Key Points
CTCL patients have decreased miR-29b levels and increased BRD4 binding occupancy at promoter regions of tumor-associated genes.
Therapeutic targeting of miR-29b and BRD4 in CTCL mice results in significantly decreased disease severity and progression.
Abstract
MicroRNA (miRNA) dysregulation is a hallmark of cutaneous T-cell lymphoma (CTCL), an often-fatal malignancy of skin-homing CD4+ T cells for which there are few effective therapies. The role of microRNAs (miRs) in controlling epigenetic modifier-dependent transcriptional regulation in CTCL is unknown. In this study, we characterize a novel miR dysregulation that contributes to overexpression of the epigenetic reader bromodomain-containing protein 4 (BRD4). We used patient CD4+ T cells to show diminished levels of miR-29b compared with healthy donor cells. Patient cells and miR-29b−/− mouse cells revealed an inverse relationship between miR-29b and BRD4, the latter of which is overexpressed in these cells. Chromatin immunoprecipitation and sequencing analysis revealed increased genome-wide BRD4 occupancy at promoter and enhancer regions in CD4+ T cells from CTCL patients. The cumulative result of BRD4 binding was increased expression of tumor-associated genes such as NOTCH1 and RBPJ, as well as the interleukin-15 (IL-15) receptor complex, the latter enhancing IL-15 autocrine signaling. Furthermore, we confirm the in vivo relevance of this pathway in our IL-15 transgenic mouse model of CTCL by showing that interference with BRD4-mediated pathogenesis, either by restoring miR-29b levels via bortezomib treatment or by directly inhibiting BRD4 binding via JQ1 treatment, prevents progression of CTCL. We describe a novel oncogenic pathway featuring IL-15, miR-29b, and BRD4 in CTCL and suggest targeting of these components as a potentially effective therapy for CTCL patients.
Introduction
Cutaneous T-cell lymphoma (CTCL) is a non-Hodgkin lymphoma characterized by progressive infiltration and proliferation of mature skin-homing CD4+ T cells in the skin, followed by systemic spread to lymph nodes, blood, and viscera in a significant fraction of patients.1-3 Recent whole-genome sequencing studies highlighted the central role of somatic mutations that affect cellular signaling and epigenetic regulation in the pathogenesis of CTCL.4-8 Although epigenetic modifiers such as histone deacetylase (Hdac) inhibitors are approved for treatment of CTCL, responses occur in approximately 30% of patients with high relapse rates,9,10 which highlights the need for better understanding the epigenetic aberrations and regulatory pathways that promote disease pathogenesis and sustain survival and proliferation of malignant T cells in CTCL.
Tumor suppressive microRNAs (miRNAs) that downregulate epigenetic modifiers and lead to effects on tumor suppressor gene expression are termed “epi-miRs.” miR-29b targets DNA methyltransferases resulting in global DNA hypomethylation in malignant cells.11-13 Experimental overexpression of miR-29b in multiple myeloma,14 acute myeloid leukemia,15 and rhabdomyosarcoma induces apoptosis and suppresses cell growth.16,17 Expression of miR-29b can be increased pharmacologically with bortezomib, a proteasome inhibitor, which leads to dose-dependent increases of miR-29b and apoptosis.14
An additional mechanism of epigenetic regulation is posttranslational modification of histones recognized by epigenetic reader proteins, which direct assembly of transcription factors at gene promoter regions. Bromodomain-containing protein 4 (BRD4), a bromodomain and extra-terminal (BET) protein, binds acetylated lysine residues on histones and regulates many genes involved in cellular proliferation, cell-cycle progression, and apoptosis.18 Numerous models have demonstrated BRD4 dependence for tumor cell proliferation and survival. A small molecule inhibitor of BET, JQ1, which displaces BRD4 from chromatin, was recently found to be a potent inducer of apoptosis in B-cell leukemia,19,20 acute myeloid leukemia,21,22 glioblastoma,23 and lung cancer.24
Expression of chronic inflammatory cytokine interleukin-15 (IL-15) is increased in neoplastic T cells from CTCL patients, and autocrine IL-15 signaling results in epigenetically driven induction of oncogene expression.25 These oncogenic drivers of T-cell malignancies include NOTCH1 and its effector cofactor recombination signal binding protein for immunoglobulin-kappa-J region (RBPJ).26 Aberrant NOTCH1 activity is oncogenic in several malignancies and results in accelerated tumor growth.27 In CTCL, NOTCH1 expression is increased in advanced stages, and in vitro blockade of NOTCH1 signaling induces apoptosis in CTCL-derived cell lines.28
In this study, we show decreased expression of miR-29b associated with increased expression of BRD4. In addition, increased genomic BRD4 occupancy in CTCL cells results in increased expression of NOTCH1 and RBPJ as well as IL-15 receptor complex, suggesting that the miR-29b-BRD4 axis may be an important targetable pathway in CTCL.
Materials and methods
Mouse strains
All mice were bred and maintained in pathogen-free conditions, and all studies were approved by the Ohio State University Institutional Animal Care and Use Committee. FVB/N mice with global overexpression of IL-15 have been described.25 All mice were age and gender matched. C57Bl6 miR-29b−/− mice (kindly provided by Carlo M. Croce, The Ohio State University) have been previously described.29
Human T-cell isolation
All samples from patients and normal donors were processed per protocol approved by the Institutional Review Board of The Ohio State University Comprehensive Cancer Center. Fresh peripheral blood from patients was obtained through The Ohio State University Leukemia Tissue Bank. Normal donor peripheral blood was obtained through the American Red Cross (Columbus, OH). CD4+ T-cell isolation was performed as described,25 and full details for the method are available in the supplemental Data, available on the Blood Web site.
Mouse tissue isolation
Mouse tissue isolation was performed as described,25 and full details regarding the methods can be found in the supplemental Data.
In vivo drug treatments
Three- to 4-week-old IL-15 transgenic mice were dosed intraperitoneally with 50 mg/kg JQ1 or vehicle control (10% cyclodextrin in phosphate-buffered saline [PBS]) 5 times per week for 4 weeks and with 1 mg/kg bortezomib or vehicle control (50% PBS/50% dimethyl sulfoxide) twice per week for 5 weeks. Cutaneous lesions were scored twice per week as described.25 Mice were euthanized by carbon dioxide inhalation followed by cervical dislocation. Skin tissues were collected in 10% neutral-buffered formalin for histology, and in 1× PBS to generate a single-cell suspension.
Chromatin immunoprecipitation and sequencing
Cell suspensions were processed for chromatin immunoprecipitation (ChIP) per Active Motif kit instructions (Active Motif, La Hulpe, Belgium). ChIP sequencing (ChIP-seq) was performed by using FactorPath ChIP-Seq technology by Active Motif. Full details regarding the methods can be found in the supplemental Data.
Immunoblotting
Cell suspensions were lysed with Bio-Rad buffer (Bio-Rad, Hercules, CA). Cell lysates were run on precast gel (Bio-Rad Criterion). Antibodies for BRD4 were obtained from Bethyl Laboratories (A301), IL-15 receptor complex (IL-15Rα [H-107], IL-2Rβ [M-20], IL-2Rγ [N-20]) from Santa Cruz Biotechnology (Dallas, TX), NOTCH1 (D3B8) from Cell Signaling (Beverly, MA), RBPJ (AB2284) from Millipore (Billerica, MA), and actin (MAB1501) from Millipore.
Isolation of RNA, complementary DNA preparation, and reverse transcription polymerase chain reaction
Silencing RNA transfection
The HuT-102 cell line was cultured at 2 × 106 cells per mL with silencing RNA (siRNA) at 0.15 nmol to BRD4 or with scrambled control. Transfections were performed by using nucleofector solution and Amaxa protocol (Lonza, Basel, Switzerland). Cells were cultured for 24 hours and then collected and analyzed for reverse transcription polymerase chain reaction (RT-PCR). Additional in vitro treatments can be found in supplemental Methods.
Statistics
Two-sample Student t test was used to compare 2 independent groups, and paired Student t test was used to compare 2 paired groups. Data transformation was performed if the original distribution was nonnormal. Analysis of variance models or generalized linear models were used to compare 3 or more groups. P values were adjusted for multiple comparisons by Holm’s procedure. A P value of < .05 was considered significant.
Results
An inverse relationship between miR-29b and BRD4 levels in CTCL patients and miR-29b−/− mice
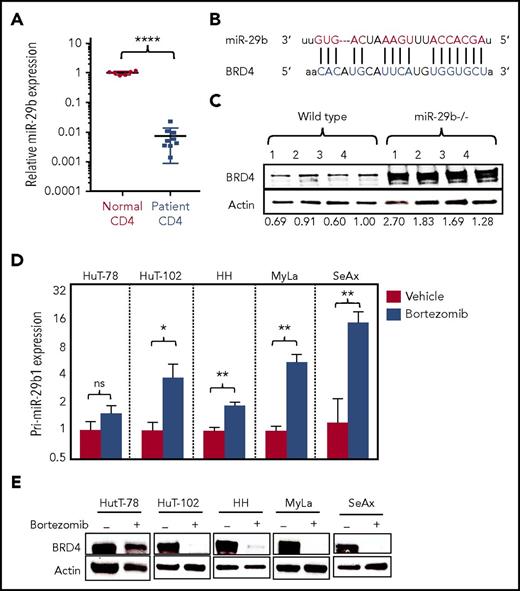
By examining purified peripheral blood CD4+ T cells from CTCL patients (supplemental Table 1) and normal donors, we found significantly decreased expression of miR-29b in CTCL patients (0.007 ± 0.002 [n = 9]) compared with normal donors (1.008 ± 0.052 [n = 6]; P < .0001) (Figure 1A). Alignment of seed sequence of miR-29b demonstrated complementarity with BRD4 3′ untranslated region (UTR) (Figure 1B). To confirm miR-29b–mediated regulation of BRD4, we analyzed isolated splenocytes from miR-29b−/− mice and found that BRD4 protein expression is significantly increased in the miR-29b−/− cells compared with wild-type (WT) mouse cells (fold change, 1.87 ± 0.29; P = .014) (Figure 1C). To investigate this potential interaction, a BRD4 3′UTR reporter assay was performed. Briefly, CD4+ T cells from normal donors were transfected with the vector construct green fluorescent protein (GFP) fused to the 3′UTR of BRD4 (BRD4 3′UTR GFP) and concurrently with miR-29b mimic or scrambled control (supplemental Figure 1A). In each of 3 normal donors, relative BRD4 activity (% GFP-expressing cells) decreased in miR-29b transfected cells compared with that of scrambled control cells (supplemental Figure 1B). Pooling data for the donors, the decrease in BRD4 activity was statistically significant (P = .0376) (data not shown). Subsequently, a BRD4 3′UTR GFP stable cell line was transfected with miR-29b mimic. A significant decrease in BRD4 activity was observed in cells transfected with miR-29b mimic (3.13 ± 0.8) vs scrambled control (69.4 ± 1.42; P < .0001) (supplemental Figure 1C).
BRD4 is inversely correlated with miR-29b in CTCL. (A) Relative expression of miR-29b in peripheral blood CD4+ T cells from CTCL patients (n = 9) and normal donors (n = 6). (B) Sequence alignment of the mature miR seed sequence of miR-29b showing complementarity to the 3′UTR of BRD4. (C) Immunoblot analysis of BRD4 protein in splenocytes from miR-29b−/− and age-matched WT mice (n = 4 each). (D) RT-PCR analysis for pri-miR-29b1 in CTCL-derived CD4+ T-cell lines treated with 10 nM bortezomib showing a significant increase in pri-miR-29b1 level after 3 hours. Data are presented as mean ± standard error of the mean (SEM) (n = 3 each). (E) Immunoblot of BRD4 protein expression in CTCL-derived CD4+ T-cell lines treated with 50 nM bortezomib for 24 hours showing decreased expression in treated cells compared with dimethyl sulfoxide–treated controls. Data are presented as mean ± standard error of the mean (SEM). *P ≤ .05; **P ≤ .01; ***P ≤ .001; ****P ≤ .0001; unpaired 2-tailed Student t test. ns, not significant.
BRD4 is inversely correlated with miR-29b in CTCL. (A) Relative expression of miR-29b in peripheral blood CD4+ T cells from CTCL patients (n = 9) and normal donors (n = 6). (B) Sequence alignment of the mature miR seed sequence of miR-29b showing complementarity to the 3′UTR of BRD4. (C) Immunoblot analysis of BRD4 protein in splenocytes from miR-29b−/− and age-matched WT mice (n = 4 each). (D) RT-PCR analysis for pri-miR-29b1 in CTCL-derived CD4+ T-cell lines treated with 10 nM bortezomib showing a significant increase in pri-miR-29b1 level after 3 hours. Data are presented as mean ± standard error of the mean (SEM) (n = 3 each). (E) Immunoblot of BRD4 protein expression in CTCL-derived CD4+ T-cell lines treated with 50 nM bortezomib for 24 hours showing decreased expression in treated cells compared with dimethyl sulfoxide–treated controls. Data are presented as mean ± standard error of the mean (SEM). *P ≤ .05; **P ≤ .01; ***P ≤ .001; ****P ≤ .0001; unpaired 2-tailed Student t test. ns, not significant.
Bortezomib increases miR-29b level and thus decreases BRD4 expression
Bortezomib, a 26S proteasome inhibitor, has been previously shown to increase miR-29b expression in neoplastic cells.14 Treatment of 5 patient-derived CTCL cell lines (HuT-78, HuT-102, HH, MyLa, and SeAx) with 10 nM bortezomib for 3 hours in vitro resulted in significantly increased pri-miR-29b1 expression level above that of dimethyl sulfoxide–treated cells for 4 of the 5 lines, whereas HuT-78 had a nonsignificant increase in miR-29b level, likely due to the presence of an NF-κB/p65 truncation mutation in that line (Figure 1D). Similarly, treatment of purified CD4+ cells from a CTCL patient in vitro with 100 nM bortezomib for 2 hours resulted in a significant increase in the pri-miR-29b1 expression level (P = .0334) (supplemental Figure 2A). Because miR-29b levels are regulated by recruitment of c-Myc and Hdac1,30 we measured these transcripts in bortezomib-treated CTCL cell lines. Significant decreases were seen in c-Myc for HH (P = .0157), HuT-78 (P = .0059), and HuT-102 (P < .0001), and were also seen in Hdac1 for HH (P = .0002), HuT-78 (P < .0001), and HuT-102 (P < .0001) (supplemental Figure 2B-C). These data demonstrate that bortezomib regulates miR-29b in CTCL, at least in part by downregulation of c-Myc and Hdac1. In addition, treatment of CTCL lines with 50 nM bortezomib for 24 hours resulted in profound decreases in BRD4 protein expression in the same 4 lines showing reversal of pri-miR-29b1 levels (Figure 1E). We demonstrated that effects of bortezomib on BRD4 expression are miR-29b dependent by treating T cells from WT and miR-29b−/− mice with 50 nM bortezomib for 24 hours. BRD4 protein decreased in WT mice, as expected; however, there was no change in BRD4 in cells from miR-29−/− mice (supplemental Figure 2D). Furthermore, we transfected normal donor CD4+ T cells (which have higher levels of miR-29b) with miR-29b inhibitor, and we subsequently treated these cells with bortezomib. We demonstrated that in scrambled control transfected cells, bortezomib results in a decrease of ∼80% in BRD4 expression. However, miR-29b inhibitor–transfected cells had no change in BRD4 expression in response to bortezomib (supplemental Figure 2E). These data demonstrate a novel regulatory association between BRD4 and miR-29b, and furthermore, that treatment with bortezomib results in increased miR-29b levels with a resultant decrease in BRD4 in vitro.
Genome-wide increase in BRD4 occupancy in genome of CTCL patients compared with that of healthy donors with reversal of occupancy by treatment with BET inhibitor JQ1
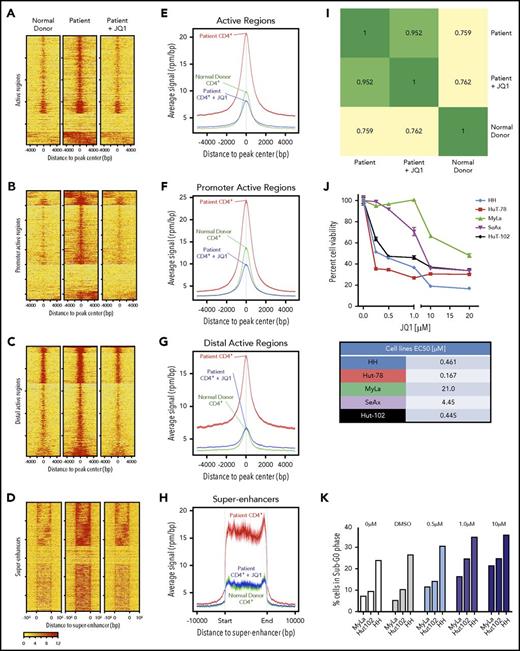
To determine the overall occupancy of BRD4 in a CTCL patient, we used ChIP-seq. CD4+ T cells from CTCL patients demonstrated increased BRD4 binding at gene-regulatory regions (Figure 2A), promoter-active regions (Figure 2B), distal-active regions (Figure 2C), and super-enhancer regions (Figure 2D) compared with normal donor CD4+ T cells. Enhanced binding was reversed after CTCL patient cells were treated with 100 nM JQ1. Presented as average signal intensity, BRD4 binding in patient cells was increased over normal donor and JQ1-treated patient cells (Figure 2E-H). Importantly, treating CTCL patient cells with JQ1 returns the level of BRD4 binding to approximately that in normal donor cells. The correlation coefficient (R) between patient and normal donor is 0.759, whereas R = 0.762 when comparing normal donor and patient cells treated with JQ1 (Figure 2I). Gene ontology analysis performed by using the GREAT functional annotation tool31 revealed enrichment of BRD4-regulated genes in CD4+ cells from patients and healthy donors, and they were analyzed on the basis of biological process, molecular function, and signaling pathways (supplemental Table 2). These data demonstrate the extensive binding activity of BRD4 at regulatory regions in CTCL patients and that this binding profile can be reversed to that of a normal donor with JQ1 treatment.
BRD4 binds regulatory regions in CTCL patients. (A) ChIP analysis of BRD4 binding to active regions of the genome in CD4+ T cells from a normal donor (left), CTCL patient (middle), and CTCL patient cells treated with 100 nM JQ1 in vitro for 24 hours. (B-D) ChIP analysis of BRD4 binding to (B) promoter regions, (C) distal active regions, and (D) super-enhancer regions. (E) Average signal intensity plot of BRD4 binding to active regions of the genome in CD4+ T cells from a normal donor, CTCL patient, and CTCL patient cells treated with 100 nM JQ1 in vitro for 24 hours. (F-H) Average signal intensity plot of BRD4 binding to (F) promoter regions, (G) distal active regions, and (H) super-enhancer regions. (I) Pearson correlation map demonstrating correlation coefficients of BRD4 binding across all genomic regulatory regions between normal donor, CTCL patient, and patient treated with JQ1. (J) MTS assay showing viability of CTCL-derived cell lines exposed to increasing doses of JQ1 from 0.25 µM to 20 µM. All cell lines demonstrate <50% cell viability at high doses of JQ1. Fifty percent effective concentration (EC50) values are presented as a table. (K) Cell cycle analysis of CTCL-derived cell lines exposed to increasing doses of JQ1 from 0.5 µM to 10 µM demonstrating the percentage of cells in sub-G0/G1 phase. Data are presented as mean ± SEM unless otherwise specified.
BRD4 binds regulatory regions in CTCL patients. (A) ChIP analysis of BRD4 binding to active regions of the genome in CD4+ T cells from a normal donor (left), CTCL patient (middle), and CTCL patient cells treated with 100 nM JQ1 in vitro for 24 hours. (B-D) ChIP analysis of BRD4 binding to (B) promoter regions, (C) distal active regions, and (D) super-enhancer regions. (E) Average signal intensity plot of BRD4 binding to active regions of the genome in CD4+ T cells from a normal donor, CTCL patient, and CTCL patient cells treated with 100 nM JQ1 in vitro for 24 hours. (F-H) Average signal intensity plot of BRD4 binding to (F) promoter regions, (G) distal active regions, and (H) super-enhancer regions. (I) Pearson correlation map demonstrating correlation coefficients of BRD4 binding across all genomic regulatory regions between normal donor, CTCL patient, and patient treated with JQ1. (J) MTS assay showing viability of CTCL-derived cell lines exposed to increasing doses of JQ1 from 0.25 µM to 20 µM. All cell lines demonstrate <50% cell viability at high doses of JQ1. Fifty percent effective concentration (EC50) values are presented as a table. (K) Cell cycle analysis of CTCL-derived cell lines exposed to increasing doses of JQ1 from 0.5 µM to 10 µM demonstrating the percentage of cells in sub-G0/G1 phase. Data are presented as mean ± SEM unless otherwise specified.
Inhibition of BRD4 activity results in cytotoxicity to CTCL cells, partially by induction of sub-G0/G1 phase
BET inhibitor JQ1 has been investigated as a treatment strategy in many hematologic malignancies,19,32 but whether JQ1 has therapeutic efficacy in CTCL is not known. In our study, 3 CTCL cell lines were highly sensitive to JQ1 treatment, with 50% effective concentration (EC50) values at 0.461 µM for the HH cell line, 0.167 µM for HuT-78, and 0.445 µM for HuT-102. Two lines were moderately resistant to JQ1 with EC50 values of 4.45 µM for SeAx and 21 µM for MyLa cell lines (Figure 2J). JQ1 treatment of cell lines resulted in a dose-dependent increase in population of cells in sub-G0/G1 phase of the cell cycle (Figure 2K). To further address the specificity of BRD4 in cellular survival, we transfected HuT-102 cells with siRNA to BRD4 (siBRD4). We demonstrated significant decrease in BRD4 expression (0.47 ± 0.01 compared with 1.0 ± 0.01 for control RNA; P < .0001) (supplemental Figure 3A) as well as decreased cell count (supplemental Figure 3B) and significantly decreased proliferation (P = .0316) (supplemental Figure 3C) in siBRD4-transfected cells compared with control cells. These data demonstrate the specificity of BRD4 inhibition in cytotoxicity of malignant T cells.
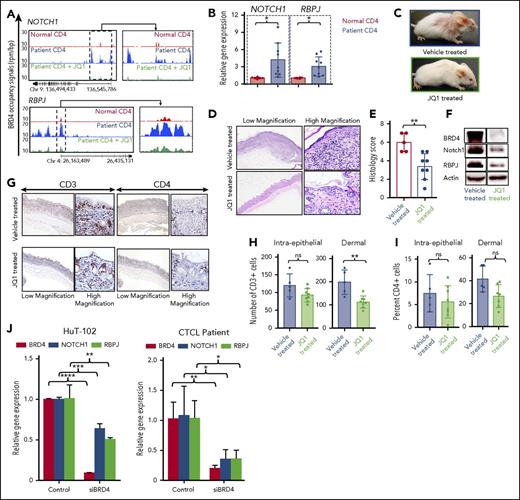
BRD4 binding at promoter regions is associated with increased transcriptional activity of tumor-associated NOTCH1 gene and its cofactor RBPJ
It was previously shown that increased BRD4 activity modulates induction of oncogene expression.33 Here we describe the role of BRD4 in regulating expression of 2 tumor-associated genes in CTCL: NOTCH1 and RBPJ. NOTCH1 signaling acts through its effector and co-activator RBPJ,34 allowing transcriptional activation of NOTCH1 target genes,35 and is oncogenic in T-cell leukemias.26,36 ChIP-seq analysis revealed that BRD4 binding at NOTCH1 promoter was increased in CTCL patient CD4+ T cells compared with normal donor cells. Binding was reversed by treating patient cells with 100 nM JQ1 (Figure 3A, top panel). BRD4 binding was also increased at the promoter of RBPJ in CTCL patients and was similarly decreased by treatment with JQ1 to the same level of binding as that in normal donor cells (Figure 3A, bottom panel). Furthermore, transcript levels of both NOTCH1 (4.16 ± 0.99 [n = 9]; P = .024) and RBPJ (3.03 ± 0.56; P = .0122) were significantly increased in CD4+ cells from patients compared with normal donors (1.01 ± 0.06 and 1.01 ± 0.05, respectively [n = 6]) (Figure 3B). These data demonstrate that increased binding of BRD4 to regulatory regions in CTCL is associated with enhanced expression of NOTCH1 and RBPJ.
JQ1 reverses BRD4 binding–mediated enhanced oncogene expression in vivo. (A) Gene tracks depicting genomic occupancy of BRD4 (rpm per bp) at regulatory regions of NOTCH1 and RBPJ in CD4+ T cells from a normal donor, a CTCL patient, and patient cells treated with 100 nM JQ1 for 24 hours in vitro. (B) RT-PCR of transcript levels of NOTCH1 and RPBJ in peripheral blood CD4+ T cells from normal donors (n = 6) and CTCL patients (n = 9). (C) Clinical images of representative IL-15 transgenic FVB/N mice treated intraperitoneally with 10% cyclodextran vehicle (above) or 50 mg/kg JQ1 (below) for 4 weeks. (D) Photomicrographs of skin from representative IL-15 transgenic CTCL mice treated with vehicle or JQ1. Hematoxylin and eosin stain, 40× (left), 400× (right). (E) Histology lesion severity score of skin tissue from mice treated with vehicle (n = 5) or JQ1 (n = 7). (F) Immunoblot of BRD4, NOTCH1, and RBPJ from skin tissue of IL-15 transgenic CTCL mice treated with vehicle or 50 mg/kg JQ1 with actin as an internal control. (G) Photomicrographs of skin from representative IL-15 transgenic mice treated with vehicle or JQ1. Immunohistochemistry for mouse CD3 (40×, left; 400×, right) and immunohistochemistry for mouse CD4 (40×, left; 400×, right). (H) Counts of CD3+ cells within surface and follicular epithelium (left) and within the superficial dermis (right) in vehicle-treated and JQ1-treated mice. (I) Counts of CD4+ cells within surface and follicular epithelium (left) and within the superficial dermis (right) in vehicle-treated and JQ1-treated mice. (J) RT-PCR of BRD4, NOTCH1, and RBPJ transcript levels in CTCL cell line HuT-102 transfected with 0.15 nmol siRNA to BRD4 for 24 hours (left). RT-PCR of BRD4, NOTCH1, and RBPJ transcript levels in CTCL patient CD4+ T cells transfected with 0.15 nmol siRNA to BRD4 for 24 hours (right). Data are presented as mean ± SEM unless otherwise specified. *P ≤ .05; **P ≤ .01; ***P ≤ .001; ****P ≤ .0001; unpaired 2-tailed Student t test.
JQ1 reverses BRD4 binding–mediated enhanced oncogene expression in vivo. (A) Gene tracks depicting genomic occupancy of BRD4 (rpm per bp) at regulatory regions of NOTCH1 and RBPJ in CD4+ T cells from a normal donor, a CTCL patient, and patient cells treated with 100 nM JQ1 for 24 hours in vitro. (B) RT-PCR of transcript levels of NOTCH1 and RPBJ in peripheral blood CD4+ T cells from normal donors (n = 6) and CTCL patients (n = 9). (C) Clinical images of representative IL-15 transgenic FVB/N mice treated intraperitoneally with 10% cyclodextran vehicle (above) or 50 mg/kg JQ1 (below) for 4 weeks. (D) Photomicrographs of skin from representative IL-15 transgenic CTCL mice treated with vehicle or JQ1. Hematoxylin and eosin stain, 40× (left), 400× (right). (E) Histology lesion severity score of skin tissue from mice treated with vehicle (n = 5) or JQ1 (n = 7). (F) Immunoblot of BRD4, NOTCH1, and RBPJ from skin tissue of IL-15 transgenic CTCL mice treated with vehicle or 50 mg/kg JQ1 with actin as an internal control. (G) Photomicrographs of skin from representative IL-15 transgenic mice treated with vehicle or JQ1. Immunohistochemistry for mouse CD3 (40×, left; 400×, right) and immunohistochemistry for mouse CD4 (40×, left; 400×, right). (H) Counts of CD3+ cells within surface and follicular epithelium (left) and within the superficial dermis (right) in vehicle-treated and JQ1-treated mice. (I) Counts of CD4+ cells within surface and follicular epithelium (left) and within the superficial dermis (right) in vehicle-treated and JQ1-treated mice. (J) RT-PCR of BRD4, NOTCH1, and RBPJ transcript levels in CTCL cell line HuT-102 transfected with 0.15 nmol siRNA to BRD4 for 24 hours (left). RT-PCR of BRD4, NOTCH1, and RBPJ transcript levels in CTCL patient CD4+ T cells transfected with 0.15 nmol siRNA to BRD4 for 24 hours (right). Data are presented as mean ± SEM unless otherwise specified. *P ≤ .05; **P ≤ .01; ***P ≤ .001; ****P ≤ .0001; unpaired 2-tailed Student t test.
JQ1 treatment significantly reduces disease severity in the IL-15 transgenic mouse model of CTCL
To confirm the effects of JQ1 on inhibition of BRD4 as a potential anti-tumor agent, we used our recently characterized IL-15 transgenic mouse model.25 This mouse develops spontaneous cutaneous disease with 100% penetrance by 8 weeks of age, which is phenotypically and morphologically comparable to human CTCL.25 After a 4-week treatment with 50 mg/kg JQ1 injected intraperitoneally into 4-week-old mice, development of CTCL was apparent only in vehicle-treated IL-15 transgenic mice, whereas JQ1-treated mice were less affected (Figure 3C). Gross lesion scores represent a scale of cutaneous disease severity from mild erythema (score of 1) to severe extensive ulceration (score of 5) (supplemental Figure 4A). Gross lesion severity scores for JQ1-treated mice (n = 7) averaged 1.0 ± 0.12 compared with 2.4 ± 0.23 of 5 possible (P = .0318) for vehicle-treated mice (n = 5) (supplemental Figure 4B). Histologically, cutaneous lesions in vehicle-treated mice were characterized by intense intraepidermal and dermal infiltrate of neoplastic T cells with a formation of intraepidermal Pautrier’s microabscesses, a histologic hallmark of CTCL (Figure 3D). The severity of histologic lesions was significantly decreased in JQ1-treated mice (n = 7) (3.37 ± 0.49; P = .0041) compared with vehicle-treated controls (n = 5) (6.0 ± 0.45, with 7 as the highest possible score) (Figure 3E). Skin from JQ1-treated mice also showed significantly reduced protein expression of BRD4, NOTCH1, and RBPJ compared with vehicle-treated mice (Figure 3F). Infiltrating T cells in the skin were CD3+CD4+ (Figure 3G). The numbers of infiltrating CD3+ and CD4+ T cells were decreased in JQ1-treated mice compared with vehicle-treated mice (Figure 3H-I). In the dermis where high numbers of CD3+-infiltrating cells were present, JQ1-treated mice (n = 7) had a mean of 112.5 ± 9.19 CD3+ cells per high-power field (400×) compared with 198.4 ± 23.61 (P = .002) CD3+ cells in vehicle-treated mice (n = 5). After further evaluating CD4+ cells in the dermis, the mean number of CD4+ cells per high-power field (400×) in JQ1-treated cells was 28.6 ± 4.9 compared with 41 ± 5.26 in vehicle-treated mice.
Inhibition of BRD4 binding by JQ1 significantly reduces expression of tumor-associated NOTCH1 and RBPJ genes
To determine whether the expression effects are specific to BRD4, we used siRNA to BRD4 in HuT-102 cells, which significantly decreased BRD4 expression (80% to 90% transfection efficiency; P < .0001). In these transfected cells, expression of NOTCH1 (P = .0007) and RBPJ (P = .0065) were also significantly reduced (Figure 3J, left). This is confirmed in patient CD4+ T cells in which siBRD4 results in a significant decrease in BRD4 level (P = .0057) as well as NOTCH1 (P = .0260) and RBPJ (P = .0138) transcript levels (Figure 3J, right). By using ChIP-PCR, we demonstrated that treatment of CTCL patient–derived cell lines with 1 µM JQ1 for 24 hours resulted in a decrease in BRD4 binding at the promoter regions of NOTCH1 and RBPJ (supplemental Figure 5A). JQ1 treatment also resulted in significant dose-dependent decreases in transcript levels of both tumor-associated genes in vitro (supplemental Figure 5B-C). Altogether, these data show that the broad genomic and gene-specific effects of pharmacologic inhibition of BRD4 with JQ1 are associated with clinical efficacy in an in vivo mouse model of CTCL, suggesting that BRD4 should be explored as a therapeutic target in CTCL.
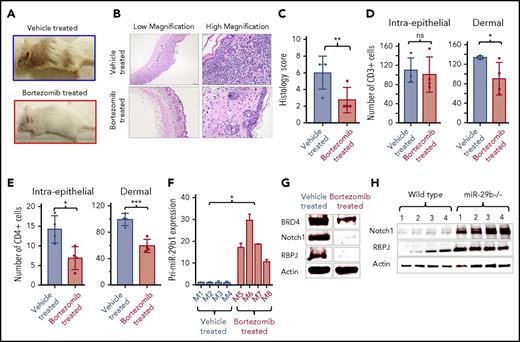
Proteasome inhibitor bortezomib decreases BRD4 expression and reduces disease severity in CTCL mice
To validate the finding in CTCL-derived cell lines that bortezomib treatment decreased BRD4 expression (Figure 1D-E) and to further support the roles of BRD4 and miR-29b in the pathogenesis of CTCL, we treated 3-week-old IL-15 transgenic mice for 5 weeks with 1 mg/kg of intraperitoneal bortezomib. Bortezomib-treated mice failed to develop clinical disease (Figure 4A), and histologic lesions were mild (Figure 4B-C) compared with vehicle-treated mice. Grossly, lesion severity scores for bortezomib-treated mice (n = 4) averaged 0.5 ± 0.14 compared with 2.5 ± 0.14 of 5 possible (P = .0027) for vehicle-treated mice (n = 4) (supplemental Figure 4C). Average histologic lesion score for bortezomib-treated mice (n = 4) was 2.75 ± 0.75 compared with the severity in the tissue of vehicle-treated mice (6.0 ± 1.0 of 7 possible; P = .04). The numbers of infiltrating malignant CD3+ and CD4+ T cells were significantly decreased in bortezomib-treated mice compared with vehicle-treated mice (Figure 4D-E). Bortezomib-treated mice (n = 4) had a mean of 90.4 ± 16.69 infiltrating dermal CD3+ cells compared with 133.8 ± 1.65 (P = .04) in vehicle-treated mice (n = 4) (Figure 4D). Bortezomib-treated mice had 59.1 ± 5.08 dermal CD4+ T cells per high-power field (400×) compared with 99.2 ± 4.55 (P = .001) in vehicle-treated mice (Figure 4E). We also analyzed transcript levels of pri-miR-29b1 in peripheral blood mononuclear cells from bortezomib- and vehicle-treated mice as well as protein expression of BRD4, NOTCH1, and RBPJ in the skin. As expected, pri-miR-29b1 expression was significantly higher in bortezomib-treated mice (n = 4) (18.95 ± 3.65; P = .02) compared with vehicle-treated mice (n = 4) (1.07 ± 0.19) (Figure 4F), whereas BRD4, NOTCH1, and RBPJ protein expression was decreased (Figure 4G). These data demonstrate that treatment with bortezomib results in decreased expression of NOTCH1 and RBPJ in vivo, in part by upregulation of miR-29b, which leads to reduced activity and expression of BRD4. As a further confirmation of the role of miR-29b in downstream tumor-associated gene expression, we assessed NOTCH and RBPJ protein levels and found levels for both to be elevated in splenocytes of miR-29b−/− mice compared with WT mice (Figure 4H).
Bortezomib increases miR-29b and decreases oncogene expression in vivo. (A) Clinical images of representative IL-15 transgenic CTCL mice treated intraperitoneally with PBS vehicle (above) or 1 mg/kg bortezomib (below) for 5 weeks. (B) Photomicrographs of skin from representative IL-15 transgenic CTCL mice treated with vehicle (PBS/dimethyl sulfoxide) or bortezomib. Hematoxylin and eosin stain (40×, left; 400×, right). (C) Histology lesion severity score of skin tissue from mice treated with vehicle (n = 4) or bortezomib (n = 4). (D) Counts of CD3+ cells within surface and follicular epithelium (left) and within the superficial dermis (right) in vehicle-treated and bortezomib-treated mice. (E) Counts of CD4+ cells within surface and follicular epithelium (left) and within the superficial dermis (right) in vehicle-treated and bortezomib-treated mice. (F) RT-PCR of pri-miR-29b1 in blood of bortezomib-treated mice (n = 4) compared with vehicle-treated mice (n = 4). (G) Immunoblot analysis of BRD4, NOTCH1, and RBPJ from skin tissue of IL-15 transgenic CTCL mice treated with vehicle or 1 mg/kg bortezomib, with actin as an internal control. (H) Immunoblot of NOTCH1 and RBPJ protein expression in splenocytes from miR-29b−/− mice compared with that of age-matched WT mice. Data are presented as mean ± SEM unless otherwise specified. *P ≤ .05; **P ≤ .01; ***P ≤ .001; ****P ≤ .0001; unpaired 2-tailed Student t test.
Bortezomib increases miR-29b and decreases oncogene expression in vivo. (A) Clinical images of representative IL-15 transgenic CTCL mice treated intraperitoneally with PBS vehicle (above) or 1 mg/kg bortezomib (below) for 5 weeks. (B) Photomicrographs of skin from representative IL-15 transgenic CTCL mice treated with vehicle (PBS/dimethyl sulfoxide) or bortezomib. Hematoxylin and eosin stain (40×, left; 400×, right). (C) Histology lesion severity score of skin tissue from mice treated with vehicle (n = 4) or bortezomib (n = 4). (D) Counts of CD3+ cells within surface and follicular epithelium (left) and within the superficial dermis (right) in vehicle-treated and bortezomib-treated mice. (E) Counts of CD4+ cells within surface and follicular epithelium (left) and within the superficial dermis (right) in vehicle-treated and bortezomib-treated mice. (F) RT-PCR of pri-miR-29b1 in blood of bortezomib-treated mice (n = 4) compared with vehicle-treated mice (n = 4). (G) Immunoblot analysis of BRD4, NOTCH1, and RBPJ from skin tissue of IL-15 transgenic CTCL mice treated with vehicle or 1 mg/kg bortezomib, with actin as an internal control. (H) Immunoblot of NOTCH1 and RBPJ protein expression in splenocytes from miR-29b−/− mice compared with that of age-matched WT mice. Data are presented as mean ± SEM unless otherwise specified. *P ≤ .05; **P ≤ .01; ***P ≤ .001; ****P ≤ .0001; unpaired 2-tailed Student t test.
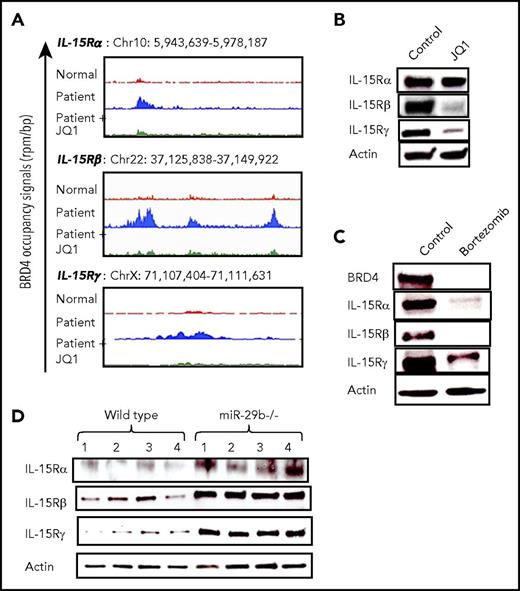
BRD4 binding enhances expression of IL-15 receptor complex in CTCL
IL-15 overexpression alone can induce development of CTCL in vivo25 ; however, the mechanisms underlying autocrine signaling in the progression of CTCL are poorly understood. To determine the regulatory effect of BRD4 on IL-15 signaling, we analyzed ChIP-seq data and found that BRD4 binding was increased in CTCL patient cells at promoter regions of all 3 components of the IL-15 receptor complex (IL-15Rα, IL-15Rβ, and IL-15Rγ) (Figure 5A). This binding is reversed upon treatment with 100 nM JQ1 to a level similar to that in normal donor cells. Furthermore, treatment of the HuT-102 cell line with 1 µM JQ1 for 48 hours resulted in a significant decrease in IL-15Rβ and IL-15Rγ, with a modest decline in the IL-15Rα subunit (Figure 5B). Additional in vitro treatment of HuT-102 cells with 50 nM bortezomib for 24 hours resulted in significant reductions in all IL-15 receptor constituents, and BRD4 protein expression was also decreased in bortezomib-treated cells (Figure 5C). Finally, splenocytes from miR-29b−/− mice demonstrated significantly higher IL-15 receptor complex protein expression than cells from WT mice (Figure 5D).
BRD4 binding enhances IL-15 receptor complex expression in CTCL patient cells. (A) Gene tracks depicting genomic occupancy of BRD4 (rpm per bp) at regulatory regions of IL-15 receptor complex loci in CD4+ T cells from a normal donor, a CTCL patient, and patient cells treated with 100 nM JQ1 for 24 hours in vitro. (B) Immunoblot of IL-15 receptor complex subunits (α, β, and γ) in HuT-102 cell line treated with vehicle or 1 µM JQ1 for 48 hours, with actin as internal control. (C) Immunoblot of BRD4 and IL-15 receptor complex in HuT-102 cells treated in vitro with vehicle or 50 nM bortezomib for 24 hours with actin as internal control. (D) Immunoblot of IL-15 receptor constituent protein expression in splenocytes from miR-29b−/− mice and age-matched WT mice (n = 4 each). Data are presented as mean ± SEM unless otherwise specified.
BRD4 binding enhances IL-15 receptor complex expression in CTCL patient cells. (A) Gene tracks depicting genomic occupancy of BRD4 (rpm per bp) at regulatory regions of IL-15 receptor complex loci in CD4+ T cells from a normal donor, a CTCL patient, and patient cells treated with 100 nM JQ1 for 24 hours in vitro. (B) Immunoblot of IL-15 receptor complex subunits (α, β, and γ) in HuT-102 cell line treated with vehicle or 1 µM JQ1 for 48 hours, with actin as internal control. (C) Immunoblot of BRD4 and IL-15 receptor complex in HuT-102 cells treated in vitro with vehicle or 50 nM bortezomib for 24 hours with actin as internal control. (D) Immunoblot of IL-15 receptor constituent protein expression in splenocytes from miR-29b−/− mice and age-matched WT mice (n = 4 each). Data are presented as mean ± SEM unless otherwise specified.
IL-15 signaling increases BRD4 expression in human and mouse T cells
It was previously shown that IL-15 drives a decrease in miR-29b levels in natural killer cells.30 To examine the potential role of this pathway in CTCL, CD4+ T cells from healthy donors were stimulated with 100 ng IL-15 in vitro for 48 hours. IL-15 treatment significantly decreased pri-miR-29b1 levels compared with untreated cells (P = .0001) (supplemental Figure 6A), whereas BRD4 protein expression was increased (supplemental Figure 6B, top). Consistent with our hypothesis, IL-15 transgenic mice have increased expression of BRD4 (supplemental Figure 6B, bottom), NOTCH, and RBPJ (supplemental Figure 6C). Our working model for the regulation of BRD4 and miR-29b by IL-15 is presented in supplemental Figure 6D.
IL-15 signaling results in repression of miR-29b and increased BRD4 activity
To examine this proposed pathway further, we analyzed normal donor T cells stimulated with IL-15 for 24 hours by immunoblot and by ChIP-PCR at the miR-29b promoter. We show increased expression of components of the repressor complex c-Myc and Hdac1 in stimulated cells (supplemental Figure 7A). This result corresponds to increased binding of repressor complex constituents c-Myc and Hdac-1 at the miR-29b promoter (supplemental Figure 7B). Next we transfected normal donor CD4+ cells with BRD4 3′UTR GFP or control GFP vector and subsequently stimulated transfected cells with 50 ng/mL IL-15 or control PBS. IL-15 signaling resulted in increased BRD4 activity (percentage of GFP-expressing cells) compared with unstimulated CD4+ T cells, which is consistent with repression of miR-29b (supplemental Figure 7C).
IL-15 and lack of miR-29b cooperate to drive cellular transformation
To determine whether there may be cooperation between IL-15 signaling and decreased miR-29b that would drive malignant transformation in CD4+ T cells, we performed a cell transformation assay using total splenocytes and thymocytes from WT and miR-29b−/− mice, with or without IL-15. We observed significantly increased cell transformation in miR-29b−/− cells when compared with WT cells (relative percentage of transformation, 112.9 ± 2.004 vs 100 ± 3.472 in spleen [P = .0325]; 129.8 ± 4.884 vs 100 ± 7.895 in thymus [P = .0325]). With IL-15 stimulation, we observed significantly increased transformation over unstimulated cells (relative percentage of transformation in WT spleen, 127.3 ± 6.561 [P = .0213]; WT thymus, 133.3 ± 3.163 [P = .0173]; miR-29b−/− spleen, 159.8 ± 7.461 [P = .0037]; and miR-29b−/− thymus, 183.3 ± 9.768 [P = .008]) (supplemental Figure 8A-B). Altogether, these data show that IL-15 repressed miR-29b levels in CD4+ cells through recruitment of Myc/Hdac1 at the miR-29b regulatory region and that repressed miR-29b levels contribute to increased BRD4 activity and ultimately CTCL disease pathogenesis.
miR-29b regulates BRD4 and IL-15 receptor complex expression in primary cells
To confirm the function of our proposed pathway in primary human cells, we purified CD4+ T cells from CTCL patients (n = 2) and transfected them with increasing doses of miR-29b mimic (100 ng and 250 ng) and scrambled control (250 ng). We achieved significant increase in miR-29b expression in these primary cells (P < .0001) (supplemental Figure 9A) and subsequently noted significantly decreased expression of BRD4 (100 ng: 0.75 ± 0.02 and 0.56 ± 0.02; 250 ng: 0.78 ± 0.02 and 0.62 ± 0.01) (supplemental Figure 9B) and all 3 components of the IL-15 receptor complex (IL-15Rα: 0.43 ± 0.03 and 0.59 ± 0.1745 for 100 ng and 0.29 ± 0.01 and 0.29 ± 0.03 for 250 ng; IL-15Rβ: 0.36 ± 0.01 and 0.68 ± 0.01 for 100 ng and 0.24 ± 0.01 and 0.62 ± 0.02 for 250 ng; IL-15Rγ: 0.28 ± 0.006 and 0.44 ± 0.02 for 100 ng and 0.24 ± 0.009 and 0.43 ± 0.02 for 250 ng) (supplemental Figure 9C-E). We also demonstrated decreased survival in miR-29b mimic–transfected cells (supplemental Figure 9F). These data further demonstrate that the activity of our proposed regulatory pathway occurs in primary human cells.
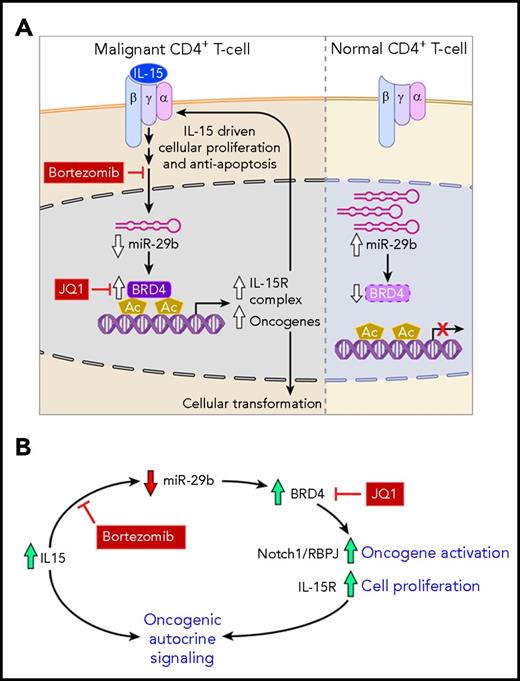
Our working model demonstrates that in malignant cells, the IL-15–driven decrease in miR-29b allows increased binding of BRD4, which results in tumor-associated gene expression, as well as expression of the IL-15 receptor complex (Figure 6A). This regulatory scheme promotes the formation of an autocrine feedback loop, which further promotes proliferation and cell survival in malignant cells (Figure 6B). Finally, we showed that strategic targeting of several components in this pathway led to disruption of this loop and resulted in a significant therapeutic benefit in our preclinical model.
Working model. (A) IL-15 receptor signaling in malignant cells (left) and normal CD4+ T cells (right). IL-15 cytokine binds its receptor complex α-β-γ to drive intracellular signaling, which includes reducing the level of miR-29b in cells. Decreased miR-29b releases negative regulation on BRD4 which then binds acetylated lysine residues on chromatin to direct gene expression. Upregulated genes include oncogenes and IL-15 receptor complex constituents, which drive cellular proliferation and resistance to apoptosis. Bortezomib inhibits the pathways that lead to decreased miR-29b expression, whereas JQ1 reduces BRD4 binding of chromatin, and both of these therapeutic strategies reduce oncogene and IL-15 receptor complex expression. In nonmalignant CD4+ T cells, miR-29b basal levels are higher, and BRD4 binding is relatively low. (B) IL-15 autocrine signaling loop. The regulatory pathway described facilitates formation of a self-sustaining autocrine loop that drives oncogene activation and cellular proliferation. This loop can be disrupted through use of bortezomib and JQ1.
Working model. (A) IL-15 receptor signaling in malignant cells (left) and normal CD4+ T cells (right). IL-15 cytokine binds its receptor complex α-β-γ to drive intracellular signaling, which includes reducing the level of miR-29b in cells. Decreased miR-29b releases negative regulation on BRD4 which then binds acetylated lysine residues on chromatin to direct gene expression. Upregulated genes include oncogenes and IL-15 receptor complex constituents, which drive cellular proliferation and resistance to apoptosis. Bortezomib inhibits the pathways that lead to decreased miR-29b expression, whereas JQ1 reduces BRD4 binding of chromatin, and both of these therapeutic strategies reduce oncogene and IL-15 receptor complex expression. In nonmalignant CD4+ T cells, miR-29b basal levels are higher, and BRD4 binding is relatively low. (B) IL-15 autocrine signaling loop. The regulatory pathway described facilitates formation of a self-sustaining autocrine loop that drives oncogene activation and cellular proliferation. This loop can be disrupted through use of bortezomib and JQ1.
Discussion
The role of epigenetic dysregulation in the pathogenesis of CTCL is an area of active research and clinical interest.3,37 In addition to global changes such as promoter DNA hypomethylation, mutations in epigenetic modifiers have been identified in CTCL patients and have been suggested to play a role in the development of disease.6,7,38,39 Epigenetic reader protein BRD4 has been a therapeutic target of interest in several tumor types because BRD4 inhibitors such as JQ1 have shown efficacy in early-phase clinical trials.40 The role and regulation of BRD4 in CTCL pathogenesis is not yet known. Further defining the mechanisms of epigenetic dysregulation in CTCL will be critical in developing effective treatment strategies to modify epigenetic landscapes and influence favorable clinical outcomes for CTCL patients.
In this study, we sought to assess the expression and regulation of BRD4 activity in CTCL cells as well as genome-wide and gene-specific patterns of BRD4 binding in CTCL patients. Previously studied mechanisms of the regulation of BRD4 focused on its subcellular localization41 or interaction with transcriptional co-activators such as Mediator.42 We identified IL-15 signaling and downstream miR-29b as novel regulators of BRD4 protein expression in CTCL. The significance of these findings is highlighted by demonstration of the critical role of IL-15 in CTCL pathogenesis,25 our observation of significantly lower expression of miR-29b in CD4+ T cells from CTCL patients, and increased binding of BRD4 to regulatory regions in these cells. The specificity of the effect of miR-29b on BRD4 expression was confirmed by using a GFP-3′UTR BRD4 vector construct in normal donor CD4+ T cells and by evaluating tissues from miR-29b−/− mice. We performed short-term in vitro treatment of CTCL cell lines with bortezomib and quantified pri-miR-29b1 to evaluate early effects. HuT-78 was the only cell line that did not increase miR-29b, and we suspect that this was a result of the NF-kB–truncating mutation harbored by HuT-78 cells.43,44 Bortezomib acts in part by inhibiting NF-kB, which also results in increased miR-29b,30 and thus the response in HuT-78 cells may be due to altered NF-kB signaling.
Decreased miR-29b results in increased BRD4 expression and subsequent binding at regulatory regions throughout the genome in CTCL patients. Molecular targeting of BRD4 by JQ1, which competitively binds the acetyl-lysine binding motif to displace BRD4 from chromatin,45 reverses BRD4-mediated effects on expression of NOTCH1 and RBPJ in patients and CTCL cell lines, as well as in vivo in the IL-15 transgenic mouse. We showed a slight decrease in BRD4 protein expression after in vitro and in vivo treatment with JQ1 in CTCL-derived cell lines and IL-15 transgenic mice, respectively. The cause is not certain but may represent increased degradation of unbound protein through physiologic cellular recycling.
We used a multi-tiered approach to demonstrate downstream effects of IL-15 signaling in CTCL. IL-15 expression is increased in CTCL cells, in part because of IL-15 promoter hypermethylation that results in reduced binding of transcriptional repressor Zeb1.30 Enhanced IL-15 signaling in CTCL is thought to form an autocrine loop with increased IL-15 receptor complex expression on tumor cells thus promoting cellular proliferation.30 A similar signaling loop is described in adult T-cell lymphocytic leukemia,46 but the mechanisms are poorly understood. We used the proteasome inhibitor bortezomib to indirectly decrease BRD4 function via increased levels of miR-29b, and a specific inhibitor, JQ1, to directly disrupt BRD4 chromatin binding to study the downstream effects of IL-15 signaling. Both approaches resulted in decreased expression of BRD4-regulated genes NOTCH1 and RBPJ and a decrease in IL-15 receptor complex constituents. The availability of several drugs targeting the 26S proteasome and current investigation of BRD4 inhibitors in clinical trials provides an opportunity to explore the therapeutic value of inhibiting this newly described miR-29b/BRD4 oncogenic loop in CTCL.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors acknowledge Donna Bucci from leukemia tissue bank, the comparative pathology and mouse phenotyping service, and Tim Vojt for assistance with image graphics. The authors also thank Carlo M. Croce, The Ohio State University for kindly providing miR-29b−/− mice.
This study was supported by the Spatz Foundation, American Skin Association, Cutaneous Lymphoma Foundation, Pelotonia Research Award, The Ohio State University Lymphoma Research Funds (A.M.) and a National Institutes of Health T32 fellowship from the Office of the Director (OD010429) (R.K.), and in part by National Institutes of Health, National Cancer Institute grant CA016058.
Authorship
Contribution: R.K. and A.M. designed the project; R.K., J.W., B.M.-B., K.M., A.K., L.G., A.H., B.M., N.C., and M.Y. performed experiments; B.M.-B., P.P., B.W., J.E.B., and M.A.C. provided mouse colony, patient samples, and research materials; R.K. wrote the manuscript; and all authors critically read and approved of the draft of the manuscript.
Conflict-of-interest disclosure: J.E.B. is now a shareholder and executive at Novartis AG. The remaining authors declare no competing financial interests.
Correspondence: Anjali Mishra, Ohio State University, 460 W. 12th Ave, Room 886, BRT, Columbus, OH 43210; e-mail: anjali.mishra@osumc.edu.
References
Author notes
P.P. and A.M. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal