Key Points
HLA-DPB1 alleles diverged into 2 major groups according to highly conserved DNA sequences Ex3-3′UTR.
Two evolutionarily different HLA-DPB1 gene regions complementarily affect aGVHD in HLA-DPB1 mismatch UR-HCT.
Abstract
HLA-DPB1 T-cell epitope (TCE) mismatching algorithm and rs9277534 SNP at the 3′ untranslated region (3′UTR) in the HLA-DPB1 gene are key factors for transplant-related events in unrelated hematopoietic cell transplantation (UR-HCT). However, the association of these 2 mechanisms has not been elucidated. We analyzed 19 frequent HLA-DPB1 alleles derived from Japanese healthy subjects by next-generation sequencing of the entire HLA-DPB1 gene region and multi-SNP data of the HLA region in 1589 UR-HCT pairs. The risk of acute graft-versus-host disease (aGVHD) was analyzed in 1286 patients with single HLA-DPB1 mismatch UR-HCT. The phylogenetic tree constructed using the entire gene region demonstrated that HLA-DPB1 alleles were divided into 2 groups, HLA-DP2 and HLA-DP5. Although a phylogenetic relationship in the genomic region from exon 3 to 3′UTR (Ex3-3′UTR) obviously supported the division of HLA-DP2 and HLA-DP5 groups, which in exon 2 showed intermingling of HLA-DPB1 alleles in a non–HLA-DP2 and non–HLA-DP5-group manner. Multi-SNP data also showed 2 discriminative HLA-DPB1 groups according to Ex3-3′UTR. Risk of grade 2-4 aGVHD was significantly higher in patient HLA-DP5 group mismatch than patient HLA-DP2 group mismatch (hazard ratio, 1.28; P = .005), regardless of donor mismatch HLA-DP group. Regarding TCE mismatch, increasing risk of aGVHD in patient HLA-DP5 group mismatch and TCE-nonpermissive mismatch were observed only in patients with TCE-permissive mismatch and patient HLA-DP2 group mismatch, respectively. Evolutionary analysis revealed that rs9277534 represented a highly conserved HLA-DPB1 Ex3-3′UTR region and may provoke aGVHD differently to TCE mismatching algorithm, reflecting exon 2 polymorphisms. These findings enrich our understanding of the mechanism of aGVHD in HLA-DPB1 mismatch UR-HCT.
Introduction
Hematopoietic cell transplantation (HCT) from an unrelated (UR) donor is now an established mode of curative therapy for hematologic malignancies and other hematological or immunologic disorders. Patient-donor HLA locus mismatch induces strong alloreactivity, and affects graft-versus-host disease (GVHD), engraftment failure, and transplant-related mortality after UR-HCT.1-3 In the practical clinical setting, HLA-A, HLA-B, HLA-C, and HLA-DRB1 matched or HLA-A, HLA-B, HLA-C, HLA-DRB1, and HLA-DQB1 (HLA-10/10)- matched donors are selected as optimal donors. Although HLA-DPB1 typing is not routinely performed for donor selection, retrospective clinical studies for HLA-DPB1 matching provide clear evidence that HLA-DPB1 mismatch in UR-HCT is associated with an increased risk of acute GVHD and reduced risk of leukemia relapse.3-5
The HLA-DPB1 gene consists of 5 coding exons, 1 noncoding exon (3′ untranslated region [3′UTR]) and 5 introns and is located at the most centromere site in the HLA region. A recombination hot spot between the HLA-DQ and DP loci makes it difficult to find HLA-DPB1–matched unrelated donors even among HLA-10/10–matched patient-donor pairs.6,7
HLA-DP molecules were originally defined using the primed lymphocyte test, which indicated the distinctive cellular immune reactivity derived from this molecule.8,9 Based on the in vitro cross-reactivity pattern of the T-cell clone obtained from a patient at the time of rejection after HLA-DPB1–mismatched allogeneic transplantation, an algorithm for the prediction of HLA-DPB1 permissive mismatches (PMs) and nonpermissive mismatches (non-PMs) was constructed using the presence of T-cell epitope (TCE) mismatching.10 Retrospective analyses using large UR-HCT data demonstrated that non-PM HLA-DPB1 TCE were associated with higher risk of acute GVHD, mortality, or both.11,12
A recent report noted that the risk of acute GVHD associated with HLA-DPB1 mismatch was influenced by the SNP rs9277534 in the 3′UTR of the HLA-DPB1 gene.13 This variant is also known as marker for HLA-DP expression level.
A structural study demonstrated that the crystal structure of HLA-DP5/peptide complex is distinct from HLA-DP2/peptide complex, and an evolutional analysis showed that the HLA-DP5 and HLA-DP2 groups represent 2 major groups of the HLA-DP family.14
Based partly on these findings, we hypothesized that evolutionary differences in HLA-DP molecules are related to transplant-related immunological events and may induce alloreactivity by a different mechanism from the TCE mismatching algorithm, which reflects polymorphisms in exon 2 of HLA-DPB1 gene.
To facilitate precise HLA typing without phase ambiguity, we previously developed the super high-resolution single molecule sequence-based typing method, which combines long-range polymerase chain reaction (PCR) amplification and next-generation sequencing (NGS) technologies for 8 classical HLA loci. This method provides extremely high-resolution typing to a field 4 level, including nucleotide differences in both the coding and noncoding regions of HLA genes.15
Here, to better understand the influence of HLA-DPB1 gene regions in acute GVHD in HLA-DPB1 mismatch UR-HCT, we determined the genomic sequences of 19 major HLA-DPB1 alleles in healthy Japanese that cover the entire HLA-DPB1 gene region, ranging from the promoter-enhancer region to 3′UTR, using the super high-resolution single molecule sequence-based typing method. We also clarified the phylogenetic relationships among the HLA-DPB1 alleles and analyzed multi-SNP data in the HLA region of a large number of unrelated individuals. Finally, we analyzed the clinical data of 1286 unrelated bone marrow transplantation (UR-BMT) pairs to examine the GVHD risk associated with the HLA-DP group as defined by phylogenetic analysis; we further examined the association between the HLA-DP group and HLA-DPB1 TCE mismatching algorithm.
Methods
Study population
For sequencing analysis of the entire region of the HLA-DPB1 gene by NGS, we used 19 genomic samples from healthy Japanese subjects who represented a distribution of more than 99.5% of HLA-DPB1 alleles by field 2 level typing.16 We analyzed multi-SNP data of the HLA region in 1589 pairs (3178 individuals) following UR-BMT through the Japan Marrow Donor Program (JMDP) from 1993 to 2005 and performed genome-wide association studies.17
Transplant outcomes were assessed in 1286 UR-BMT pairs from the JMDP database who met the following criteria and were included in the analysis: (1) patients transplanted from donors who were HLA-10/10–matched and had only 1 HLA-DPB1 mismatch in the graft-versus-host (GVH) direction; (2) transplantation pairs were retyped for HLA-10/10 and HLA-DPB1 alleles; (3) transplanted non–T cell-depleted marrow without in vivo use of antithymocyte globulin for GVHD prophylaxis; (4) first transplantation; (5) Japanese ethnicity; and (6) survival >7 days after transplantation. Patient and donor characteristics are shown in Table 1. A final clinical survey of the patients was completed by September 2012. Informed consent was obtained from patients and donors in accordance with the Declaration of Helsinki, and approval of the study was obtained from the Institutional Review Board of Aichi Cancer Center, Fujita Health University, Tokai University, and JMDP.
Patient and transplant characteristics
| . | Patient mismatch HLA-DP group . | . | |||
|---|---|---|---|---|---|
| . | HLA-DP2 group (N = 657) . | HLA-DP5 group (N = 629) . | P . | ||
| Median patient age (range), y | 36 | (0-77) | 36 | (1-68) | .41 |
| Median donor age (range), y | 35 | (20-55) | 34 | (20-58) | .71 |
| Years of transplantation, no. (%) | |||||
| 1993-2000 | 204 | (31.1) | 197 | (31.3) | .92 |
| 2001-2005 | 247 | (37.6) | 230 | (36.6) | |
| 2006-2010 | 206 | (31.4) | 202 | (32.1) | |
| Patient-donor sex, no. (%) | |||||
| Male-male | 275 | (41.9) | 269 | (42.8) | .62 |
| Female-female | 115 | (17.5) | 109 | (17.3) | |
| Male-female | 167 | (25.4) | 143 | (22.7) | |
| Female-male | 100 | (15.2) | 108 | (17.2) | |
| Disease, no. (%) | |||||
| ALL | 201 | (30.6) | 189 | (30.0) | .99 |
| AML | 282 | (42.9) | 269 | (42.8) | |
| MDS | 85 | (12.9) | 82 | (13.0) | |
| CML | 89 | (13.5) | 89 | (14.1) | |
| Leukemia risk, no. (%) | |||||
| Standard | 383 | (58.3) | 324 | (51.5) | .05 |
| High | 242 | (36.8) | 272 | (43.2) | |
| Other | 32 | (4.9) | 33 | (5.2) | |
| GVHD prophylaxis, no. (%) | |||||
| Cyclosporine based | 294 | (44.7) | 294 | (46.7) | .56 |
| Tacrolimus based | 348 | (53.0) | 325 | (51.7) | |
| Other | 15 | (2.3) | 10 | (1.6) | |
| Conditioning regimen, no. (%) | |||||
| Nonmyeloablative | 83 | (12.6) | 92 | (14.6) | .39 |
| Myeloablative | 541 | (82.3) | 499 | (79.3) | |
| NA | 33 | (5.0) | 38 | (6.0) | |
| HLA-DPB1 TCE mismatching, no. (%) | |||||
| Permissive | 566 | (90.7) | 388 | (61.7) | <.001 |
| Nonpermissive | 91 | (13.9) | 241 | (38.3) | |
| . | Patient mismatch HLA-DP group . | . | |||
|---|---|---|---|---|---|
| . | HLA-DP2 group (N = 657) . | HLA-DP5 group (N = 629) . | P . | ||
| Median patient age (range), y | 36 | (0-77) | 36 | (1-68) | .41 |
| Median donor age (range), y | 35 | (20-55) | 34 | (20-58) | .71 |
| Years of transplantation, no. (%) | |||||
| 1993-2000 | 204 | (31.1) | 197 | (31.3) | .92 |
| 2001-2005 | 247 | (37.6) | 230 | (36.6) | |
| 2006-2010 | 206 | (31.4) | 202 | (32.1) | |
| Patient-donor sex, no. (%) | |||||
| Male-male | 275 | (41.9) | 269 | (42.8) | .62 |
| Female-female | 115 | (17.5) | 109 | (17.3) | |
| Male-female | 167 | (25.4) | 143 | (22.7) | |
| Female-male | 100 | (15.2) | 108 | (17.2) | |
| Disease, no. (%) | |||||
| ALL | 201 | (30.6) | 189 | (30.0) | .99 |
| AML | 282 | (42.9) | 269 | (42.8) | |
| MDS | 85 | (12.9) | 82 | (13.0) | |
| CML | 89 | (13.5) | 89 | (14.1) | |
| Leukemia risk, no. (%) | |||||
| Standard | 383 | (58.3) | 324 | (51.5) | .05 |
| High | 242 | (36.8) | 272 | (43.2) | |
| Other | 32 | (4.9) | 33 | (5.2) | |
| GVHD prophylaxis, no. (%) | |||||
| Cyclosporine based | 294 | (44.7) | 294 | (46.7) | .56 |
| Tacrolimus based | 348 | (53.0) | 325 | (51.7) | |
| Other | 15 | (2.3) | 10 | (1.6) | |
| Conditioning regimen, no. (%) | |||||
| Nonmyeloablative | 83 | (12.6) | 92 | (14.6) | .39 |
| Myeloablative | 541 | (82.3) | 499 | (79.3) | |
| NA | 33 | (5.0) | 38 | (6.0) | |
| HLA-DPB1 TCE mismatching, no. (%) | |||||
| Permissive | 566 | (90.7) | 388 | (61.7) | <.001 |
| Nonpermissive | 91 | (13.9) | 241 | (38.3) | |
Leukemia risk: standard risk indicates the first or second complete remission of AML; first complete remission of ALL; and first chronic phase of CML, refractory anemia, and refractory anemia with ringed sideroblasts. Advanced risk indicates all other leukemias. Conditioning regimen: Myeloablative regimen indicates conditioning regimens with total body irradiation >8 Gy, oral busulfan ≥9 mg/kg, intravenous busulfan ≥7.2 mg/kg, or melphalan >140 mg/m2; reduced-intensity regimen indicates all other conditioning regimens.
ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia; CML, chronic myeloid leukemia; MDS, myelodysplastic syndrome; NA, not available.
HLA typing and SNP genotyping in patients and donors
All donor-patient pairs were retrospectively genotyped for HLA-10/10 and HLA-DPB1 alleles at the field 1 and field 2 level using either or both the PCR sequence-specific oligonucleotide and PCR-sequencing-based typing methods, as described elsewhere.3
A total of 1589 donor and patient pairs were genotyped for 500 568 SNPs using the Affymetrix Human Mapping 500K Array Set. Genotypes of unobserved SNPs were imputed on the basis of the published HapMap data as described previously.17 In this study, we analyzed 3246 SNPs in the HLA-DPB1 gene region. Schematic diagram of multi-SNP analysis of HLA-DPB1 haplotypes is shown in supplemental Figure 1, available on the Blood Web site.
Nucleotide sequence determination of HLA-DPB1 gene for 19 healthy individuals
HLA-DPB1 specific primers, long-range PCR, and purification and quantitation of the PCR products were performed according to a previous report.15 Preparation of barcoded-library measurement of DNA size and quantitation for each library, mixture of each barcoded library, emulsion PCR, Ion Personal Genome Machine sequencing, data processing, and allele assignment were performed according to previously published reports.16,18
Phylogenic analysis
Multiple sequence alignments were created using the ClustalW Sequence Alignment program of the Molecular Evolution Genetics Analysis software 6.19 Nucleotide alignments were separately created for 3 datasets, namely 12 149 bp of the entire HLA-DPB1 region, 2668 bp from the exon 3 to 3′UTR region, and 264 bp of the exon 2 region.
A detailed description of the methods of phylogenetic analysis is available in the supplemental Methods.
Statistical analysis of transplant outcomes
First, we grouped patient and donor HLA-DPB1 alleles into 2 HLA-DP groups according to the results of phylogenic analysis. The effects of patient mismatch HLA-DP group (HLA-DP group of nonshared HLA-DPB1 allele with donor) on transplant outcomes were assessed (supplemental Figure 2). Patient-, disease-, and transplantation-related factors were compared between patient mismatch HLA-DP groups using the χ2 test for categorical variables and the Student t test for continuous variables. HLA-DPB1 PM or non-PM (TCE mismatching) were defined according to the previously published algorithm.10,11 Multivariable competing risk regression analyses20 were conducted to evaluate the impact of acute GVHD, leukemia relapse, and transplant-related mortality. Confounders considered were sex (donor-recipient pair), patient age (linear), donor age (linear), disease, risk of leukemia relapse (standard and high), GVHD prophylaxis (cyclosporine-based regimen, tacrolimus-based regimen, and the other regimen without cyclosporine or tacrolimus), and conditioning regimen (reduced-intensity conditioning and myeloablative conditioning). Conditioning regimens were classified as myeloablative when total body irradiation was >8 Gy, oral busulfan was ≥9 mg/kg, intravenous busulfan was ≥7.2 mg/kg, or melphalan was >140 mg/m2, in accordance with Giralt et al.21 A competing event regarding acute GVHD and leukemia relapse was defined as death without acute GVHD and death without relapse, respectively. For transplant-related mortality, relapse was the competing event. An adjusted comparison of the groups with regard to overall survival was performed using Cox’s proportional hazards regression model.22 The cumulative incidence of acute GVHD was assessed by a method described elsewhere to eliminate the effect of competing risk.23
Results
HLA typing and evolutional analysis of HLA-DPB1 genes
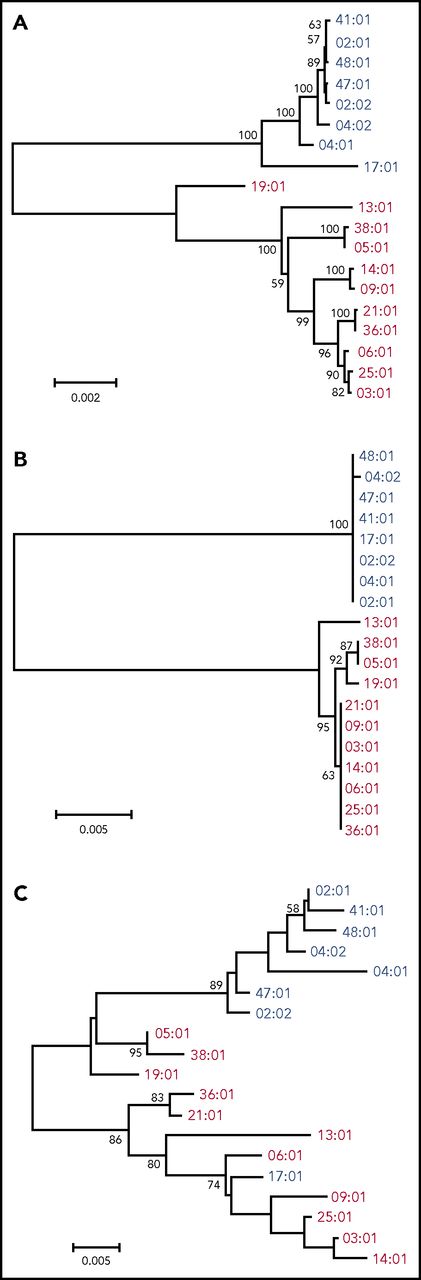
Nineteen kinds of HLA allele sequence to the field 4 level were determined by NGS of the entire gene region (Table 2). First, the phylogenetic trees were constructed with the entire gene sequencing data (aligned length: 12 149 bp, Figure 1A). HLA-DPB1 alleles diverged into 2 major groups, namely HLA-DP5 and HLA-DP2 groups, as described in a previous report.14 We also constructed a phylogenetic tree constructed using phylogenetic trees from exon 2 and from exon 3 to 3′UTR (Ex3-3′UTR) separately, because the great majority of nucleotide polymorphisms occur in exon 2 of the HLA class II genes. The phylogenetic tree from Ex3-3′UTR (aligned length, 2668 bp; Figure 1B) showed a similar structure to that of the tree of the whole gene, and the grouping pattern of the 2 HLA-DP groups was more obvious in Ex3-3′UTR than in the whole gene. The phylogenetic tree of allelic lineages for HLA-DPB1 exon 2 also diverged into 2 major groups, although the phylogenetic relationship among HLA-DPB1 alleles was ambiguous compared with that of Ex3-3′UTR (aligned length, 264 bp; Figure 1C). We confirmed allelic relationships for exon 2 using the maximum likelihood method and Bayesian inference method (supplemental Figure 3), which showed similar structures. These data indicated that each of the HLA-DP2 and HLA-DP5 groups was highly conserved in Ex3-3′UTR, whereas exon 2 showed more diversity and was not as conserved as Ex3-3′UTR.
HLA DPB1 allele information used for this study
| Allele name at field 2 level . | Allele frequency (n = 2966 alleles),* % . | Allele name at field 4 level . | Accession no. . | Predicted immunogenicity† . | rs9277534 allele . | Grouping for DP2 or DP5‡ . |
|---|---|---|---|---|---|---|
| DPB1*05:01 | 38.40 | DPB1*05:01:01:01 | LC257912 | Group 3 | G | DP5 |
| DPB1*02:01 | 24.11 | DPB1*02:01:02:01 | LC257894 | Group 3 | A | DP2 |
| DPB1*09:01 | 9.95 | DPB1*09:01:01 | LC257921 | Group 1 | G | DP5 |
| DPB1*04:02 | 9.78 | DPB1*04:02:01:02 | LC257910 | Group 3 | A | DP2 |
| DPB1*04:01 | 5.06 | DPB1*04:01:01:01 | LC257908 | Group 3 | A | DP2 |
| DPB1*03:01 | 3.98 | DPB1*03:01:01 | LC257905 | Group 2 | G | DP5 |
| DPB1*02:02 | 3.41 | DPB1*02:02:01:05 | LC257902 | Group 3 | A | DP2 |
| DPB1*13:01 | 1.96 | DPB1*13:01:01 | LC257922 | Group 3 | G | DP5 |
| DPB1*14:01 | 1.48 | DPB1*14:01:01 | LC257924 | Group 2 | G | DP5 |
| DPB1*19:01 | 0.74 | DPB1*19:01 | LC257926 | Group 3 | G | DP5 |
| DPB1*06:01 | 0.57 | DPB1*06:01:01 | LC257920 | Group 3 | G | DP5 |
| DPB1*17:01 | 0.14 | DPB1*17:01 | LC257925 | Group 1 | A | DP2 |
| DPB1*36:01 | 0.14 | DPB1*36:01 | LC257929 | Unknown | G | DP5 |
| DPB1*41:01 | 0.10 | DPB1*41:01:01 | LC257931 | Group 3 | A | DP2 |
| DPB1*38:01 | 0.07 | DPB1*38:01 | LC257930 | Group 3 | G | DP5 |
| DPB1*25:01 | 0.03 | DPB1*25:01 | LC257928 | Unknown | G | DP5 |
| DPB1*47:01 | 0.03 | DPB1*47:01 | LC257932 | Group 3 | A | DP2 |
| DPB1*21:01 | Unknown | DPB1*21:01 | LC257927 | Unknown | G | DP5 |
| DPB1*48:01 | Unknown | DPB1*48:01 | LC257933 | Group3 | A | DP2 |
| Allele name at field 2 level . | Allele frequency (n = 2966 alleles),* % . | Allele name at field 4 level . | Accession no. . | Predicted immunogenicity† . | rs9277534 allele . | Grouping for DP2 or DP5‡ . |
|---|---|---|---|---|---|---|
| DPB1*05:01 | 38.40 | DPB1*05:01:01:01 | LC257912 | Group 3 | G | DP5 |
| DPB1*02:01 | 24.11 | DPB1*02:01:02:01 | LC257894 | Group 3 | A | DP2 |
| DPB1*09:01 | 9.95 | DPB1*09:01:01 | LC257921 | Group 1 | G | DP5 |
| DPB1*04:02 | 9.78 | DPB1*04:02:01:02 | LC257910 | Group 3 | A | DP2 |
| DPB1*04:01 | 5.06 | DPB1*04:01:01:01 | LC257908 | Group 3 | A | DP2 |
| DPB1*03:01 | 3.98 | DPB1*03:01:01 | LC257905 | Group 2 | G | DP5 |
| DPB1*02:02 | 3.41 | DPB1*02:02:01:05 | LC257902 | Group 3 | A | DP2 |
| DPB1*13:01 | 1.96 | DPB1*13:01:01 | LC257922 | Group 3 | G | DP5 |
| DPB1*14:01 | 1.48 | DPB1*14:01:01 | LC257924 | Group 2 | G | DP5 |
| DPB1*19:01 | 0.74 | DPB1*19:01 | LC257926 | Group 3 | G | DP5 |
| DPB1*06:01 | 0.57 | DPB1*06:01:01 | LC257920 | Group 3 | G | DP5 |
| DPB1*17:01 | 0.14 | DPB1*17:01 | LC257925 | Group 1 | A | DP2 |
| DPB1*36:01 | 0.14 | DPB1*36:01 | LC257929 | Unknown | G | DP5 |
| DPB1*41:01 | 0.10 | DPB1*41:01:01 | LC257931 | Group 3 | A | DP2 |
| DPB1*38:01 | 0.07 | DPB1*38:01 | LC257930 | Group 3 | G | DP5 |
| DPB1*25:01 | 0.03 | DPB1*25:01 | LC257928 | Unknown | G | DP5 |
| DPB1*47:01 | 0.03 | DPB1*47:01 | LC257932 | Group 3 | A | DP2 |
| DPB1*21:01 | Unknown | DPB1*21:01 | LC257927 | Unknown | G | DP5 |
| DPB1*48:01 | Unknown | DPB1*48:01 | LC257933 | Group3 | A | DP2 |
These data are available at the HLA laboratory Web page (http://www.hla.or.jp/haplo/haplonavi).
These data are based on Kusano et al14 and our study.
Phylogenetic analysis for HLA-DPB1 alleles. Phylogenetic trees were constructed using the Neighbor-Joining method. Numbers on the branches are bootstrap support values. Blue and red letters indicate DP2 group (rs9277534: A) and DP5 group (rs9277534: G) alleles, respectively. (A) The 12 149-bp nucleotide alignment for HLA-DPB1 whole gene was used. (B) The 2668-bp nucleotide alignment for the region from exon 3 to 3′UTR was used. (C) The 264-bp nucleotide alignment for exon 2 was used.
Phylogenetic analysis for HLA-DPB1 alleles. Phylogenetic trees were constructed using the Neighbor-Joining method. Numbers on the branches are bootstrap support values. Blue and red letters indicate DP2 group (rs9277534: A) and DP5 group (rs9277534: G) alleles, respectively. (A) The 12 149-bp nucleotide alignment for HLA-DPB1 whole gene was used. (B) The 2668-bp nucleotide alignment for the region from exon 3 to 3′UTR was used. (C) The 264-bp nucleotide alignment for exon 2 was used.
All HLA-DPB1 alleles in the HLA-DP2 group had the previously published HLA-DP expression marker rs9277534A allele (low expression marker), whereas those in HLA-DP5 group had the rs9277534G allele (high expression marker) (Table 2).
Multi-SNP analysis of HLA-DPB1 haplotypes
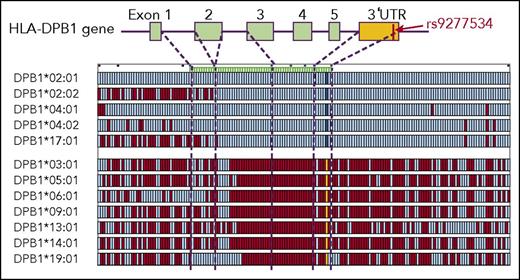
We previously demonstrated that the common Japanese HLA haplotypes (HP-P1; HLA-A*24:02 -Cw*12:02 -B*52:01 -DRB1*15:02 -DQB1*06:01 -DPB1*09:01, HP-P2; HLA-A*3303 -Cw*14:03 -B*44:03 -DRB1*13:02 -DQB1*06:04 -DPB1*04:01, HP-P3; HLA-A*24:02 -Cw*07:02 -B*07:02 -DRB1*01:01 -DQB1*05:01 -DPB1*04:02) were essentially conserved at least in the 3.3-Mb HLA region from HLA-A to HLA-DPB1 using multi-SNP data.24 Individuals with homozygous HP-P1, HLA-P2 and HLA-P3 had consecutive homozygous SNPs in the HLA region, and the consensus sequences of these haplotypes were easily determined. A total of 1604 individuals carried at least 1 copy of any of HP-P1, HLA-P2, or HLA-P3; that is, they shared the same HLA-10/10 and HLA-DPB1 alleles of these HLA haplotypes, and had identical alleles for more than 99.5% of 3246 consecutive SNPs across the 2 Mb HLA class II region as consensus sequences of these haplotypes. We identified SNP sequences of opposite haplotypes in these individuals and made haplotype groups that had identical HLA-DPB1. We found 12 HLA-DPB1 haplotype groups with more than 5 individual haplotypes (HLA-DPB1*02:01, n = 369; *02:02, n = 68; *03:01, n = 60; *04:01, n = 79; *04:02, n = 194; *05:01, n = 585; *06:01, n = 16; *09:01, n = 183; *13:01, n = 18; *14:01, n = 16; *17:01, n = 6; *19:01, n = 5). To evaluate the homogeneity of each HLA-DPB1 haplotype group, the SNP sequence of individual haplotypes was compared with the consensus sequence of the HLA-DPB1 haplotype group. Supplemental Figure 4 compares HLA-DPB1*02:01 haplotypes and the consensus sequence of this haplotype group. The HLA-DPB1*02:01 haplotype group had a highly conserved region around the HLA-DPB1 gene that extended beyond 3′UTR. Similarly, other HLA-DPB1 haplotype groups also had highly conserved regions (data not shown).
To compare HLA-DPB1 haplotype groups, 1 representative individual haplotype from each haplotype group was compared with the HLA-DPB1*02:01 haplotype (Figure 2). HLA-DPB1 haplotypes were clearly divided into 2 groups according to SNP pattern around the HLA-DPB1 gene. The HLA-DP2 group, which included HLA-DPB1*02:01, HLA-DPB1*02:02, HLA-DPB1*04:01, HLA-DPB1*04:02, and HLA-DPB1*17:01, had completely identical SNP sequences from intron 2 to the centromere region of HLA-DPB1. The HLA-DP5 group, which included HLA-DPB1*03:01, HLA-DPB1*05:01, HLA-DPB1*06:01, HLA-DPB1*09:01, HLA-DPB1*13:01, HLA-DPB1*14:01, and HLA-DPB1*19:01, also had identical SNP sequences from intron 2 to the centromere region of HLA-DPB1. The HLA-DP2 and HLA-DP5 groups had completely different SNP sequences. HLA-DPB1 expression marker rs9277534 was 1 tag SNP in the highly conserved region (ie, the HLA-DP2 group had rs9277534A, whereas the HLA-DP5 group had rs9277534G).
Comparison of SNP sequence among HLA-DPB1 haplotypes. One representative individual haplotype from each haplotype group was compared with the HLA-DPB1*02:01 haplotype. Each row indicates 1 haplotype. Aqua, identical SNPs to the alleles of HLA-DPB1*02:01 haplotype; red, SNPs that differ from them; cobalt blue, SNP and yellow SNP at 3′UTR of HLA-DPB1 gene indicate the rs9277534 A allele and G alleles, respectively; dashed lines, location of SNPs within the HLA-DPB1 gene.
Comparison of SNP sequence among HLA-DPB1 haplotypes. One representative individual haplotype from each haplotype group was compared with the HLA-DPB1*02:01 haplotype. Each row indicates 1 haplotype. Aqua, identical SNPs to the alleles of HLA-DPB1*02:01 haplotype; red, SNPs that differ from them; cobalt blue, SNP and yellow SNP at 3′UTR of HLA-DPB1 gene indicate the rs9277534 A allele and G alleles, respectively; dashed lines, location of SNPs within the HLA-DPB1 gene.
Association between patient mismatch HLA-DP group and clinical outcome
Because the HLA-DP2 group and HLA-DP5 group corresponded to rs9277534A and rs9277534G in HLA-DPB1 gene 3′UTR, respectively, we examined the effect of patient mismatch HLA-DP group on various clinical outcomes after transplantation (Table 3). In this analysis, we restricted the patient-donor pairs with only 1 HLA-DPB1 mismatch in the GVH direction (patient and donor had 1 matched HLA-DPB1 and 1 mismatched HLA-DPB1 allele in the GVH direction) to avoid any effect of disparity at other HLA loci. We excluded pairs with HLA-DPB1 mismatch only in the host-versus-graft direction. The patient mismatch HLA-DP5 group (n = 629) showed significantly higher risk of grade 2-4 acute GVHD than the HLA-DP2 group (n = 657) (hazard ratio [HR], 1.28; 95% confidence interval [CI], 1.07-1.52; P = .005). Cumulative incidence of grade 2-4 acute GVHD in patients in the mismatch DP2 group and the mismatch DP5 group was 36.3% (95% CI, 33.0-39.6) and 43.7% (95% CI, 40.3-47.1), respectively. On the other hand, there were no significant differences between the 2 groups in grade 3-4 acute GVHD (HR, 1.18; 95% CI, 0.88-1.57; P = .268), leukemia relapse (HR, 0.83; 95% CI, 0.66-1.05; P = .121), transplant-related mortality (HR,.1.01; 95% CI, 0.81-1.28; P = .905), or overall mortality (HR, 0.99; 95% CI, 0.83-1.17; P = .877).
Effect of patient mismatch HLA-DP group on transplant outcome
| Clinical outcome . | Patient mismatch HLA-DP group . | HR . | 95% CI . | P . | N . |
|---|---|---|---|---|---|
| Acute GVHD 2-4 | |||||
| All patients | DP2 | 1.00 | 657 | ||
| DP5 | 1.28 | 1.07-1.52 | .005 | 629 | |
| Donor HLA-DP2 mismatch | DP2 | 1.00 | 241 | ||
| DP5 | 1.32 | 1.01-1.72 | .040 | 378 | |
| Donor HLA-DP5 mismatch | DP2 | 1.00 | 416 | ||
| DP5 | 1.35 | 1.06-1.72 | .013 | 251 | |
| Acute GVHD 3-4 | |||||
| All patients | DP2 | 1.00 | 657 | ||
| DP5 | 1.18 | 0.88-1.57 | .268 | 629 | |
| Donor HLA-DP2 mismatch | DP2 | 1.00 | 241 | ||
| DP5 | 1.26 | 0.80-1.99 | .312 | 378 | |
| Donor HLA-DP5 mismatch | DP2 | 1.00 | 416 | ||
| DP5 | 1.20 | 0.81-1.78 | .357 | 251 | |
| Relapse | |||||
| All patients | DP2 | 1.00 | 657 | ||
| DP5 | 0.83 | 0.66-1.05 | .121 | 629 | |
| Donor HLA-DP2 mismatch | DP2 | 1.00 | 241 | ||
| DP5 | 0.83 | 0.58-1.17 | .288 | 378 | |
| Donor HLA-DP5 mismatch | DP2 | 1.00 | 416 | ||
| DP5 | 0.86 | 0.62-1.20 | .381 | 251 | |
| Transplant-related mortality | |||||
| All patients | DP2 | 1.00 | 657 | ||
| DP5 | 1.01 | 0.81-1.28 | .905 | 629 | |
| Donor HLA-DP2 mismatch | DP2 | 1.00 | 241 | ||
| DP5 | 1.10 | 0.78-1.56 | .589 | 378 | |
| Donor HLA-DP5 mismatch | DP2 | 1.00 | 416 | ||
| DP5 | 0.92 | 0.66-1.29 | .625 | 251 | |
| Overall mortality | |||||
| All patients | DP2 | 1.00 | 657 | ||
| DP5 | 0.99 | 0.83-1.17 | .877 | 629 | |
| Donor HLA-DP2 mismatch | DP2 | 1.00 | 241 | ||
| DP5 | 1.08 | 0.84-1.39 | .560 | 378 | |
| Donor HLA-DP5 mismatch | DP2 | 1.00 | 416 | ||
| DP5 | 0.90 | 0.70-1.15 | .393 | 251 |
| Clinical outcome . | Patient mismatch HLA-DP group . | HR . | 95% CI . | P . | N . |
|---|---|---|---|---|---|
| Acute GVHD 2-4 | |||||
| All patients | DP2 | 1.00 | 657 | ||
| DP5 | 1.28 | 1.07-1.52 | .005 | 629 | |
| Donor HLA-DP2 mismatch | DP2 | 1.00 | 241 | ||
| DP5 | 1.32 | 1.01-1.72 | .040 | 378 | |
| Donor HLA-DP5 mismatch | DP2 | 1.00 | 416 | ||
| DP5 | 1.35 | 1.06-1.72 | .013 | 251 | |
| Acute GVHD 3-4 | |||||
| All patients | DP2 | 1.00 | 657 | ||
| DP5 | 1.18 | 0.88-1.57 | .268 | 629 | |
| Donor HLA-DP2 mismatch | DP2 | 1.00 | 241 | ||
| DP5 | 1.26 | 0.80-1.99 | .312 | 378 | |
| Donor HLA-DP5 mismatch | DP2 | 1.00 | 416 | ||
| DP5 | 1.20 | 0.81-1.78 | .357 | 251 | |
| Relapse | |||||
| All patients | DP2 | 1.00 | 657 | ||
| DP5 | 0.83 | 0.66-1.05 | .121 | 629 | |
| Donor HLA-DP2 mismatch | DP2 | 1.00 | 241 | ||
| DP5 | 0.83 | 0.58-1.17 | .288 | 378 | |
| Donor HLA-DP5 mismatch | DP2 | 1.00 | 416 | ||
| DP5 | 0.86 | 0.62-1.20 | .381 | 251 | |
| Transplant-related mortality | |||||
| All patients | DP2 | 1.00 | 657 | ||
| DP5 | 1.01 | 0.81-1.28 | .905 | 629 | |
| Donor HLA-DP2 mismatch | DP2 | 1.00 | 241 | ||
| DP5 | 1.10 | 0.78-1.56 | .589 | 378 | |
| Donor HLA-DP5 mismatch | DP2 | 1.00 | 416 | ||
| DP5 | 0.92 | 0.66-1.29 | .625 | 251 | |
| Overall mortality | |||||
| All patients | DP2 | 1.00 | 657 | ||
| DP5 | 0.99 | 0.83-1.17 | .877 | 629 | |
| Donor HLA-DP2 mismatch | DP2 | 1.00 | 241 | ||
| DP5 | 1.08 | 0.84-1.39 | .560 | 378 | |
| Donor HLA-DP5 mismatch | DP2 | 1.00 | 416 | ||
| DP5 | 0.90 | 0.70-1.15 | .393 | 251 |
HR indicates comparison of the patient mismatch HLA-DP5 group to HLA-DP2 group.
Because an increasing risk of grade 2-4 acute GVHD was observed in the patient mismatch HLA-DP5 group, we analyzed whether this effect was consistently observed regardless of donor mismatch HLA-DP group. We compared the effect of patient mismatch HLA-DP group on acute GVHD separately in the donor mismatch HLA-DP5 group and patient mismatch HLA-DP2 group (Table 3). The increasing risk of grade 2-4 acute GVHD in the patient mismatch HLA-DP5 group compared with patient mismatch HLA-DP2 group was apparent in both patients receiving transplants from donors with the mismatch DP2 group (HR, 1.32; 95% CI, 1.01-1.72; P = .040) and donors with the mismatch DP5 group (HR, 1.35; 95% CI, 1.06-1.72; P = .013). There were no significant differences between patient mismatch HLA-DP2 and HLA-DP5 groups in other clinical outcomes regardless of donor HLA-DP mismatch group.
Relation of patient mismatch HLA-DP group and TCE mismatching
Recent analyses suggest that PM and non-PM donor-recipient HLA-DPB1 can be defined by the TCE mismatching algorithm, and non-PM is considered to be associated with increased overall morality after unrelated hematopoietic stem cell transplantation.10-12 In our present study, among 1286 donor-recipient pairs with only 1 HLA-DPB1 mismatch in the GVH direction, 954 pairs were HLA-DPB1 PM, whereas the remaining 332 pairs were non-PM. Non-PM was associated with a higher risk of grade 2-4 acute GVHD (HR, 1.24; 95% CI, 1.02-1.49; P = .028) and grade 3-4 acute GVHD (HR, 1.37; 95% CI, 1.00-1.86; P = .047) than PM, whereas no significant difference was seen in leukemia relapse, transplant-related mortality, or overall mortality between the non-PM and PM (supplemental Table 1).
Among 657 pairs with patient HLA-DP2 group mismatch, only 91 pairs (13.9%) were non-PM. On the other hand, among 629 pairs with patient HLA-DP5 mismatch, 241 pairs (38.3%) were non-PM, showing a significant correlation between patient mismatch HLA-DP group and HLA-DPB1 TCE mismatching (P < .001).
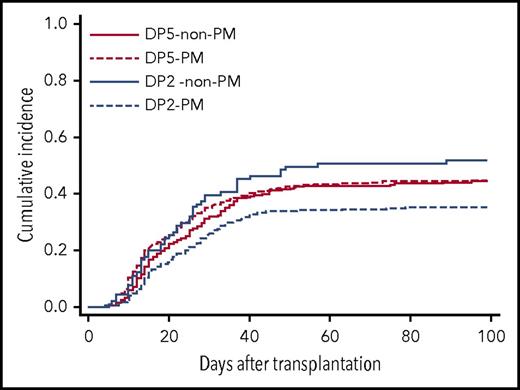
To clarify the effect of patient mismatch HLA-DP group on acute GVHD in relation to TCE mismatching, the patients were divided into 4 groups (Table 4). An increasing risk of acute GVHD with patient mismatch HLA-DP5 was apparent in the TCE PM group (HR, 1.42; 95% CI, 1.15-1.75; P = .001), whereas this effect was not observed in the TCE non-PM group (HR, 0.76; 95% CI, 0.53-1.09; P = .138). Among the patient mismatch HLA-DP2 group, the risk of acute GVHD in TCE non-PM was significantly higher than in TCE PM (HR, 1.67; 95% CI, 1.20-2.31; P = .002), whereas there was no significant difference in the risk of acute GVHD between TCE PM and TCE non-PM in the patient mismatch HLA-DP5 group. The cumulative incidence of grade 2-4 acute GVHD in these groups was as follows: patient mismatch HLA-DP2 group and TCE-PM (DP2-PM), 35.2% (95% CI, 31.3-39.1); patient mismatch HLA-DP2 group and TCE non-PM (DP2 non-PM), 51.6% (95% CI, 41.0-61.3); patient mismatch HLA-DP5 group and TCE-PM (DP5-PM), 44.4% (95% CI, 39.4-49.2); and patient mismatch HLA-DP5 group and TCE non-PM (DP5 non-PM), 44.4% (95% CI, 38.0-50.5) (Figure 3). These data suggested that an increasing risk of acute GVHD induced by patient mismatch HLA-DP5 would become apparent when patients received transplants from donors with HLA-DPB1 TCE PM. Likewise, the risk of HLA-DPB1 TCE non-PM might become apparent when patient mismatch HLA-DPB1 is the HLA-DP2 group. On the other hand, patient mismatch HLA-DP5 and HLA-DPB1 TCE non-PM might have no increasing risk effect for acute GVHD in transplantation with HLA-DPB1 TCE non-PM and patient mismatch HLA-DP5, respectively.
Association of patient mismatch HLA-DP group and HLA-DPB1 TCE mismatching
| . | HR . | 95% CI . | P . | N . | |
|---|---|---|---|---|---|
| Patient mismatch HLA-DP group effect | |||||
| PM | DP2 group | 1.00 | 566 | ||
| DP5 group | 1.42* | 1.15-1.75 | .001 | 388 | |
| Non-PM | DP2 group | 1.00 | 91 | ||
| DP5 group | 0.76* | 0.53-1.09 | .138 | 241 | |
| HLA-DPB1 TCE mismatching effect | |||||
| DP2 group | PM | 1.00 | 566 | ||
| Non-PM | 1.67† | 1.20-2.31 | .002 | 91 | |
| DP5 group | PM | 1.00 | 388 | ||
| Non-PM | 0.94† | 0.74-1.20 | .638 | 241 | |
| . | HR . | 95% CI . | P . | N . | |
|---|---|---|---|---|---|
| Patient mismatch HLA-DP group effect | |||||
| PM | DP2 group | 1.00 | 566 | ||
| DP5 group | 1.42* | 1.15-1.75 | .001 | 388 | |
| Non-PM | DP2 group | 1.00 | 91 | ||
| DP5 group | 0.76* | 0.53-1.09 | .138 | 241 | |
| HLA-DPB1 TCE mismatching effect | |||||
| DP2 group | PM | 1.00 | 566 | ||
| Non-PM | 1.67† | 1.20-2.31 | .002 | 91 | |
| DP5 group | PM | 1.00 | 388 | ||
| Non-PM | 0.94† | 0.74-1.20 | .638 | 241 | |
HR indicates comparison of the patient mismatch HLA-DP5 group to HLA-DP2 group in patients with TCE PM and non-PM transplant.
HR indicates comparison of TCE non-PM to PM in patients with patient HLA-DP5 group mismatch and HLA-DP2 group mismatch.
Cumulative incidence of grade 2-4 acute GVHD by patient mismatch HLA-DP group and T-cell epitope mismatching. Patients were divided into 4 groups according to patient mismatch HLA-DP group and HLA-DPB1 T-cell epitope mismatching. DP5–non-PM, patient mismatch HLA-DP5 group and TCE nonpermissive mismatch; DP5-PM, patient mismatch HLA-DP5-group and TCE permissive mismatch; DP2–non-PM, patient mismatch HLA-DP2-group and TCE nonpermissive mismatch; DP2-PM, patient mismatch HLA-DP2 group and TCE permissive mismatch.
Cumulative incidence of grade 2-4 acute GVHD by patient mismatch HLA-DP group and T-cell epitope mismatching. Patients were divided into 4 groups according to patient mismatch HLA-DP group and HLA-DPB1 T-cell epitope mismatching. DP5–non-PM, patient mismatch HLA-DP5 group and TCE nonpermissive mismatch; DP5-PM, patient mismatch HLA-DP5-group and TCE permissive mismatch; DP2–non-PM, patient mismatch HLA-DP2-group and TCE nonpermissive mismatch; DP2-PM, patient mismatch HLA-DP2 group and TCE permissive mismatch.
Discussion
In this study, we demonstrated that HLA-DPB1 alleles phylogenetically diverged into 2 major groups using HLA-DPB1 sequencing data, including exon, intron, and 3′UTR. Divergence into 2 groups was obvious from exon 3 to 3′UTR of the HLA-DPB1 gene. Multi-SNP data of a large number of unrelated individuals also demonstrated that each group had a discriminative region from exon 3 to 3′UTR.
The variation in 3′UTR of the HLA-DPB1 gene was first noted as a genetic marker associated with chronic hepatitis B virus infection.25 Following this, the SNP located in the HLA-DPB1 3′UTR rs9277534G allele was reported to be associated with susceptibility to hepatitis B virus persistence and with higher levels of HLA-DP expression.26 Petersdorf et al demonstrated that the risk of acute GVHD after UR-HCT was associated with patient and donor HLA-DPB1 mismatching, in which patient mismatch HLA-DPB1 was linked to the rs9277534G allele.13 They also showed that the HLA-DPB1 allele, determined by exon 2 sequencing, was strongly linked to rs9277534. In the present study, we clearly demonstrated that rs9277534 was 1 of the tag SNPs of HLA-DPB1 haplotypes that were highly conserved from exon 3 to 3′UTR.
Because donor selection in UR-HCT is generally performed in consideration of patient and donor HLA allele matching in polymorphic exons, this identification of the effect of polymorphism in 3′UTR of the HLA-DPB1 gene on acute GVHD has led to new thinking about the genetic importance of the noncoding region of HLA genes in allogeneic HCT.27
Interestingly, HLA-DPB1 allele groups in the phylogenetic tree that was constructed using sequence data in exon 2 did not always correspond to those of the HLA-DP2 and HLA-DP5 groups (Figure 1C; supplemental Figure 3). For example, HLA-DPB1*05:01 is the most frequent HLA-DPB1 allele among Japanese and was naturally classified into the HLA-DP5 group. However, exon 2 of HLA-DPB1*05:01 showed similarity with alleles in the HLA-DP2 groups. HLA-DPB1*17:01 was classified into the HLA-DP2 group, whereas its exon 2 showed similarity with the HLA-DP5 group. HLA-DPB1 polymorphisms are clustered in exon 2, which code for the β1 extracellular domain of the HLA-DP molecule.28 This domain constitutes the peptide-binding groove together with α1 domain coded in HLA-DPA1 gene. Most of HLA alleles have been defined by sequences of polymorphic exons mainly in exon 2 and 3 in HLA class I genes and mainly in exon 2 in HLA class II genes. We speculate that recombination or selective pressure resulted in the present DPB1 alleles such as DPB1*05:01 and DPB1*17:01 after the branching of ancestral DP5 and DP2 groups during the course of evolution. Under this assumption, the following TCE grouping mainly reflects the phylogenetic tree based on exon 2 rather than that based on 3′UTR branching of the DP2 and DP5 groups (supplemental Figure 3).
In accordance with the HLA-DPB1 TCE algorithm, HLA-DPB1 alleles were classified into 3 TCE groups predicted to be immunogenic (group 1), intermediately immunogenic (group 2), and poorly immunogenic (group 3).10 HLA-DPB1 alleles in the same TCE group share similar amino acid sequences in the hypervariable regions of the HLA-DP molecule. Recently, the HLA-DPB1 classification algorithm has been updated to predict patient and donor HLA-DPB1 non-PM based on functional distance scores of 10 polymorphic amino acids located within the hypervariable region of the HLA-DPB1 gene.29,30 The Immuno Polymorphism Database-ImMunoGeneTics/HLA Web site provides both original and updated TCE algorithm tools to identify PM and non-PM for HLA-DPB1 for unrelated-donor hematopoietic stem cell transplantation (http://www.ebi.ac.uk/ipd/imgt/hla/dpb.html). The National Marrow Donor Program also provides a Web-based application that incorporates the HLA-DPB1 TCE algorithm for transplant centers to search unrelated donors and cord blood units.31 Although HLA-DPB1 non-PM was associated with an increased risk of grade 3-4 acute GVHD, it did not affect overall survival after UR-BMT in the current JMDP analysis. We speculate that the discrepancy in transplant outcomes associated with HLA-DPB1 TCE mismatching between Caucasian and Japanese patients might be partly attributed to ethnic backgrounds.11,32
Given the relationship identified here between patient mismatch HLA-DP group and T-cell epitope mismatching group in the risk of acute GVHD, we suspect the presence of discrete acute GVHD induction by these 2 groups and their hierarchy model. For patients in the HLA-DP2 mismatch group, non-PM of TCE was associated with a higher risk of acute GVHD than PM. Also, when patients were in the TCE PM group, patient mismatch HLA-DP5 was associated with a higher risk of acute GVHD than HLA-DP2. An additive effect of patient mismatch HLA-DP-5 and TCE non-PM on the risk of acute GVHD was not observed. The relation of patient mismatch HLA-DP group and updated TCE algorithm (difference in functional distance of mismatched HLA-DPB1 allele)30 was consistent with the original TCE algorithm. Increasing risk of acute GVHD in patient HLA-DP5-group mismatch and high difference in functional distance score were observed only in patients with low difference in functional distance score group and patient HLA-DP2 group mismatch, respectively (supplemental Tables 2 and 3).
Given that the HLA-DPB1 TCE mismatch algorithm is considered based on the difference in polymorphic amino acids in a hypervariable region in the HLA-DP molecule between patient and donor, it might reflect the peptide-binding groove structure of HLA-DP molecules. On the other hand, rs9277534 is represented as the region from exon 3 to 3′UTR of HLA-DPB1 gene, and the structure of this region might be associated with the expression level of HLA-DP molecules. Consequently, 2 recently published HLA-DPB1 grouping algorithms for the risk of transplant outcomes,10,11,13 which reflect an association with the HLA-DP expression marker rs9277534 in 3′UTR and HLA-DPB1 TCE mismatching algorithm, might reflect different immunological alloreactivity after UR-HCT. We could not provide a clear explanation why TCE non-PM had no effect in patient mismatch HLA-DP5 group or why patient mismatch HLA-DP5 had no effect in TCE non-PM group in the present study. We were also unable to clearly explain why an increasing risk effect of patient mismatch HLA-DP5 group was evident on grade 2-4 but not grade 3-4 acute GVHD. Further experimental and clinical studies are needed to elucidate these questions and clarify the association and interaction of the 2 algorithms.
Our findings are limited to the Japanese population. Because most DPB1 alleles are shared with other ethnic populations, it is important to analyze the international HCT database with HLA-DPB1 allele typing in other ethnic populations and to confirm the common phylogenic structure of HLA-DPB1 and its effect on allogeneic immune response in HCT.
In conclusion, using NGS sequence data and multi-SNP data of HLA-DPB1, we have conclusively demonstrated that the HLA-DP5 group and HLA-DP2 group represent 2 major groups of HLA-DPB1, and that each group has a highly conserved region from exon 3 to 3′UTR. The patient mismatch HLA-DP5 group showed significantly higher risk of acute GVHD than the HLA-DP2 group. Furthermore, through combined analysis with the HLA-DPB1 TCE algorithm, discrete and/or prioritized induction of acute GVHD by the patient mismatch HLA-DP group and TCE mismatching was observed. These findings suggest that acute GVHD after UR-HCT might be induced by structural differences between patient and donor in HLA-DP molecules, and that these differences might be mainly attributed to 2 evolutionarily different regions in the HLA-DPB1 gene.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Miho Matsubara for technical assistance with data management, the staff members of the transplantation centers and donor centers, the Japan Marrow Donor Program office, and the Japanese Data Center for Hematopoietic Cell Transplantation for their generous cooperation.
This work was supported by the Japan Society for the Promotion of Science Grants-in-Aid for Scientific Research (KAKENHI) (grant 17H05797) (S.M.) and by the Practical Research Project for Allergic Diseases and Immunology (Research on Technology of Medical Transplantation) from the Japan Agency for Medical Research and Development (grants 15ek0510002h0602 [S.M.], 15ek0510002h0202 [T. Shiina], 16ek0510002s0303 [S.M.], 16ek0510002s0103 [T. Shiina], 17ek0510022s0401 [S.M.], and 17ek0510022h0001 [T. Shiina]).
Authorship
Contribution: S.M., T. Shiina, M.S., T. Sasazuki, and Y.M. participated in the design of the study; T. Shiina and S.S. performed next-generation sequencing analysis; S.O. and A.S.-O. performed multi-SNP analysis; K.K., F.A., and T.Y. performed the histocompatibility analysis in transplantation pairs; S.K., Y.K., and Y.M. organized and collected clinical data and samples for transplantation; S.M. and Y.M. performed statistical data analysis; and S.M., T. Shiina, and Y.M. performed analysis and wrote the paper. All authors checked the final version of the paper.
Conflict-of interest disclosure: The authors declare no competing financial interests.
The research group of the Japan Marrow Donor Program consists of the authors of this article.
Correspondence: Satoko Morishima, Division of Endocrinology, Diabetes and Metabolism, Hematology, Rheumatology (Second Department of Internal Medicine), Graduate School of Medicine, University of the Ryukyus, 207 Uehara, Nishihara, Okinawa, 903-0215, Japan; e-mail: smorishi@med.u-ryukyu.ac.jp; or Takashi Shiina, Division of Basic Medical Science and Molecular Medicine, Department of Molecular Life Science, Tokai University School of Medicine, 143 Shimokasuya, Isehara, Kanagawa, 259-1193, Japan; e-mail: tshiina@is.icc.u-tokai.ac.jp.
References
Author notes
S.M. and T. Shiina contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal