquitination and thereby causing variable resistance to hepcidin-induced Fpn endocytosis and degradation.
Analysis of nonclinical hFpn-GFP mutants. Cells expressing inducible nonclinical Fpn mutants were analyzed with the same approaches as those expressing clinical mutants. (A) All mutants were displayed on the cell membrane. Membrane localization was determined as in Figure 1A. (B) All mutants exported iron. Ferritin was determined as in Figure 1B by normalizing it to the uninduced (−dox) condition for each cell line (thus, uninduced ferritin levels = 1). (C) Hepcidin binding was determined as in Figure 1D. (D) Ubiquitination was determined as in Figure 1E. (E) Relative ferritin retention after hepcidin addition was determined as in Figure 1C. Data shown are means ± standard errors of the means of 3 to 6 independent experiments. For statistical analysis the 2-tailed 1-sample Student t test was used with 1 as the comparison for panels A-D, and the 2-tailed Student t test (normally distributed data) or the Mann-Whitney rank sum test (data with nonnormal distribution) was used with WT as the comparison for panel E. After the P values were obtained, the FDR procedure was used to determine significance (*significant).
Analysis of nonclinical hFpn-GFP mutants. Cells expressing inducible nonclinical Fpn mutants were analyzed with the same approaches as those expressing clinical mutants. (A) All mutants were displayed on the cell membrane. Membrane localization was determined as in Figure 1A. (B) All mutants exported iron. Ferritin was determined as in Figure 1B by normalizing it to the uninduced (−dox) condition for each cell line (thus, uninduced ferritin levels = 1). (C) Hepcidin binding was determined as in Figure 1D. (D) Ubiquitination was determined as in Figure 1E. (E) Relative ferritin retention after hepcidin addition was determined as in Figure 1C. Data shown are means ± standard errors of the means of 3 to 6 independent experiments. For statistical analysis the 2-tailed 1-sample Student t test was used with 1 as the comparison for panels A-D, and the 2-tailed Student t test (normally distributed data) or the Mann-Whitney rank sum test (data with nonnormal distribution) was used with WT as the comparison for panel E. After the P values were obtained, the FDR procedure was used to determine significance (*significant).
Characterization of K8R Fpn mutant and evidence of its occlusion by hepcidin and minihepcidin
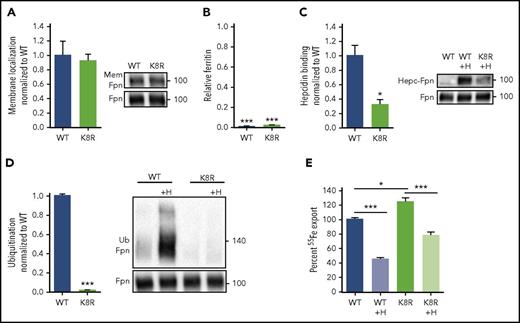
Our structural Fpn model implies that hepcidin binding deep in the Fpn central cavity should impede iron export, even if hepcidin-induced endocytosis of Fpn is disabled. To test this, we designed an Fpn variant deficient in ligand-induced endocytosis through mutations that disable ubiquitination, but have a minimal effect on the intramembrane domains of Fpn. The cytoplasmic loop that connects the two 6-helix bundle lobes of Fpn contains lysines that undergo hepcidin-dependent ubiquitination,3 so we generated a cell line stably expressing inducible K8R Fpn-GFP, in which 8 lysines of the loop were mutated to arginines (supplemental Figure 7E). K8R localized on the cell membrane (Figure 4A), exported iron similarly to WT as measured by the loss of intracellular ferritin after Fpn induction (Figure 4B), and bound hepcidin, though at about 30% of WT Fpn binding (Figure 4C). K8R had severely impaired hepcidin-induced ubiquitination as expected (Figure 4D), even at very high hepcidin concentrations (supplemental Figure 11). Despite the complete loss of hepcidin-dependent ubiquitination, K8R 55Fe export was still decreased by hepcidin treatment (Figure 4E). We compared ferritin retention and Fpn degradation in WT, C326S (a mutant that cannot bind hepcidin), and K8R in response to a range of hepcidin concentrations. After hepcidin addition, K8R Fpn retained ferritin in a dose-dependent manner, and higher hepcidin concentrations achieved maximal iron retention comparable with WT (Figure 5A). The C326S mutant was resistant to hepcidin-induced iron retention. The profound block of iron export through K8R was achieved despite impaired Fpn degradation, as determined by western blotting (Figure 5B), and despite the lack of degradation or punctate structures characteristic of endocytosis, as shown by microscopy (Figure 5C). PR73,41 a minihepcidin that was engineered to bind to Fpn more strongly than hepcidin, was 10 times more potent than hepcidin in inducing ferritin retention without endocytic degradation of K8R Fpn (supplemental Figure 12).
Iron export by the K8R mutant is inhibited by hepcidin despite the absence of ligand-induced ubiquitination. (A) Cells were treated as in Figure 1A. K8R localized to the cell membrane similarly to WT. (B) Cells were treated as in Figure 1B. K8R exported iron and decreased ferritin similarly to WT. For statistical analysis, the 1-sample Student t test with 1 as the comparison (normally distributed data) or the 1-sample signed rank test (data with nonnormal distribution) was used. (C) HEK293T cells expressing WT and mutant Fpn-GFP were treated with N-terminally biotinylated hepcidin for 30 minutes, immunoprecipitated with anti-GFP Ab, run under nonreducing conditions, and immunoblotted with streptavidin-HRP or anti-GFP Abs. Hepcidin bound less to K8R compared with WT. (D) Cells were treated as in Figure 1E. The K8R mutant was not ubiquitinated after hepcidin addition. For the statistical analysis in panels A, C, and D, the 2-tailed Student t test was used with WT as the comparison. (E) Cells were loaded with 2 mM 55Fe-NTA for 48 hours, washed, replated, induced overnight, washed again, and then ±3 μg/ml hepcidin was added. Extracellular radioactivity was measured at 0, 2, 4, and 8 hours. The “uninduced” measurement at each time point was subtracted as background, and the slope for each sample was determined and used to calculate the percentage of iron export by normalizing the slopes to the untreated WT. For the statistical analysis in panel E, the 2-tailed Student t test (normally distributed data) and Mann-Whitney rank sum test (data with nonnormal distribution) were used with WT as the comparison. Data shown are means ± standard errors of the means of 3 to 4 biological replicates. ***P < .001; **P < .01; *P < .05. Hepc-Fpn, hepcidin complexed with Fpn; Mem, membrane.
Iron export by the K8R mutant is inhibited by hepcidin despite the absence of ligand-induced ubiquitination. (A) Cells were treated as in Figure 1A. K8R localized to the cell membrane similarly to WT. (B) Cells were treated as in Figure 1B. K8R exported iron and decreased ferritin similarly to WT. For statistical analysis, the 1-sample Student t test with 1 as the comparison (normally distributed data) or the 1-sample signed rank test (data with nonnormal distribution) was used. (C) HEK293T cells expressing WT and mutant Fpn-GFP were treated with N-terminally biotinylated hepcidin for 30 minutes, immunoprecipitated with anti-GFP Ab, run under nonreducing conditions, and immunoblotted with streptavidin-HRP or anti-GFP Abs. Hepcidin bound less to K8R compared with WT. (D) Cells were treated as in Figure 1E. The K8R mutant was not ubiquitinated after hepcidin addition. For the statistical analysis in panels A, C, and D, the 2-tailed Student t test was used with WT as the comparison. (E) Cells were loaded with 2 mM 55Fe-NTA for 48 hours, washed, replated, induced overnight, washed again, and then ±3 μg/ml hepcidin was added. Extracellular radioactivity was measured at 0, 2, 4, and 8 hours. The “uninduced” measurement at each time point was subtracted as background, and the slope for each sample was determined and used to calculate the percentage of iron export by normalizing the slopes to the untreated WT. For the statistical analysis in panel E, the 2-tailed Student t test (normally distributed data) and Mann-Whitney rank sum test (data with nonnormal distribution) were used with WT as the comparison. Data shown are means ± standard errors of the means of 3 to 4 biological replicates. ***P < .001; **P < .01; *P < .05. Hepc-Fpn, hepcidin complexed with Fpn; Mem, membrane.
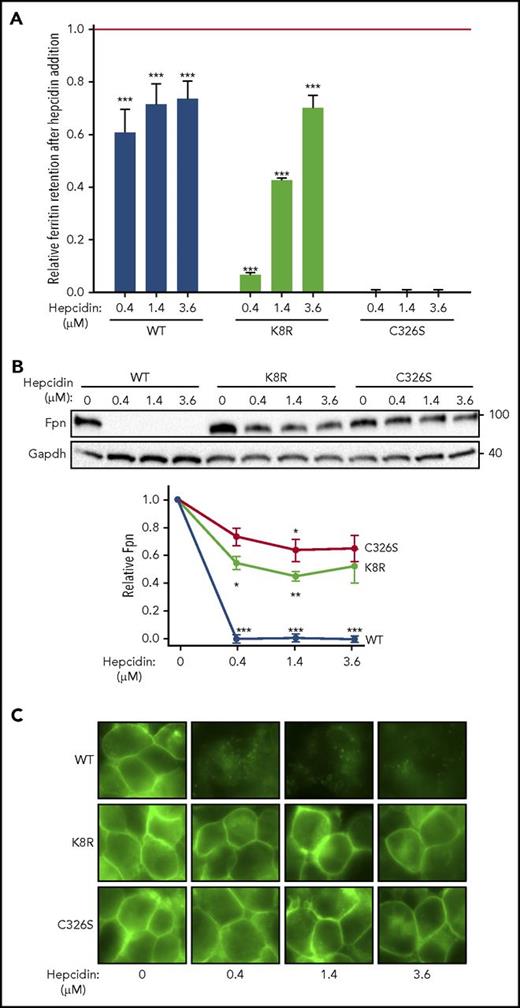
Evidence for Fpn occlusion by hepcidin. Cells expressing inducible nonubiquitinating K8R mutant were compared with those expressing WT Fpn or C326S mutant, which does not bind hepcidin. (A) Ferritin retention after hepcidin addition was determined as in Figure 1C. For the statistical analysis the 2-tailed Student t test was used with the respective untreated control for comparison. ***P < .001; **P < .01; *P < .05. (B) Lysates from panel A were analyzed by western blotting. Top: representative western blot. Bottom: densitometry of triplicate western blots. The Fpn signal was first normalized to glyceraldehyde-3-phosphate dehydrogenase, then expressed as a fraction of its respective untreated control (no hepcidin treatment). For the statistical analysis, the 2-tailed 1-sample Student t test was used with 1 as the comparison. ***P < .001; **P < .01; *P < .05. C) Microscopy of live cells from panel A after 24 hours (original magnification ×40). Data shown are the mean ± standard errors of the mean of 3 independent experiments.
Evidence for Fpn occlusion by hepcidin. Cells expressing inducible nonubiquitinating K8R mutant were compared with those expressing WT Fpn or C326S mutant, which does not bind hepcidin. (A) Ferritin retention after hepcidin addition was determined as in Figure 1C. For the statistical analysis the 2-tailed Student t test was used with the respective untreated control for comparison. ***P < .001; **P < .01; *P < .05. (B) Lysates from panel A were analyzed by western blotting. Top: representative western blot. Bottom: densitometry of triplicate western blots. The Fpn signal was first normalized to glyceraldehyde-3-phosphate dehydrogenase, then expressed as a fraction of its respective untreated control (no hepcidin treatment). For the statistical analysis, the 2-tailed 1-sample Student t test was used with 1 as the comparison. ***P < .001; **P < .01; *P < .05. C) Microscopy of live cells from panel A after 24 hours (original magnification ×40). Data shown are the mean ± standard errors of the mean of 3 independent experiments.
We next tested whether hepcidin-mediated occlusion occurs in those clinical Fpn mutants that bind hepcidin strongly, but have impaired ubiquitination (Y64N and H507R). However, unlike in K8R Fpn, we could not dissociate occlusion from internalization in these mutants: Fpn-GFP underwent endocytosis and degradation when treated with high concentrations of hepcidin (supplemental Figure 13A) or minihepcidin (supplemental Figure 13B-C). Although it remains to be explained why these clinical nonubiquitinating mutants are less severe than the K8R mutant, the finding has important implications because it suggests that patients carrying these mutations may benefit from treatment with pharmacological doses of hepcidin.
Hepcidin occludes both WT and K8R Fpn in Xenopus oocytes
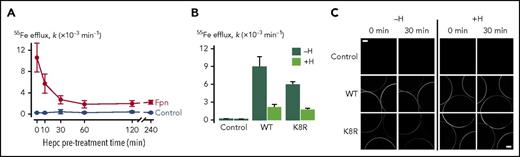
To further confirm that hepcidin can inhibit iron export without causing Fpn endocytosis but by occluding Fpn, we used Xenopus oocytes expressing WT or K8R Fpn-GFP. Oocytes expressing WT Fpn-GFP were pretreated with hepcidin for up to 4 hours to determine the time course of the hepcidin effect on iron export. In contrast to mammalian cells, in which hepcidin treatment results in a progressively greater inhibition of iron export for ≤10 hours, maximal inhibition of iron export in oocytes was observed after only 30 minutes of pretreatment (Figure 6A). We found that 30 minutes of hepcidin treatment inhibited 55Fe efflux from oocytes expressing K8R Fpn to the same degree as it did for WT Fpn (Figure 6B). Live imaging of oocytes revealed that the 30-minute hepcidin treatment did not induce endocytosis of either WT or K8R Fpn (Figure 6C). Whereas some reduction in the intensity of GFP fluorescence at the oocyte perimeter was observed in WT and K8R oocytes over time (presumably due to photobleaching), hepcidin did not stimulate a loss of fluorescence from the oocyte perimeter either for WT or K8R. These data reveal that hepcidin can directly inhibit Fpn-mediated iron transport activity independently of Fpn internalization not only in the K8R mutant but also in WT Fpn.
Effect of hepcidin on WT and mutant Fpn expressed in Xenopus oocytes. (A) First-order rate constants (k) describing 55Fe efflux (assayed over 30 minutes) from control oocytes (gray) and oocytes expressing WT Fpn (black) pretreated with 10 µM hepcidin for 0 to 240 minutes (n = 8-12 per group). Two-way analysis of variance (ANOVA) revealed an interaction (P < .001); within Fpn, the 0- and 10-minute time points differed from all other time points (P < .001), and the 30- to 240-minute time points did not differ from one another (P ≥ .35). (B) 55Fe efflux in control oocytes and oocytes expressing WT or K8R Fpn that were untreated (−H) or pretreated for 30 minutes with 10 µM hepcidin (+H) (n = 9-12 per group). Two-way ANOVA revealed an interaction (P < .001). Percent inhibition of 55Fe efflux by hepcidin did not differ between WT (76% ± 6%) and K8R (71% ± 3%) (means ± standard errors of the means) (P = .47). (C) Live-cell imaging of control oocytes and oocytes expressing WT or K8R Fpn before and after 30 minutes of treatment without hepcidin (−H) or with 10 µM hepcidin (+H) in the same oocyte preparation as used in panel B. Each frame captures portions of 3 oocytes, and the image plane approximately bisects the oocytes. Scale bars, 0.2 mm. Two-way ANOVA of the change in fluorescence intensity (ΔF) over time revealed a greater loss of fluorescence in untreated oocytes (−H) compared with hepcidin-treated (+H) (P = .005) and that ΔF did not differ between WT and K8R (P = .75).
Effect of hepcidin on WT and mutant Fpn expressed in Xenopus oocytes. (A) First-order rate constants (k) describing 55Fe efflux (assayed over 30 minutes) from control oocytes (gray) and oocytes expressing WT Fpn (black) pretreated with 10 µM hepcidin for 0 to 240 minutes (n = 8-12 per group). Two-way analysis of variance (ANOVA) revealed an interaction (P < .001); within Fpn, the 0- and 10-minute time points differed from all other time points (P < .001), and the 30- to 240-minute time points did not differ from one another (P ≥ .35). (B) 55Fe efflux in control oocytes and oocytes expressing WT or K8R Fpn that were untreated (−H) or pretreated for 30 minutes with 10 µM hepcidin (+H) (n = 9-12 per group). Two-way ANOVA revealed an interaction (P < .001). Percent inhibition of 55Fe efflux by hepcidin did not differ between WT (76% ± 6%) and K8R (71% ± 3%) (means ± standard errors of the means) (P = .47). (C) Live-cell imaging of control oocytes and oocytes expressing WT or K8R Fpn before and after 30 minutes of treatment without hepcidin (−H) or with 10 µM hepcidin (+H) in the same oocyte preparation as used in panel B. Each frame captures portions of 3 oocytes, and the image plane approximately bisects the oocytes. Scale bars, 0.2 mm. Two-way ANOVA of the change in fluorescence intensity (ΔF) over time revealed a greater loss of fluorescence in untreated oocytes (−H) compared with hepcidin-treated (+H) (P = .005) and that ΔF did not differ between WT and K8R (P = .75).
Hepcidin occludes endogenous Fpn in mature red blood cells
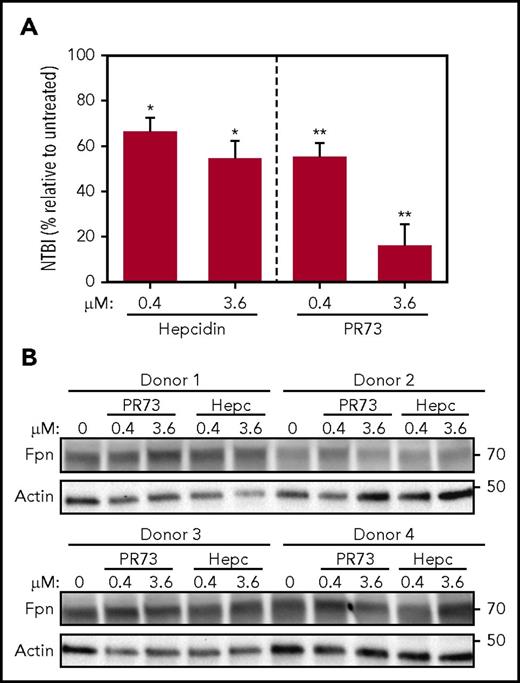
In vivo, occlusion by hepcidin may be the critical mechanism of regulation of iron efflux in Fpn-expressing cells that lack endocytic machinery, such as mature red blood cells (RBCs).42 To test this, we treated PRBCs ex vivo in a transferrin-free medium with hepcidin or minihepcidin for 24 hours. Although Fpn was not degraded even with high concentrations (3.6 µM) of hepcidin or PR73, iron export was dose-dependently decreased, as evidenced by the lower concentration of NTBI in the medium (Figure 7), confirming that hepcidin-mediated occlusion modulates iron export by endogenous Fpn in RBCs.
Hepcidin blocks iron export from human RBCs without degrading Fpn. Human PRBCs from 4 different donors were treated with hepcidin or PR73 for 24 hours at 37°C. (A) Iron export from PRBCs was assessed by measuring NTBI in the medium. For the statistical analysis, the 2-tailed 1-sample Student t test was used with 100% as the comparison. (B) Western blotting of Fpn and actin levels in PBRCs.
Hepcidin blocks iron export from human RBCs without degrading Fpn. Human PRBCs from 4 different donors were treated with hepcidin or PR73 for 24 hours at 37°C. (A) Iron export from PRBCs was assessed by measuring NTBI in the medium. For the statistical analysis, the 2-tailed 1-sample Student t test was used with 100% as the comparison. (B) Western blotting of Fpn and actin levels in PBRCs.
How much occlusion contributes to the regulation of iron fluxes in vivo is difficult to determine in the absence of a mouse model expressing a nonendocytosing Fpn. Because injection of synthetic full-length hepcidin in mice was previously shown to cause ferroportin degradation in the spleen and duodenum,43-46 we probed for the presence of occlusion in vivo by injecting mice with minihepcidins, because these peptides occlude Fpn more efficiently than does native hepcidin. Minihepcidins were injected into WT or thalassemic mice, and serum iron and Fpn levels were assessed. Treatment of WT mice with 2 60-nmol injections of PR73 over 16 hours resulted in a dramatic decrease in serum iron (supplemental Figure 14A), however, no decrease in Fpn protein levels in the duodenum or the spleen were observed (supplemental Figure 14B-E). Likewise, a single injection of another minihepcidin, M009,39 did not decrease spleen Fpn levels at 6, 12, 24, or 36 hours postinjection (supplemental Figure 14F), but caused a prolonged decrease of serum iron (supplemental Figure 14G). Transient suppression of endogenous hepcidin production was also observed in response to hypoferremia (supplemental Figure 14H). Thus, in the absence of detectable Fpn degradation, the profound effect of hepcidin agonists on serum iron was consistent with the mechanism of occlusion in both mouse models. Interestingly, in contrast to the lack of Fpn degradation in vivo, minihepcidins readily caused Fpn degradation in vitro (supplemental Figure 12B-C). The reasons for this differential effect of minihepcidins on Fpn endocytosis are not clear. It is possible that Fpn glycosylation and/or Fpn-interacting proteins differ between in vitro cellular models and tissues in vivo, which may differentially affect minihepcidin binding to Fpn, Fpn conformational change, and its endocytosis. The data suggest that serum iron rather than Fpn levels should be used as a more accurate measure of the activity of hepcidin and its agonists.
Discussion
In this study, we characterized the mechanisms by which known clinical Fpn mutations (Table 1) cause nonclassical FD and extended these findings to gain insight into the structural basis of Fpn function and its regulation by hepcidin. Our cellular models revealed that nonclassical FD mutations cause resistance to hepcidin either by impairing the binding of hepcidin to Fpn or by altering hepcidin-induced conformational change and thereby impairing the ubiquitination required for endocytosis. Although all of the mutations were associated with a clinical phenotype of nonclassical FD, the mutants varied considerably in their degree of hepcidin resistance. In particular, V72F, D270V, and S338R showed only very mild hepcidin resistance in our cellular model, suggesting that additional factors may modify the phenotype of FD caused by these mutations. One such factor could be the decreased stability of some mutants, including Y64N, H507R, D270V, and S338R (supplemental Figure 3A,C). Such instability could decrease Fpn cell surface expression and iron export at baseline, perhaps accounting for the frequent reports of a mixed form of FD (Table 1) manifesting restriction of iron export leading to iron accumulation in Kupffer cells with simultaneous resistance to hepcidin leading to systemic and hepatic iron overload. The structural model also suggests that D270V and S338R may be mild because of their peripheral position on the Fpn structure: S338R is predicted to be located on an extracellular loop rather than at a helix-helix interface, and D270V in the long intracellular loop, where it may act by interfering with ubiquitination of a nearby lysine residue. Lastly, important modifiers that affect disease severity include sex, age, and comorbidities, such as alcohol abuse, obesity, and metabolic syndrome, which are all known to affect the iron status of patients with FD.20,47 Although our cellular model faithfully documents profound hepcidin resistance in the more severe mutations, it lacks the complex cellular and systemic regulatory factors that could amplify the effects of the weakest mutations.
Our structural models indicate that hepcidin binding occurs within the main cavity of Fpn, where hepcidin interacts with as many as 4 different helices. Additionally, we conclude that peripheral amino acids, positioned at helix-helix interfaces, influence the hepcidin-induced conformational change that leads to the ubiquitination of Fpn. They may even affect Fpn conformation in the absence of hepcidin, which explains how some of the ubiquitination-impaired mutants (Y64N, H507R, D270V, S338R, and F324A), but none of the hepcidin binding-impaired mutants, are unstable. Although the alternative structure generated by adding BdFPN to the threading set displayed small differences from our earlier model (supplemental Figure 7), it did not alter the conclusions of our analysis.
Using the model as a guide to design informative nonclinical mutants, we examined F508Y, the nearest residue that could contribute to the binding site based on its location on helix 11 immediately above Y501 and D504. F508Y was a milder hepcidin-resistant mutant than Y501C and D504N, likely because the amino acid substitution we chose was very conservative. We previously showed that hepcidin binding to Fpn involves a disulfide-thiol interaction between hepcidin and Fpn C326 residue, because the isosteric C326S substitution results in the complete loss of hepcidin binding.27,40 However, our current study implicates several additional residues on multiple helices in stabilizing the binding between the ligand and its receptor. We also examined F324 and Y333 Fpn residues, because we had previously noted that mutants at these locations did not internalize radiolabeled hepcidin.40 In the present study, we found that these mutants bound hepcidin, but displayed impaired ubiquitination and endocytosis. Our proposed Fpn structural model is consistent with all our experimental data classifying residues as those involved in hepcidin binding versus those that affect the conformational change leading to the ubiquitination of Fpn. After we completed the cellular analyses and modeling, Praschberger et al described a partially hepcidin-resistant clinical Fpn mutant, A69T,48 in a patient with iron-overloaded hepatocytes. Another group described a patient with the A69T mutation as having severe hyperferritinemia (6242 µg/L) and elevated serum transferrin saturation (95.4%).49 According to our hFpn computational model, this mutation would be expected to affect hepcidin binding based on its location in the central cavity (supplemental Figure 7).
Chung et al previously observed that hepcidin treatment inhibited iron efflux in Caco-2 cells, but failed to decrease Fpn protein levels, and hypothesized that hepcidin could directly block iron export through Fpn in these cells.44 Taniguchi et al.33 similarly proposed Fpn occlusion by hepcidin based on the structure of the prokaryotic homolog BbFPN. Our refined models also predict that hepcidin binds to Fpn in the main cavity, and our mutagenesis data experimentally identified a patch of Fpn residues critical for hepcidin binding. Furthermore, we provide the first experimental evidence that hepcidin binding occludes Fpn and directly blocks iron export even without causing Fpn internalization. We report that the engineered Fpn mutant, K8R, shows severely impaired hepcidin-induced Fpn internalization and degradation, but its ability to export iron is completely inhibited by hepcidin, although requiring higher concentrations than WT. Furthermore, using Xenopus oocytes, which do not endocytose Fpn in response to hepcidin, we show that hepcidin blocks iron export by occlusion of not only K8R, but also WT Fpn.
We surmise that high concentrations of hepcidin are necessary to demonstrate the occlusion of iron transport, because at lower concentrations, many Fpn molecules may not be stably occupied by hepcidin. If so, hepcidin-induced endocytosis serves to amplify the effect of transiently bound hepcidin. The relative contribution of Fpn occlusion and endocytosis may differ depending on the cell type, its endocytic machinery, and even Fpn glycosylation, perhaps explaining the reported differences between tissues in sensitivity to hepcidin and the discrepancies between the effects of hepcidin on iron export compared with endocytosis.43 Indeed, we observed the effect of occlusion in mature human RBCs, a cell type that expresses high levels of Fpn,42 but lacks endocytic machinery: hepcidin and minihepcidin decreased iron export from RBCs without affecting Fpn levels in these cells. Similarly, injection of WT or thalassemic mice with minihepcidins profoundly decreased serum iron without lowering Fpn protein levels in the duodenum or the spleen, suggesting that serum iron rather than Fpn degradation should be the primary readout of the activity of hepcidin and its agonists in vivo. The high potency of minihepcidin in inhibiting iron export by Fpn K8R suggests that assays of Fpn iron export and FPN endocytosis measure potentially distinct activities of hepcidin agonists, and that engineered hepcidin agonists may exert a therapeutic effect even in patients exhibiting Fpn resistance to hepcidin.
The online version of this article contains a data supplement
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors would like to thank Tracey Rouault and Deliang Zhang for illuminating discussions related to ferroportin function in RBCs.
This work was supported by National Institutes of Health awards R01 DK107309 (E.N., M.J., and B.M.), R01 DK090554 (E.N. and T.G.), F31 HL129760 and T32 GM007185 (S.A.), and R01 DK095112 and R01 DK090554 (S.R.).
Authorship
Contribution: S.A., M.J., B.M., S.R., T.G., and E.N. designed the study; S.A., B.Q., E.V.V., A.C.S., T.A.R., K.R.V., D.S., G.J., C.C., S.R., and B.M. conducted the experiments and analyzed the data; S.A., M.J., B.M., T.G., and E.N. wrote the manuscript; and all authors contributed to the editing of the final manuscript.
Conflict-of-interest disclosure: E.N. and T.G. are consultants and shareholders of Intrinsic LifeSciences and Silarus Therapeutics and consultants for La Jolla Pharmaceuticals. T.G. is a consultant for Keryx Pharmaceuticals. B.M. is a recipient of a Vifor Pharma grant. S.R. is a member of the scientific advisory board and the recipient of a grant from Ionis Pharmaceuticals. The remaining authors declare no competing financial interests.
Correspondence: Elizabeta Nemeth, Department of Medicine, David Geffen School of Medicine, University of California, Los Angeles, 10833 LeConte Ave, Center for Health Sciences 37-131, Los Angeles, CA 90095; e-mail: enemeth@mednet.ucla.edu.