Key Points
The MLL fusion partner ENL engages PAF1 to induce antisilencing H2B ubiquitination.
YEATS domain mutations or fusion with MLL increases affinity for PAF1 and transforms hematopoietic cells.
Abstract
Eleven-nineteen leukemia (ENL) is a chromatin reader present in complexes stimulating transcriptional elongation. It is fused to mixed-lineage leukemia (MLL) in leukemia, and missense mutations have been identified in Wilms tumor and acute myeloid leukemia. Here we demonstrate that ENL overcomes polycomb silencing through recruitment of PAF1 via the conserved YEATS domain, which recognizes acetylated histone H3. PAF1 was responsible for antirepressive activities of ENL in vitro, and it determined the transforming potential of MLL-ENL. MLL-ENL target loci showed supraphysiological PAF1 binding, hyperubiquitination of histone H2B and hypomodification with H2AUb, resulting in accelerated transcription rates. YEATS mutations induced a gain of function, transforming primary hematopoietic cells in vitro and in transplantation assays through aberrant transcription and H2B ubiquitination of Hoxa9 and Meis1. Mechanistically, H3 and PAF1 competed for ENL interaction, with activating mutations favoring PAF1 binding, whereas the MLL moiety provided a constitutive PAF1 tether allowing MLL fusions to circumvent H3 competition.
Introduction
The gene for the eleven-nineteen leukemia (ENL, or MLLT1) protein was initially discovered in leukemia as a mixed-lineage leukemia (MLL) fusion partner.1 More recently, somatic mutations of ENL have been discovered in a subset of Wilms tumors2 and in a case of pediatric acute myeloid leukemia (AML) (http://cancer.sanger.ac.uk/cosmic).
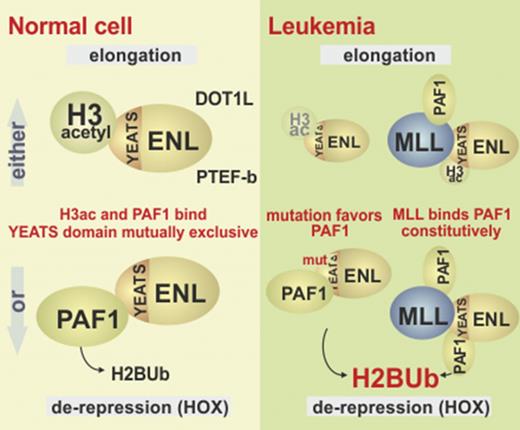
ENL copurifies with a large assembly of proteins called ENL associated proteins (EAP), which are necessary for efficient transcription by RNA polymerase II.3,4 EAP can be separated into 3 entities: the superelongation complex comprising positive transcription elongation factor b (P-TEFb) and several other MLL fusion partners such as AF4 (alternative name, AFF1)5 ; DOT1L, a histone H3K79 methyltransferase3,6 whose catalytic activity opposes epigenetic silencing by excluding histone deacetylase activities7 ; and paradoxically, polycomb repressive complex I (PRC1).8 As is implied by the name, PRC1 has a repressive function that renders chromatin less permissive for transcription. Biochemically, ENL acts as a chromatin reader through the highly conserved N-terminal YEATS domain. This moiety has a preference for acetylated histone H3,9-11 but experimentally it will also bind to unmodified H312 (for an overview of the various ENL interactions, see Figure 1A). Hence, ENL recruits its associated factors preferentially to active, open chromatin.
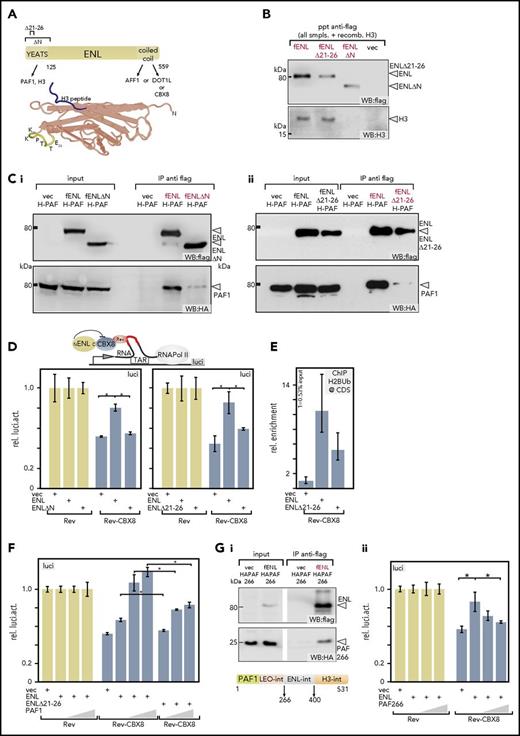
PAF1 binding is necessary for neutralization of polycomb mediated repression. (A) Schematic overview of ENL and crystal structure of a YEATS domain. Known binding partners of ENL are listed. Numbers denote amino acids. The mutations introduced for this study are indicated. The crystal structure (PDB: 4TMP) represents the YEATS domain of the close ENL homolog AF9 with a cocrystalized H3 tail peptide (blue). Highlighted amino acids are from ENL. (In AF9 the sequence reads KKPTVE). (B) Interactions of YEATS-domain mutants with histone H3. Extracts of 293T cells expressing flag-tagged versions of wild-type (wt)-ENL, ENL∆21-26, or ENL∆N were supplemented with recombinant histone H3, followed by flag-specific immunoprecipitation and immunoblotting. Flag- and H3-specific western blots are displayed. Because equal amounts of H3 were added manually, no separate input sample is shown. (C) Binding of PAF1 to YEATS mutants. Extracts from cells expressing HA-tagged PAF1 and flag-labeled wt-ENL, ENL∆N (Ci), or ENL∆21-26 (Cii) were precipitated with antiflag and analyzed on western blots, as indicated. (D) Elongation reporter assay. The impact of regulators on transcriptional elongation can be detected by a reporter system based on a HIV LTR that regulates transcription by elongation control. Proteins can be recruited selectively to a short RNA trailing behind a stalled RNA polymerase II by fusing them to the RNA binding protein Rev. In this way their impact on elongation can be measured by luciferase output. Either Rev-only as control (light tan bars) or Rev-CBX8 (dark blue bars) was recruited, and the impact of addition of wt-ENL or a YEATS mutant was recorded, as indicated. Values were normalized to the Rev-only control. The bar chart shows averages and standard deviations of triplicate experiments. (E) Chromatin immunoprecipitation on reporter plasmids. Elongation reporter plasmid, Rev-CBX8, and wt-ENL, ENL∆21-26, or a vector control were coexpressed and cells subjected to crosslinking and H2Bub-specific immunoprecipitation. Quantitation was done by qPCR, with primers spanning the luciferase coding sequence. Results shown are averages and standard deviations of PCR triplicates and represent a typical example of a duplicate. (F) Effect of increasing PAF1 concentration on ENL and ENL∆21-26-mediated derepression. Rev-CBX8 was coexpressed with limiting amounts of wt-ENL or ENL∆21-26 and increasing concentrations of PAF1, as indicated. The impact on elongation was measured and depicted as described for panel D. (G) Disruption of the PAF1 ENL interaction. An internal part of PAF1 encompassing amino acids 266 to 400 (PAF266) is sufficient for interaction with ENL in cells (Gi). A flag-specific immunoprecipitation of flag-ENL and HA-tagged PAF266 is shown. The ENL interaction domain is located between the previously identified18 LEO and histone H3-binding regions, as schematically depicted. Results of a reporter assay performed as described in panel F, exchanging wt-PAF1 for PAF266 (Gii). *P < .05. CDS, luciferase coding sequence; f, flag; H-PAF, HA-tagged PAF1; IP, immunoprecipitation; luci, luciferase; ppt, precipitation; recomb., recombinant; rel., relative; smpls., samples; TAR, transactivation response element; vec, vector; WB, western blot.
PAF1 binding is necessary for neutralization of polycomb mediated repression. (A) Schematic overview of ENL and crystal structure of a YEATS domain. Known binding partners of ENL are listed. Numbers denote amino acids. The mutations introduced for this study are indicated. The crystal structure (PDB: 4TMP) represents the YEATS domain of the close ENL homolog AF9 with a cocrystalized H3 tail peptide (blue). Highlighted amino acids are from ENL. (In AF9 the sequence reads KKPTVE). (B) Interactions of YEATS-domain mutants with histone H3. Extracts of 293T cells expressing flag-tagged versions of wild-type (wt)-ENL, ENL∆21-26, or ENL∆N were supplemented with recombinant histone H3, followed by flag-specific immunoprecipitation and immunoblotting. Flag- and H3-specific western blots are displayed. Because equal amounts of H3 were added manually, no separate input sample is shown. (C) Binding of PAF1 to YEATS mutants. Extracts from cells expressing HA-tagged PAF1 and flag-labeled wt-ENL, ENL∆N (Ci), or ENL∆21-26 (Cii) were precipitated with antiflag and analyzed on western blots, as indicated. (D) Elongation reporter assay. The impact of regulators on transcriptional elongation can be detected by a reporter system based on a HIV LTR that regulates transcription by elongation control. Proteins can be recruited selectively to a short RNA trailing behind a stalled RNA polymerase II by fusing them to the RNA binding protein Rev. In this way their impact on elongation can be measured by luciferase output. Either Rev-only as control (light tan bars) or Rev-CBX8 (dark blue bars) was recruited, and the impact of addition of wt-ENL or a YEATS mutant was recorded, as indicated. Values were normalized to the Rev-only control. The bar chart shows averages and standard deviations of triplicate experiments. (E) Chromatin immunoprecipitation on reporter plasmids. Elongation reporter plasmid, Rev-CBX8, and wt-ENL, ENL∆21-26, or a vector control were coexpressed and cells subjected to crosslinking and H2Bub-specific immunoprecipitation. Quantitation was done by qPCR, with primers spanning the luciferase coding sequence. Results shown are averages and standard deviations of PCR triplicates and represent a typical example of a duplicate. (F) Effect of increasing PAF1 concentration on ENL and ENL∆21-26-mediated derepression. Rev-CBX8 was coexpressed with limiting amounts of wt-ENL or ENL∆21-26 and increasing concentrations of PAF1, as indicated. The impact on elongation was measured and depicted as described for panel D. (G) Disruption of the PAF1 ENL interaction. An internal part of PAF1 encompassing amino acids 266 to 400 (PAF266) is sufficient for interaction with ENL in cells (Gi). A flag-specific immunoprecipitation of flag-ENL and HA-tagged PAF266 is shown. The ENL interaction domain is located between the previously identified18 LEO and histone H3-binding regions, as schematically depicted. Results of a reporter assay performed as described in panel F, exchanging wt-PAF1 for PAF266 (Gii). *P < .05. CDS, luciferase coding sequence; f, flag; H-PAF, HA-tagged PAF1; IP, immunoprecipitation; luci, luciferase; ppt, precipitation; recomb., recombinant; rel., relative; smpls., samples; TAR, transactivation response element; vec, vector; WB, western blot.
ENL links to PRC1 through a direct contact with its subunit CBX8.13 CBX8 is a chromatin reader recognizing methylated histone H3K27. This mark is deposited by the catalytic activity of the companion polycomb repressive complex 2. Because PRC1 contains the histone H2A-specific ubiquitin ligases RING1/2, H2AUb is a hallmark of repressed chromatin. Binding of CBX8 and other PRC1 components can physically compact chromatin also in the absence of any catalytic activity, supposedly by forming multimeric aggregates.14,15 Previously, we have found that ENL acts epistatic to CBX8, because ENL neutralizes CBX8-associated repressive activity, thus explaining the paradoxical interaction of 2 factors with opposite effects on transcription.8
In addition to EAP proteins, transcriptional activation by ENL has been linked to polymerase-associated factor 1 (PAF1). Although ENL does not readily copurify with PAF1, in certain contexts the elongation-stimulating properties of ENL are dependent on a direct interaction with this protein.16,17 PAF1 binds to chromatin with preference for methylated H3K4, but in vitro interaction was also possible with unmodified full-length H3.18,19 PAF1 is the central constituent of a larger complex (PAFc) that coordinates various stages of transcription from initiation to termination. A hallmark of PAFc activity is the deposition of monoubiquitin on histone H2B, catalyzed by the associated RNF20/RNF40 ubiquitin ligase. H2BUb modification is a prerequisite for H3K4- and H3K79 methylation, with H2BUb posing a steric obstruction to chromatin compaction20 with HOX genes as sentinel loci that react first to a perturbation of H2BUb deposition.21 PAF1 also binds to MLL and MLL fusion proteins.22,23 Whereas it was first believed that PAF1 mainly aids the targeting of MLL fusions, a recent report24 rather ascribes a role for PAF1 in MLL-fusion-induced gene activation.
Here, we show that an interaction of ENL with PAF1 is key to the relief from PRC1 repression, and we provide information on how this process can be controlled by explaining the gain of oncogenic properties by ENL mutants and MLL-ENL fusion proteins.
Methods
Detailed methods can be found in the supplemental Methods, available on the Blood Web site.
Transplantation assays
For transplantation experiments, sublethally irradiated (Balb/C; 7Gy) syngenic recipients were injected with 1 × 106 transduced cells and 1 × 106 total bone marrow cells for radioprotection.
Elongation reporter assays
Elongation was assessed by a special reporter described by Gold and Rice.25 In essence, this system is based on a HIV long terminal repeat (LTR) promoter that is known to be controlled at the elongation step at which effects on pausing/elongation of RNA polymerase can then be directly read out as luciferase activity.
Chromatin immunoprecipitation sequence (ChIP-seq) and RNA seq
ChIP was done on formaldehyde crosslinked cells according to Milne et al.26 Antibodies used are described in the supplemental Methods. Raw data were submitted to the European Bioinformatics Institute under accession number E-MTAB-5569. Nascent RNA seq was done exactly as described by Garcia-Cuellar et al.27
Results
ENL requires PAF1 to counteract CBX8-induced repression
Previously, we have shown that ENL counteracts polycomb-mediated repression upon binding the PRC1 component CBX8 through the C-terminus.8 Because deletion studies showed that this C-terminal domain is necessary but not sufficient for derepression,8 we surmised that the N-terminal YEATS domain of ENL may play an additional role for this activity. This moiety interacts with PAF116,17 and histone H3.10,12 To discriminate whether PAF1 or histone binding is essential for relief from CBX8-induced repression, we constructed 2 mutants with differential binding capabilities. Aided by the YEATS structure (Figure 1A), we deleted a small internal fragment (aa21-26, KKPTTE) opposite to the histone-interacting surface to target PAF1 interaction (ENL∆21-26). A construct deleting the entire domain up to aa125 (ENL∆N) served as a control. Although the YEATS peptide preferentially binds to acetylated histone tails, experimentally it also interacts with unmodified H3, enabling immunoprecipitations in the presence of recombinant histone (Figure 1B). The ∆21-26 mutant did not affect association with H3, whereas a loss of the entire YEATS domain abolished this interaction. In contrast, both deletions significantly reduced affinity for PAF1 (Figure 1C). YEATS mutants were consistently expressed at lower levels, supporting an involvement of this domain in transcriptional autoactivation of the plasmid-encoded promoter. Because CBX8 could simultaneously coprecipitate ENL and PAF1 (supplemental Figure 1A), CBX8 binding did not interfere with PAF1 interaction. Likewise, the ENL∆21-26 deletion mutant did not affect interaction of ENL with DOT1L, AF5, or CBX8 (supplemental Figure 1B). Next, the capability of ENL∆N and ENL∆21-26 to overcome CBX8-induced repression was tested in an elongation reporter system (Figure 1D). In this assay, CBX8 is recruited via the Rev-RNA binding protein to nascent RNA that trails behind a stalled RNA polymerase on a HIV LTR promotor. Whereas wt-ENL counteracted CBX8-induced repression, both mutants were largely ineffective, with only spurious activity remaining for the ∆21-26 deletion. Because ENL∆21-26 loses most of PAF1 affinity but still binds to H3, this suggested PAF1 as the critical component mediating antirepressive activity. Because PAF1 is associated with monoubiquitination of H2B, ChIP assays were done on the reporter plasmid. Even though this was a transient assay, H2BUb could be detected on reporter chromatin, with ENL∆21-26 sustaining much lower modification levels than did wt-ENL (Figure 1E). Further support for an involvement of PAF1 in antirepression was seen in cotransfection experiments (Figure 1F). Increasing available PAF1 boosted ENL-mediated neutralization of CBX8-induced repression. Concomitant with a residual affinity for PAF1, higher PAF1 concentrations also slightly strengthened the antirepressive activity of ENL∆21-26; however, this effect was significantly weaker than that observed with wt-ENL (Figure 1F). To specifically perturb the ENL-PAF1 interaction, we created a PAF1 fragment encompassing aa 266 to 400 (PAF266), corresponding to a minimal ENL interaction domain (Figure 1Gi). Addition of PAF266 to the elongation assay reduced CBX8 neutralization (Figure 1Gii). In summary, these experiments support an essential role of PAF1 for the antirepressive process mediated by ENL.
PAF1 and transformation by MLL-ENL
Previously, structure function analyses of MLL-ENL uncovered that the YEATS domain is actually dispensable for transformation.28 This was corroborated by clinical samples that only join the C-terminus of ENL to MLL (Figure 2A). Although at first sight this argues against a role of PAF1 for transformation, actually the MLL moiety of the fusion itself also binds to PAF1. Two independent laboratories22,23 have demonstrated that MLL needs PAF1 for efficient leukemogenesis. In these studies, PAF1 interaction could be abrogated either by introducing a point mutation (R1153A) or by deleting amino acids from position 1258 to the fusion breakpoint (∆1258). This enabled complementation experiments using flag-tagged MLL derivatives that joined wt-MLL or the PAF1-defective mutants R1153A/∆1258 to either full-length ENL or a shorter version containing the ENL C-terminus. Expression of the resulting fusion proteins was confirmed by immunoblot (Figure 2B). The oncogenic capacity of the individual constructs was assessed by replating assays of transduced hematopoietic precursors (Figure 2B-C). Whereas we could confirm that MLL-mediated PAF1 binding was crucial for oncogenic activity, this held true only for the shorter fusions. Constructs including the YEATS domain efficiently complemented for PAF1-binding deficiency of MLL mutants, and they transformed cells regardless of whether MLL-wt or a PAF1-defective MLL derivative was included. Interestingly, PAF1 binding capacity was the major determinant of colony counts. Constructs with a single PAF1 interaction domain formed significantly fewer colonies than did those that offered 2 possibilities for interaction. Juxtaposing MLL and MLL mutants with impaired PAF1 binding to ENL∆21-26, which itself has some residual PAF1 affinity (see above), reduced colony formation significantly, but not to the extent as observed with fusions that delete the complete YEATS domain (supplemental Figure 2A-B). Biochemically, MLL-ENL precipitated more PAF1 than MLL-ENL∆21-26 (supplemental Figure 2C). ENL complemented the function of PAF1-negative MLL mutants also in vivo. MLL_R1153A-ENL- and MLL-ENL-transduced cells caused lethal disease with comparable latency after injection into sublethally irradiated recipients (Figure 2D). However, animals receiving the “single PAF1 binder” MLL_R1153A-ENL had significantly lower spleen weights and showed a trend to lower white blood cell counts. Permanent cell lines could be recovered from all animals consisting of myeloid precursors with a Kitnegative/lowCD11bpositiveGr1negative phenotype and occasional differentiation toward a Gr1positive status (supplemental Figure 2D).
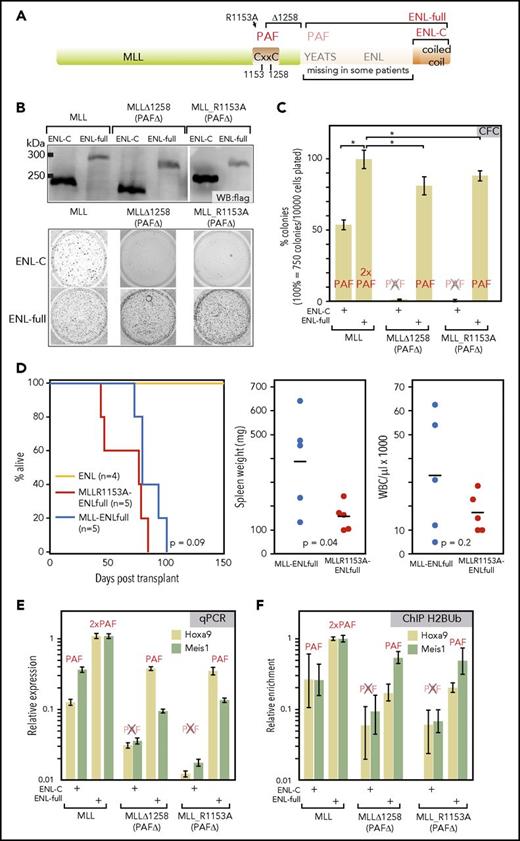
PAF1 is necessary for MLL-ENL function and can be recruited by MLL and ENL. (A) Schematic overview of the MLL-ENL fusion protein. A CxxC domain critical for MLL-ENL function has been previously identified as PAF1-binding site.22,23 Two mutations that abrogate PAF1 interaction (R1153A and a deletion of the CxxC domain from aa1258 to the breakpoint) are indicated. In patient samples, either nearly full length ENL or only the C-terminal coiled-coil can be found fused to MLL. (B) Expression and activity of combinatorial MLL-ENL fusion derivatives. Fusions of wt-MLL or the 2 PAF1-binding defective mutants MLL∆1258 and MLL R1153A with either full-length ENL or only the ENL C-terminal domain were assembled in retroviral vectors. The correct expression of the corresponding flag-tagged proteins was ascertained by western blot in extracts of retroviral packaging cell lines. Biological activity was assessed by transduction of primary hematopoietic precursors isolated from murine bone marrow, followed by a standard replating assay. The figure shows stained colonies of cells after 2 replatings that were transduced with MLL constructs as labeled. 10000 cells were plated for each well. (C) Evaluation of colony-forming cell numbers. Colony-forming assays, as above, were conducted in triplicate with averages and standard deviations given for each combination. The ability of each construct to bind PAF1 is indicated above the respective column. Colony numbers are given in relation to MLL-ENL. (D) Transplantation assays. Primary hematopoietic cells were transduced with MLL R1153A-ENLfull, which recruits PAF1 exclusively through the ENL moiety or with MLL-ENL and ENL as controls. Infected cells were selected and injected into sublethally irradiated syngenic recipients. The Kaplan-Meier plot gives survival times in days after transplant. Dot plots indicate spleen weights and white blood cell counts of diseased animals including statistical evaluation by 2-tailed t test. (E) Expression of key MLL-ENL target genes Hoxa9 and Meis1 is dependent on PAF1 binding. Primary hematopoietic precursor cells were transduced with wt and PAF1 defective MLL ENLfull/ENL-C combinations, as above. RNA was harvested after the first round of replating at a time point when all cells had not yet exhausted their replating potential. Hoxa9 and Meis1 RNA concentrations were determined by qPCR and normalized to β-actin expression. Shown are averages and standard deviations of PCR triplicates in relation to expression levels reached in cells transduced with MLL-ENLfull. (F) Histone H2B ubiquitination at Hoxa9 and Meis1 loci. Cells transduced and harvested as described for panel D were subject to crosslinking and H2Bub-specific chromatin immunoprecipitation. Material from the Hoxa9 and Meis1 loci was quantified with primers for the respective region by qPCR. Results are plotted as before. *P < .05. CFC, colony-forming cell; WBC, white blood cells.
PAF1 is necessary for MLL-ENL function and can be recruited by MLL and ENL. (A) Schematic overview of the MLL-ENL fusion protein. A CxxC domain critical for MLL-ENL function has been previously identified as PAF1-binding site.22,23 Two mutations that abrogate PAF1 interaction (R1153A and a deletion of the CxxC domain from aa1258 to the breakpoint) are indicated. In patient samples, either nearly full length ENL or only the C-terminal coiled-coil can be found fused to MLL. (B) Expression and activity of combinatorial MLL-ENL fusion derivatives. Fusions of wt-MLL or the 2 PAF1-binding defective mutants MLL∆1258 and MLL R1153A with either full-length ENL or only the ENL C-terminal domain were assembled in retroviral vectors. The correct expression of the corresponding flag-tagged proteins was ascertained by western blot in extracts of retroviral packaging cell lines. Biological activity was assessed by transduction of primary hematopoietic precursors isolated from murine bone marrow, followed by a standard replating assay. The figure shows stained colonies of cells after 2 replatings that were transduced with MLL constructs as labeled. 10000 cells were plated for each well. (C) Evaluation of colony-forming cell numbers. Colony-forming assays, as above, were conducted in triplicate with averages and standard deviations given for each combination. The ability of each construct to bind PAF1 is indicated above the respective column. Colony numbers are given in relation to MLL-ENL. (D) Transplantation assays. Primary hematopoietic cells were transduced with MLL R1153A-ENLfull, which recruits PAF1 exclusively through the ENL moiety or with MLL-ENL and ENL as controls. Infected cells were selected and injected into sublethally irradiated syngenic recipients. The Kaplan-Meier plot gives survival times in days after transplant. Dot plots indicate spleen weights and white blood cell counts of diseased animals including statistical evaluation by 2-tailed t test. (E) Expression of key MLL-ENL target genes Hoxa9 and Meis1 is dependent on PAF1 binding. Primary hematopoietic precursor cells were transduced with wt and PAF1 defective MLL ENLfull/ENL-C combinations, as above. RNA was harvested after the first round of replating at a time point when all cells had not yet exhausted their replating potential. Hoxa9 and Meis1 RNA concentrations were determined by qPCR and normalized to β-actin expression. Shown are averages and standard deviations of PCR triplicates in relation to expression levels reached in cells transduced with MLL-ENLfull. (F) Histone H2B ubiquitination at Hoxa9 and Meis1 loci. Cells transduced and harvested as described for panel D were subject to crosslinking and H2Bub-specific chromatin immunoprecipitation. Material from the Hoxa9 and Meis1 loci was quantified with primers for the respective region by qPCR. Results are plotted as before. *P < .05. CFC, colony-forming cell; WBC, white blood cells.
Complementation of the MLL PAF1-binding deficiency by the ENL YEATS domain was also seen in quantitative polymerase chain reaction (qPCR) experiments checking the 2 most crucial MLL-ENL downstream target genes Hoxa9 and Meis1 (Figure 2E). In line with the replating results, PAF1 was necessary for activation of Hoxa9 and Meis1, but it could be supplied by either MLL or ENL. This was congruent with ChIP results detecting H2BUb, the major histone modification associated with PAF1 at Hoxa9 and Meis1 loci (Figure 2F) where H2BUb deposition depended on recruitment of PAF1.
MLL-ENL target genes are characterized by an unusual H2B ubiquitination pattern
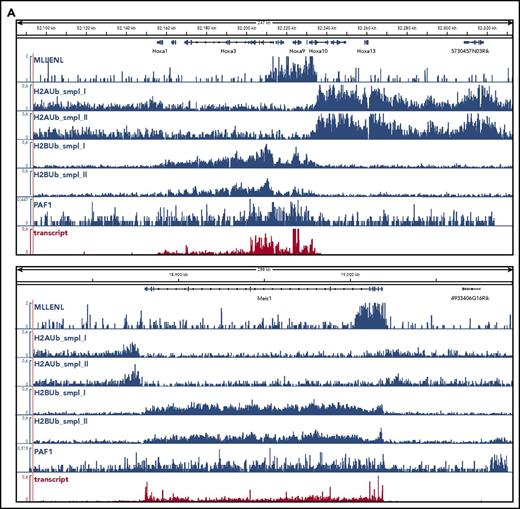
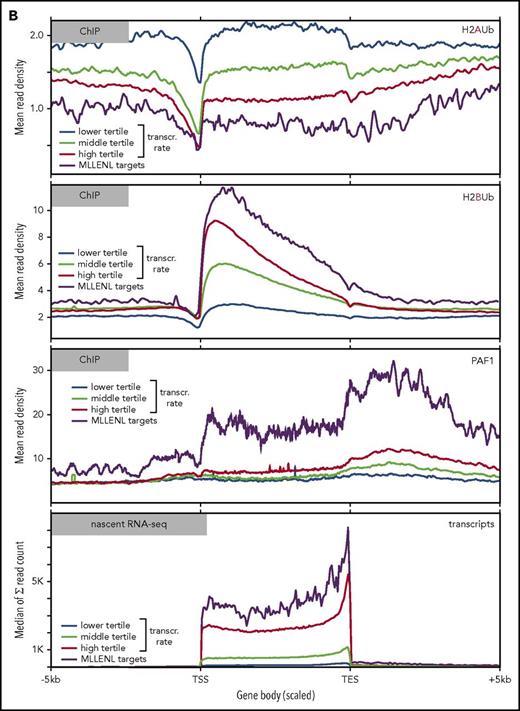
To determine the global relation of PAF1 and PRC1 activities, we performed ChIP-seq experiments in Meer cells (Figure 3). These cells carry a conditional Mll-ENL fusion under control of the endogenous murine promoter and have been extensively characterized previously.27 Precipitations for H2AUb (a hallmark activity of PRC1) and for H2BUb as well as PAF1 were performed (Figure 3). H2AUb and H2BUb were perfectly countercorrelated not only at the crucial Hox and Meis1 loci (Figure 3A) but equally well on a global scale (Figure 3B). Generally, H2AUb was organized in large domains extending upstream and downstream of transcribed units. Levels of H2AUb were inversely correlated with transcript rates, and MLL-ENL targets showed the lowest H2AUb modification of all transcribed genes.
Global analysis of H2AUb and H2BUb modification in MLL-ENL transformed cells. (A) Integrated genome viewer visualization of MLL-ENL occupancy, H2AUb and H2BUb modification (2 independent biological replicates each), PAF1 binding, and transcript landscape as determined by nascent RNA sequencing at HoxA- and Meis1 loci. Data were recorded in Meer cells, which are cells transformed by an inducible Mll-ENL derivative created by knocking in an ENL-estrogen receptor fusion into the endogenous murine Mll locus. Meer cells can be derived by cultivating bone marrow precursor cells of Meer-animals in the presence of tamoxifen and cytokines. MLL-ENL binding and transcript data were taken from Garcia-Cuellar et al.27 (B) Global distribution of H2AUb, H2Bub, and PAF1 in Meer cells. Plots depict the relative distribution of the 2 ubiquitin marks, PAF1 occupancy, as well as transcript density with respect to transcribed genes. MLL-ENL target genes, as defined by Garcia-Cuellar et al,27 were separately analyzed with all other genes binned into 3 groups according to their transcription rates. Transcr, transcription.
Global analysis of H2AUb and H2BUb modification in MLL-ENL transformed cells. (A) Integrated genome viewer visualization of MLL-ENL occupancy, H2AUb and H2BUb modification (2 independent biological replicates each), PAF1 binding, and transcript landscape as determined by nascent RNA sequencing at HoxA- and Meis1 loci. Data were recorded in Meer cells, which are cells transformed by an inducible Mll-ENL derivative created by knocking in an ENL-estrogen receptor fusion into the endogenous murine Mll locus. Meer cells can be derived by cultivating bone marrow precursor cells of Meer-animals in the presence of tamoxifen and cytokines. MLL-ENL binding and transcript data were taken from Garcia-Cuellar et al.27 (B) Global distribution of H2AUb, H2Bub, and PAF1 in Meer cells. Plots depict the relative distribution of the 2 ubiquitin marks, PAF1 occupancy, as well as transcript density with respect to transcribed genes. MLL-ENL target genes, as defined by Garcia-Cuellar et al,27 were separately analyzed with all other genes binned into 3 groups according to their transcription rates. Transcr, transcription.
H2BUb spread across the complete transcribed region, with the highest density recorded immediately downstream of the transcription initiation site. Similarly to DOT1L-catalyzed H3K79 methylation,29 H2BUb domains extended well beyond narrow MLL-ENL peaks. Genes in the upper tertile of transcription rates were more densely decorated with H2BUb than were those in the middle and lower ranges. MLL-ENL targets were characterized by the highest concentration of H2BUb of all active genes correlating to their accelerated transcription rates. A similar pattern was seen for PAF1. In line with previous results published by Roeder and colleagues30 and reflecting the multiple functions of PAF1 during transcript processing, binding was detectable across the whole transcribed region, with a particularly strong signal beyond the transcription end sites.
Wilms tumor–specific ENL alterations create gain-of-function mutants that transform myeloid cells
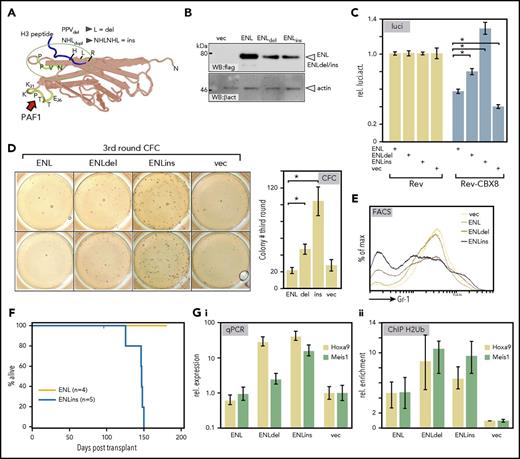
Mutations in the YEATS domain of ENL have been reported in a subset of Wilms tumors marked by a relative overexpression of HOX genes, particularly HOXA13.2 Superimposing the Wilms mutations on the YEATS structure showed that these alterations cluster in a region mediating histone contacts (Figure 4A; for sequence, see supplemental Figure 3A). To examine the effect of these pathogenic ENL derivatives on transcriptional elongation, we introduced 2 patient-specific mutations into the ENL framework. A deletion changing a PPV peptide to a single leucine (ENLdel) and a duplication of a NHL tripeptide (ENLins) were created. These alterations led to a slight reduction in protein expression levels; nonetheless, ENLdel and in particular ENLins had a considerably stronger output in HIV-LTR reporter elongation assays overcoming CBX8-mediated repression more efficiently than did wt-ENL (Figure 4B-C).
Wilms tumor–specific mutations in ENL are gain-of-function alterations. (A) Superimposition of somatic mutations identified in a subgroup of Wilms tumors onto the YEATS crystal structure of the ENL homolog AF9. These mutations affect a loop close to the histone-binding surface and either change a PPV peptide to a single leucine (ENLdel) or duplicate an immediately adjacent NHL tripeptide (ENLins). (B) Expression of Wilms tumor–specific mutations. Equal amounts of wt-ENL, ENLdel, and ENLins were transfected into 293T cells, and protein expression was detected by flag-specific western blot. (C) Derepressive activity of Wilms ENL mutants. Upon cotransfection with the elongation reporter plasmid and Rev-CBX8, the ability of ENL derivatives to counteract CBX8-induced repression was recorded in luciferase assays. Constructs were transfected as indicated with bars representing averages and standard deviation of triplicates. The figure is a typical example of a duplicate experiment. (D) Wilms-specific ENL mutations induce colony-forming cell activity. Primary hematopoietic precursor cells were isolated from bone marrow and transduced with vector wt-ENL, ENLdel, and ENLins, as indicated. Cells were replated twice into growth factor–supplemented methylcellulose. The figure shows stained colonies arising in third round of growth after 2 replatings (left; duplicate samples), and the bar chart depicts average colony numbers and standard deviations of an independent biological triplicate. (E) ENL Wilms mutants retard differentiation. FACS surface marker staining for the differentiation marker Gr1 (Ly6G) on cells transduced with ENL and derivatives as indicated. Cells recovered after the primary plating round were used before control cells exhausted their replicative potential. (F) Kaplan-Meier survival plot of animals transplanted with primary hematopoietic cells expressing either ENL or the Wilms tumor–derived mutant ENLins. Note that the experiment was terminated after 175 days with all ENL-injected animals still healthy. (G) Effect of ENL Wilms mutants on Hoxa9 and Meis1 loci. RNA and crosslinked chromatin was prepared from cells, as described for panel E, which were recovered after 1 round of replating. Hoxa9 and Meis1 expression was tested by qPCR (Gi) and H2Bub- specific ChIP was performed (Gii). Bar charts show averages and standard deviations of PCR triplicates and are representative of a duplicate experiment.
Wilms tumor–specific mutations in ENL are gain-of-function alterations. (A) Superimposition of somatic mutations identified in a subgroup of Wilms tumors onto the YEATS crystal structure of the ENL homolog AF9. These mutations affect a loop close to the histone-binding surface and either change a PPV peptide to a single leucine (ENLdel) or duplicate an immediately adjacent NHL tripeptide (ENLins). (B) Expression of Wilms tumor–specific mutations. Equal amounts of wt-ENL, ENLdel, and ENLins were transfected into 293T cells, and protein expression was detected by flag-specific western blot. (C) Derepressive activity of Wilms ENL mutants. Upon cotransfection with the elongation reporter plasmid and Rev-CBX8, the ability of ENL derivatives to counteract CBX8-induced repression was recorded in luciferase assays. Constructs were transfected as indicated with bars representing averages and standard deviation of triplicates. The figure is a typical example of a duplicate experiment. (D) Wilms-specific ENL mutations induce colony-forming cell activity. Primary hematopoietic precursor cells were isolated from bone marrow and transduced with vector wt-ENL, ENLdel, and ENLins, as indicated. Cells were replated twice into growth factor–supplemented methylcellulose. The figure shows stained colonies arising in third round of growth after 2 replatings (left; duplicate samples), and the bar chart depicts average colony numbers and standard deviations of an independent biological triplicate. (E) ENL Wilms mutants retard differentiation. FACS surface marker staining for the differentiation marker Gr1 (Ly6G) on cells transduced with ENL and derivatives as indicated. Cells recovered after the primary plating round were used before control cells exhausted their replicative potential. (F) Kaplan-Meier survival plot of animals transplanted with primary hematopoietic cells expressing either ENL or the Wilms tumor–derived mutant ENLins. Note that the experiment was terminated after 175 days with all ENL-injected animals still healthy. (G) Effect of ENL Wilms mutants on Hoxa9 and Meis1 loci. RNA and crosslinked chromatin was prepared from cells, as described for panel E, which were recovered after 1 round of replating. Hoxa9 and Meis1 expression was tested by qPCR (Gi) and H2Bub- specific ChIP was performed (Gii). Bar charts show averages and standard deviations of PCR triplicates and are representative of a duplicate experiment.
ENLins and ENLdel were retrovirally transduced into hematopoietic precursors and subjected to replating assays in which ENLdel and particularly ENLins enhanced self-renewal ability. ENLins cells formed up to 5-fold more colonies after 2 rounds of replating, whereas overexpression of wt-ENL had no effect (Figure 4D). Despite this transient enhancement of self-renewal, there was no complete transformation in vitro because the cells eventually exhausted their replating potential. The partial differentiation block accompanying the increased self-renewal could be detected by fluorescence-activated cell sorter (FACS) (Figure 4E). Cell populations transduced with ENLdel or ENLins showed lower levels of the differentiation marker Gr1 (Ly6g) than did wt-ENL or vector controls. Although Kit expression was generally low, the relative levels of this precursor-specific molecule corroborated the differentiation retardation induced by the ENL mutants (supplemental Figure 3B). Unexpectedly, ENLins caused fully malignant disease in transplantation studies. Sublethally irradiated syngenic recipients of ENLins-transduced cells succumbed to a myeloproliferative disorder/leukemia after an average latency of 139 days, whereas control animals receiving ENL-transduced cells stayed healthy until the termination of the experiment at 175 days (Figure 4F). Disease was characterized by hepato/splenomegaly and high blood counts (supplemental Figure 3C). Blood, spleen, and bone marrow showed a prominent infiltration of myeloid cells at various stages of differentiation (supplemental Figure 3D). The visual impression was supported by FACS of cells recovered from spleen that demonstrated a dominance of myeloid cells that were double-positive for CD11b and Gr-1 (supplemental Figure 3E). Corroborating the replating assays, cells reisolated from ENLins animals could not be grown permanently in vitro. In culture these cells differentiated within 2 to 3 weeks into granulocytes and macrophages, indicating that additional self-renewal signals must be present in bone marrow that were not adequately replaceable by growth medium. Because differentiation and self-renewal are strongly influenced by the Hox/Meis system, we tested the molecular changes at Hoxa9 and Meis1 loci by qPCR and ChIP. Indeed, ENLdel and ENLins cells had higher levels of Hoxa9 and to a lesser extent of Meis1. This was paralleled by a higher modification density with H2BUb at these loci in relation to vector and wt-ENL controls (Figure 4G). Cells directly reisolated from animals with ENLins-induced disease also showed an abnormally abundant transcription of Hoxa9 and Meis1 in comparison with normal bone marrow, although expression did not reach the levels induced by MLL-ENL transformed cells (supplemental Figure 3F). ChIP-seq and nascent RNA-seq experiments were performed on ENL/ENLins cells (supplemental Figure 4). Because ENL-wt by itself does not transform or retard differentiation, expansion of transduced cells was curbed by their endogenous self-renewal capacity, and thus the available number of cells was very limited. Nevertheless, ChIP and RNA-seq profiles could be obtained for genes with either high occupation density or high expression, respectively. Confirming the predominant role of Hoxa9 and Meis1 for transformation, ENLins was present at Hox/Meis1 loci, but no binding was visible in ENL-wt- transduced cells (supplemental Figure 4A). Overall the genomic pattern of ENLins was very similar to that observed for ENL-wt, but ENLins seemed to possess a generally higher chromatin affinity (supplemental Figure 4B). As was expected, nascent RNA-seq confirmed the physiological status of ENLins and ENL-wt cells (supplemental Table 1). Whereas the former were still in a proliferative state, the latter were located further along a differentiation trajectory. Gene Set Enrichment Analysis revealed a prevalent Myc signature in ENLins cells (supplemental Figure 4C). ENL-wt cells scored highly positive for a tumor necrosis factor (TNF)α-induced signal, with TNFα being the major cytokine produced by mature macrophages. Interestingly, ENLins-induced gene expression resembled an acute leukemia characterized by FLT3 internal tandem duplications (Valk, cluster 331 ), and ENL-wt cells overexpressed genes that are specifically underrepresented in Wilms tumor in comparison with normal kidney. Finally, independent corroboration for a role of ENL YEATS mutations in human hematopoietic malignancies was found in the latest version of the Cosmic database. A case of pediatric AML (COSM5487400) was identified that contained an ENL mutation strongly resembling the characteristic changes occurring in Wilms tumor. In AML the amino acids HLR within the YEATS sequence NHLR were duplicated, whereas in Wilms cases the immediately adjacent NHL tripeptide was affected (supplemental Figure 4D).
Wilms mutations change the YEATS binding properties
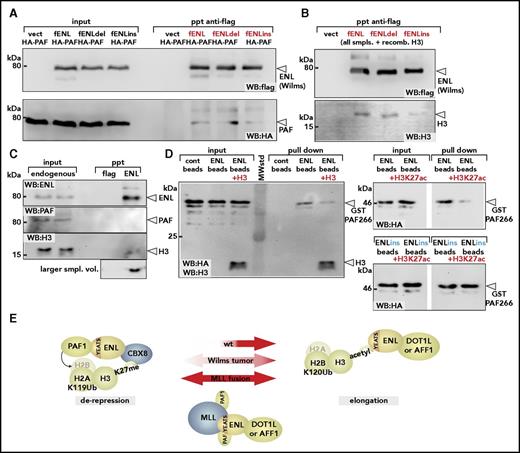
We examined the consequences of Wilms mutations for binding to PAF1 and histone H3 by coprecipitation. Flag-tagged wt-ENL and the mutants ENLdel and ENLins were each coexpressed with PAF1, and mutual interaction was probed by flag-specific immunoprecipitation. Unexpectedly, and despite the previous indications that Wilms mutants of ENL are associated with higher PAF1 activity, no difference in PAF1 affinity could be detected in these experiments (Figure 5A). Conversely, in analogous experiments, histone H3 binding was clearly reduced, particularly for the ENLins derivative (Figure 5B). This was in line with the previous results of Perlman et al,2 who noted a lower affinity of Wilms-specific ENL mutants for acetylated H3-peptides in isothermal titration calorimetry.
Wilms mutations change the YEATS-binding properties. (A) Binding of PAF1 by wt-ENL and Wilms mutants. Flag-tagged versions of wt-ENL, ENLdel, and ENLins were coexpressed together with HA-labeled PAF1, and the interaction was probed by immunoblot. (B) Affinity of ENL and mutants for histone H3. Cell extracts expressing ENL and respective mutants were supplemented with identical amounts of recombinant histone H3, and flag-specific immunoprecipitates were analyzed for ENL and H3 content. Note that because H3 was added exogenously, there is no separate input sample. (C) Coimmunoprecipitation of endogenous ENL, PAF1, and H3. ENL was purified from native 293T cell extracts by anti-ENL precipitation, and coprecipitating material was analyzed for PAF1 and H3 by western blotting with antibodies recognizing the respective proteins. The histone blot was done with two different sample volumes of precipitated material. (D) Pulldown of PAF1 and ENL/ENLins. The ENL N-terminus was expressed and purified as GST fusion protein (left). After cleavage from the GST moiety, ENL was covalently attached to agarose beads and incubated with a purified GST-HA-PAF266 protein, excluding the intrinsic PAF1 histone- binding domain. Pulldown was done either in the absence or after addition of recombinant histone H3. Note that the immunoblot was simultaneously developed with antihistone H3 and anti-HA antibodies. Analogous experiment done with ENL (right upper) or ENLins protein bound to beads (right lower), including H3K27 acetylated histone peptides as competitors for PAF1 binding. (E) Schematic depiction of different ENL functions. In the environment of polycomb-repressed chromatin, ENL can bind to CBX8 and recruit PAF1 through the YEATS domain. Because silenced chromatin is devoid of acetylated histone H3, the YEATS domain is free to interact with PAF1. After chromatin opening, large quantities of acetylated H3 becomes available, occupying the YEATS surface, thus supporting transfer of PAF1 to other binding partners such as RNA polymerase II. Because no CBX8 is present on active chromatin, this will free the ENL C-terminus for interaction with either DOT1L or AFF1. Wilms-specific mutations weaken interaction with histone H3, shift the balance, and favor derepression. MLL fusions permanently tether PAF1 to DOT1/AFF1 through the constitutive PAF1-binding site in the MLL N-terminus. Cont, control; vect, vector.
Wilms mutations change the YEATS-binding properties. (A) Binding of PAF1 by wt-ENL and Wilms mutants. Flag-tagged versions of wt-ENL, ENLdel, and ENLins were coexpressed together with HA-labeled PAF1, and the interaction was probed by immunoblot. (B) Affinity of ENL and mutants for histone H3. Cell extracts expressing ENL and respective mutants were supplemented with identical amounts of recombinant histone H3, and flag-specific immunoprecipitates were analyzed for ENL and H3 content. Note that because H3 was added exogenously, there is no separate input sample. (C) Coimmunoprecipitation of endogenous ENL, PAF1, and H3. ENL was purified from native 293T cell extracts by anti-ENL precipitation, and coprecipitating material was analyzed for PAF1 and H3 by western blotting with antibodies recognizing the respective proteins. The histone blot was done with two different sample volumes of precipitated material. (D) Pulldown of PAF1 and ENL/ENLins. The ENL N-terminus was expressed and purified as GST fusion protein (left). After cleavage from the GST moiety, ENL was covalently attached to agarose beads and incubated with a purified GST-HA-PAF266 protein, excluding the intrinsic PAF1 histone- binding domain. Pulldown was done either in the absence or after addition of recombinant histone H3. Note that the immunoblot was simultaneously developed with antihistone H3 and anti-HA antibodies. Analogous experiment done with ENL (right upper) or ENLins protein bound to beads (right lower), including H3K27 acetylated histone peptides as competitors for PAF1 binding. (E) Schematic depiction of different ENL functions. In the environment of polycomb-repressed chromatin, ENL can bind to CBX8 and recruit PAF1 through the YEATS domain. Because silenced chromatin is devoid of acetylated histone H3, the YEATS domain is free to interact with PAF1. After chromatin opening, large quantities of acetylated H3 becomes available, occupying the YEATS surface, thus supporting transfer of PAF1 to other binding partners such as RNA polymerase II. Because no CBX8 is present on active chromatin, this will free the ENL C-terminus for interaction with either DOT1L or AFF1. Wilms-specific mutations weaken interaction with histone H3, shift the balance, and favor derepression. MLL fusions permanently tether PAF1 to DOT1/AFF1 through the constitutive PAF1-binding site in the MLL N-terminus. Cont, control; vect, vector.
Notwithstanding a more elaborate posttranscriptional mechanism that controls YEATS-mediated protein-protein interactions and that may be affected by these mutations, the most straightforward explanation for a relative gain of function would be competitive binding of PAF1 and H3. The closely adjacent binding sites within the YEATS domain make it difficult to envisage how the 80kDa PAF1 protein and a 17kDa histone should be accommodated simultaneously. If competition occurs, most of ENL should be bound to H3 because cellular amounts of histone H3 vastly exceed those of PAF1. Indeed, precipitation of endogenous ENL in cellular extracts readily brought down detectable levels of histone H3 but not of PAF1 (Figure 5C). To probe the mutual relationship among ENL, histone H3, and PAF1, we performed pulldowns (Figure 5D). The YEATS domain of ENL was purified and attached to agarose beads. Because PAF1 also contains a histone H3-binding domain in its C-terminus, a glutathione S-transferase (GST) fusion with the previously identified minimal ENL-binding fragment (PAF266 encompassing amino acids 266 to 400 of PAF1) was used. Whereas GST-PAF266 readily bound to ENL, addition of recombinant histone H3 significantly weakened this interaction, indicating mutual competition. Because ENLdel/ins bind H3 more weakly than does wt-ENL (see Figure 5B), this would result in a relative gain of PAF1 affinity for the mutants. To keep in line with previous results2,9-11 demonstrating that histone peptides are recognized by wt but not by mutant YEATS domains if they are acetylated, we repeated the experiment with H3K27ac peptides. Similarly to full-length H3 (which does not need to be acetylated for binding to ENL), H3K27ac peptides reduced binding of GST-PAF266 to ENL but not to ENLins, explaining the relatively higher PAF1 binding affinity also in an acetylated histone context.
The amino acids of ENL affected by the Wilms mutations form a surface at the bottom of a channel accommodating the acetylated side chain of histone H3 lysines. It was shown that the single amino acid Y78 closes this channel and hence participates in H3Kac recognition11 (supplemental Figure 5A). However, no mutation at this position has been found in tumors. To test the importance of this particular residue for the derepressive activity of ENL, we tested an Y78A mutant in elongation reporter and replating assays (supplemental Figure 5B-C). In comparison with wt ENL, ENLY78A did not show any increased readout in these experiments, underscoring the close correlation between derepression and transformation/replating. ENLY78A did not affect the direct interaction with PAF1 in immunopreciptiations, but in line with Wang et al’s results,11 it slightly reduced affinity toward full-length H3 (supplemental Figure 5D). However, this was obviously not sufficient to allow a stronger PAF1 binding within a cellular environment.
Discussion
It is well known that transcription requires accessory factors to modify the packing density of chromatin. Therefore it is not surprising that the elongation factor ENL has evolved a capacity to counteract chromatin compaction by harnessing the PAFc machinery. Two major negative regulators of DNA density are polycomb repressive complexes 1 and 2. The perfect anticorrelation of H2A ubiquitination and transcription rates makes PRC1 a prime candidate that regulates “transcribability” of chromatin. The spatial distribution of H2AUb covering large domains that transgress transcriptional boundaries strongly suggests that PRC1 activity is not directly related to the known stalling or pausing of RNA polymerase shortly after initiation. Rather this initial pause is induced by dedicated antielongation factors such as negative elongation factor and 5,6-dichloro-1-β-D-ribofuranosylbenzimidazole sensitivity-inducing factor and can be overcome by the activity of P-TEFb.32 Beyond that and in particular for longer transcribed regions, additional effectors such as PAFc appear to be increasingly necessary for decompaction of chromatin during transcription. Although the supportive role for PAFc for transcriptional elongation and therefore the correlation of H2B ubiquitination and transcription rates has been noted before,33 the underlying mechanism is not completely clear. So far, no reader for H2BUb has been identified, but it was suggested that this modification affects nucleosome geometry opposing tight packing.20 Previous publications demonstrated that inhibiting H2B ubiquitination by a knock-down of the PAFc component LEO1 had profound consequences on overall transcription as well as elongation-associated H3K79 methylation, and both most strongly affected the HOX-loci.21 A role for ENL in establishing antirepression seems to be supported also by the results of Erb et al.9 Rapid, targeted destruction of ENL in vivo reduced overall traveling ratios of RNAPolII, exactly what would be expected if ENL functions in decreasing chromatin package density besides working to release RNAPolII from initial pausing through recruitment of P-TEFb.
Our results suggest that the derepressive activity of ENL would be highly self-limiting. As soon as open chromatin is accessible for histone acetyl transferases, a large number of acetylated histone H3 tails become available for binding to the YEATS domain. This would tether ENL directly to chromatin, allow the release and transfer of PAF1 onto active RNAPolII, and permit ENL to assume its “elongation mode,” now recruiting either DOT1L or P-TEFb through AFF1/AF4 (Figure 5E). Given the large number of acetylated H3 tails that cover any active transcription unit, this also would explain why all ENL is effectively trapped with either AFF1 or DOT1L in elongation complexes, whereas PAF1 has never been detected copurifying with ENL unless overexpressed. Rather, the majority of PAF1 is found to be associated with RNAPolII.
Our biochemical experiments indicate that Wilms tumor–specific mutations reduce binding affinity of ENL for acetylated histone H3 without harming PAF1 interaction. Thus it would need a concentration of acetylated histones to compete off PAF1 from these mutants higher than those from wt ENL, thus effectively strengthening their derepressive activity. This could provide a plausible explanation for why Wilms-specific ENL alterations are gain-of-function mutations. HOX-loci are the sentinel sites for polycomb-mediated repression and should respond first to perturbations of PRC function. Indeed, introduction of mutated ENL into primary hematopoietic precursors elicited increased H2B ubiquitination of Hoxa9 and Meis1, resulting in higher transcription rates and a concomitant differentiation block. The same loci showed the most pronounced differential binding of Wilms mutant and wt ENL, not only explaining the overexpression of HOX genes in this Wilms tumor subtype but also giving a rationale for the presence of analogous mutations in human AML.
The involvement of PAF1 as “antirepressor” also has repercussions with regard to the function of MLL-ENL fusions. The continuous recruitment of PAF1 by MLL derails the self-organizing derepression/elongation sequence orchestrated by ENL. Tethering PAF1 constitutively to P-TEFb, DOT1L, or both contributes to the hyperactivity of MLL-ENL, acting effectively as a “supertranscription factor” that combines derepressive and elongation functions in 1 molecule. As a consequence, MLL-ENL targets have higher transcription rates and H2BUb modification levels than does any other gene in the genome, and they are also simultaneously hypermodified with H3K79me. The fact that MLL-ENL seems to misuse the H2B ubiquitination machinery and that previous results demonstrate that a knock-down of LEO1 is incompatible with transformation by MLL-ENL34 support H2BUb as a potential therapeutic intervention point. Unfortunately, no inhibitors of H2B ubiquitin ligases are known. However, if those become available and a therapeutic window can be identified that rectifies the overshooting activity without harming the normal levels that are required for most of transcription, H2BUb may become a rewarding target for pharmaceutical development.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Renate Zimmermann and Daniel Leibinger for technical assistance.
This work was supported by research funding from Deutsche Forschungsgemeinschaft (grant SL27/7-2) and cofinanced by the Bavarian Ministry of Sciences, Research, and the Arts in the framework of the Bavarian Molecular Biosystems Research Network (R.K.S.). Additional funding was contributed by the Emerging Fields Initiative of the FAU University Erlangen-Nürnberg (R.K.S.).
Authorship
Contribution: K.H., M.-P.G.-C., and R.K.S. performed and analyzed experiments; C.B. performed high throughput sequencing and data analysis; R.K.S. conceived and supervised experiments; R.K.S. wrote the manuscript; and all authors read and discussed the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Robert K. Slany, Institute for Genetics, University Erlangen, Erwin Rommel Str 3, 91058 Erlangen, Germany; e-mail: robert.slany@fau.de.
References
Author notes
K.H. and M.-P.G.-C. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal