Key Points
A novel human CTL-based platform for comprehensive functional analysis of UNC13D variants is introduced.
Pathogenicity of a reported UNC13D variant was determined by measuring expression of the translated munc13-4 protein.
Abstract
Familial hemophagocytic lymphohistiocytosis (FHL) is the major form of hereditary hemophagocytic lymphohistiocytosis (HLH); as such, it requires prompt and accurate diagnosis. We previously reported that FHL type 3 (FHL3) can be rapidly screened by detecting munc13-4 expression in platelets using flow cytometry; however, the reliability of the munc13-4 expression assay for FHL3 diagnosis is unclear. Regardless of the type of UNC13D mutation, all reported FHL3 cases examined for the munc13-4 protein showed significantly reduced expression. However, the translated munc13-4 protein of some reportedly disease-causing UNC13D missense variants has not been assessed in terms of expression or function; therefore, their clinical significance remains unclear. The aim of this study was to determine the reliability of a munc13-4 expression assay for screening FHL3. Between 2011 and 2016, 108 HLH patients were screened by this method in our laboratory, and all 15 FHL3 patients were diagnosed accurately. To further elucidate whether munc13-4 expression analysis can reliably identify FHL3 patients harboring missense mutations in UNC13D, we developed an alloantigen-specific cytotoxic T lymphocyte (CTL) line and a CTL line immortalized by Herpesvirus saimiri derived from FHL3 patients. We then performed a comprehensive functional analysis of UNC13D variants. Transient expression of UNC13D complementary DNA constructs in these cell lines enabled us to determine the pathogenicity of the reported UNC13D missense variants according to expression levels of their translated munc13-4 proteins. Taken together with previous findings, the results presented herein show that the munc13-4 protein expression assay is a reliable tool for FHL3 screening.
Introduction
Familial hemophagocytic lymphohistiocytosis (FHL), the major form of hereditary hemophagocytic lymphohistiocytosis (HLH), is a fatal syndrome of immune dysregulation and hyperinflammation.1,2 FHL requires accurate and prompt diagnosis to initiate aggressive immunosuppressive therapy, followed by hematopoietic stem cell transplantation. To date, 4 genes have been identified as causative of FHL: PRF1 encoding perforin (FHL2),3 UNC13D encoding munc13-4 (FHL3),4 STX11 encoding syntaxin-11 (FHL4),5 and STXBP2 encoding munc18-2 (also known as syntaxin-binding protein 2) (FHL5).6,7 Perforin is an effector molecule contained within the cytolytic granules of cytotoxic T lymphocytes (CTLs) and natural killer (NK) cells. Munc13-4, syntaxin-11, and munc18-2 are involved in intracellular trafficking or fusion of these granules to the plasma membrane, followed by delivery of their contents into target cells. Thus, the NK cells and CTLs of FHL patients are intrinsically defective in cytolytic activity.8
FHL3 accounts for 20% to 40% of FHL cases, depending on ethnicity.9,10 It is functionally screened by evaluating the cytolytic activity of NK cells and CTLs, or by examining surface expression of CD107a, which reflects release of cytolytic granules.11,12 Degranulation of CD57+ CTLs can effectively identify FHL3 patients13 ; however, patients with FHL4, FHL5, and other forms of degranulation defect-associated HLH syndromes may also show the same defect.14 We previously reported that FHL3 can be rapidly screened by flow cytometric analysis of munc13-4 expression in platelets.15 This method is useful because it requires a small volume of whole blood and can be performed even for patients receiving platelet transfusion. However, no study has evaluated the reliability of the munc13-4 expression assay for FHL3 diagnosis.
In those with FHL2, most pathogenic PRF1 variants lead to perforin protein instability; therefore, flow cytometric detection of perforin expression in NK cells is a highly reliable screening method.16-18 However, some perforin proteins harboring missense mutations retain normal expression levels but cause postsynaptic functional defects that lead to reduced or absent cytolytic function.19
Various pathogenic UNC13D variants, including deep-intronic, inversion, duplication, insertion-deletion, missense, and nonsense mutations, as well as mutations affecting messenger RNA splicing, have been identified throughout the gene locus, but there is no identifiable hot spot.20-25 Regardless of the type of UNC13D variant, all reported FHL3 cases evaluated for munc13-4 expression levels showed a significant reduction in protein expression. However, neither expression of munc13-4 protein nor the function of cytolytic lymphocytes has been examined in some patients carrying UNC13D missense variants that are reported to be disease-causing; therefore, the clinical significance of these variants remains unclear.26-31
Determining the pathological significance of a particular UNC13D variant requires comprehensive characterization, including analysis of munc13-4 protein expression and functional evaluation of NK cells and/or CTLs. A platform for functional characterization of UNC13D variants (based on measuring degranulation of rat basophilic leukemia cells) has been proposed; however, the effects on cytolytic activity cannot be evaluated.32 We previously showed that the cytolytic activity of CTLs can be evaluated using alloantigen-specific CTL lines derived from HLH patients.10 Herpesvirus saimiri (HVS), a common lymphotropic virus of squirrel monkeys, can immortalize human T lymphocytes that retain normal cytolytic activity when their T-cell receptor/CD3 activation pathway is triggered.33,34 These cell lines may provide a platform for comprehensive functional characterization of UNC13D variants.
Here, we examined whether the munc13-4 expression assay is a reliable screening tool for FHL3. The assay effectively identified 15 FHL3 patients of 108 HLH patients screened in our laboratory. To further determine whether munc13-4 protein expression analysis can identify FHL3 patients carrying missense UNC13D variants reported to be disease-causing, we established an alloantigen-specific CTL line and an HVS-transformed CTL line derived from FHL3 patients. Transient expression of UNC13D complementary DNA (cDNA) constructs in these cell lines enabled us to determine the pathogenicity of the reported missense UNC13D variants according to expression levels of their translated munc13-4 proteins. Taken together with the data from previous studies, our findings support the reliability of the munc13-4 protein expression assay for FHL3 screening.
Materials and methods
Patients
Between 2011 and 2016, 143 patients suspected of having FHL by their referring physicians had blood samples screened by flow cytometry to evaluate intraplatelet munc13-4 expression. Of these, 108 patients who underwent UNC13D genetic analysis were enrolled in the study. As a control, blood obtained from healthy adults at the time of patient sampling was shipped for analysis along with the patient samples. Informed consent was obtained from the patients or their parents in accordance with the requirements of the institutional review board of Kyoto University Hospital and the Declaration of Helsinki.
UNC13D genetic analyses
Genomic DNA was isolated from patients’ peripheral blood mononuclear cells (PBMCs) using standard procedures. Primers were designed to amplify the coding exons and adjacent intronic sequences of the UNC13D gene. Primers to detect the deep-intronic mutation in the UNC13D intron 1 were also designed. The amplified products were sequenced with an Applied Biosystems ABI3130 Genetic Analyzer (Life Technologies, Carlsbad, CA) or with a GS Junior System (Roche, Basel, Switzerland).
Selection of reported missense mutations for functional analysis
Studies of FHL3 undertaken from 2003 to 2015 were reviewed, and 31 reported disease-causing missense UNC13D variants were identified (supplemental Table 1, available on the Blood Web site). Five of these variants were analyzed for munc13-4 expression, which was reduced markedly in all cases. From the remaining 26 variants, 11 were identified based on the following criteria: (A) a confirmed reduction in NK-cell or CTL degranulation, and/or decreased NK-cell cytotoxicity; (B) confirmed biallelic UNC13D mutations; and (C) no other deleterious mutation present on the same allele as the missense mutation. These 11 variants, together with 3 missense variants (found in patients P2 and P6) plus a known benign variant, were subjected to functional analyses (Table 1).
List of missense UNC13D variants analyzed in the current study
| . | Variant . | Predicted effects . | Reference . |
|---|---|---|---|
| V1 | c.175G>A | p.Ala59Thr | 22,27 |
| V2 | c.767G>A | p.Arg256Gln | 13 (found in P6) |
| V3 | c.1193C>T | p.Ser398Leu | 22 |
| V4 | c.1208T>C | p.Leu403Pro | 27 |
| V5 | c.1240C>T | p.Arg414Cys | 13 (found in P6) |
| V6 | c.1820G>C | p.Arg607Pro | 28 |
| V7 | c.1940T>C | p.Leu647Pro | 22 |
| V8 | c.2039G>C | p.Arg680Pro | 22 |
| V9 | c.2180G>A | p.Arg727Gln | 22 |
| V10 | c.2570T>G | p.Phe857Cys | 26,27 |
| V11 | c.2599A>G | p.Phe857Cys | Benign variant |
| V12 | c.2642T>C | p.Leu881Pro | 23 |
| V13 | c.2759A>G | p.Tyr920Cys | 13 (found in P2) |
| V14 | c.3049G>A | p.Glu1017Lys | 22 |
| V15 | c.3173T>C | p.Leu1058Pro | 22 |
| . | Variant . | Predicted effects . | Reference . |
|---|---|---|---|
| V1 | c.175G>A | p.Ala59Thr | 22,27 |
| V2 | c.767G>A | p.Arg256Gln | 13 (found in P6) |
| V3 | c.1193C>T | p.Ser398Leu | 22 |
| V4 | c.1208T>C | p.Leu403Pro | 27 |
| V5 | c.1240C>T | p.Arg414Cys | 13 (found in P6) |
| V6 | c.1820G>C | p.Arg607Pro | 28 |
| V7 | c.1940T>C | p.Leu647Pro | 22 |
| V8 | c.2039G>C | p.Arg680Pro | 22 |
| V9 | c.2180G>A | p.Arg727Gln | 22 |
| V10 | c.2570T>G | p.Phe857Cys | 26,27 |
| V11 | c.2599A>G | p.Phe857Cys | Benign variant |
| V12 | c.2642T>C | p.Leu881Pro | 23 |
| V13 | c.2759A>G | p.Tyr920Cys | 13 (found in P2) |
| V14 | c.3049G>A | p.Glu1017Lys | 22 |
| V15 | c.3173T>C | p.Leu1058Pro | 22 |
The 11 variants identified in the literature, together with 3 missense variants found in our patients plus a benign variant, were subjected to comprehensive functional analyses.
UNC13D cDNA constructs
FLAG-tagged cDNA carrying the wild-type UNC13D sequence was purchased from transOMIC technologies Inc (Huntsville, AL), and missense variant UNC13D sequences were constructed by polymerase chain reaction–based mutagenesis prior to further analysis. These constructs were transiently expressed in HEK293T cells (under the control of human cytomegalovirus promoter) for western blot analysis. For assays using CTL lines, FLAG-tagged UNC13D cDNA sequences were linked to T2A peptide–enhanced green fluorescent protein (EGFP) sequences using a Gibson assembly kit (New England BioLabs, Ipswich, MA). All synthetic DNA constructs were verified by Sanger sequencing.
Protein expression assays
Munc13-4 expression in patient-derived platelets was evaluated as previously described.15 To determine the effect of UNC13D missense variants on expression of munc13-4, HEK293T cells were transiently transfected with FLAG-tagged cDNA carrying wild-type or variant UNC13D sequences. Transfected cells were cultured overnight and harvested with or without additional incubation in the presence of 0.35 mM cycloheximide for 5 or 10 hours and then subjected to immunoblotting. Cell extracts were fractionated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis, and the proteins were subjected to western blotting with specific antibodies (Abs). Specific bands were visualized using the standard enhanced chemiluminescence method. For the cycloheximide chase assay, western blot signals were quantified using the ChemiDoc XRS+ System and Image Laboratory software (Bio-Rad Laboratories, Hercules, CA). Alternatively, FLAG-tagged cDNA carrying wild-type or variant UNC13D sequences linked to a T2A peptide and EGFP were transiently transfected into alloantigen-specific CTLs or HVS-transformed CD8+ T cells by electroporation with NEPA21 (Nepa Gene, Chiba, Japan). After electroporation, cells were cultured for 48 hours, harvested, and surface-stained with anti-CD3 and anti-CD8 monoclonal Abs (mAbs). Cells were then fixed, permeabilized, stained intracellularly with an anti-FLAG-polyclonal Ab, and analyzed by flow cytometry.
Establishment of FHL3 model cell lines
Alloantigen-specific CD8+ CTL lines were generated as described previously.10 Briefly, PBMCs obtained from FHL patients and healthy individuals were cocultured with a mitomycin C–treated B-lymphoblastoid cell line (KI-LCL) established from an HLA-mismatched individual. Six days later, CD8+ T lymphocytes were isolated using immunomagnetic beads (MACS beads; Miltenyi Biotec, Bergisch Gladbach, Germany). These cells were cultured in RPMI 1640 medium supplemented with 10% human serum and 10 IU/mL human recombinant interleukin-2 (rIL-2) and stimulated weekly with mitomycin C–treated KI-LCL cells for >3 weeks; the lymphocytes were then used as CD8+ alloantigen-specific CTL lines.
HVS-transformed T cell lines were established as previously described.33 Briefly, PBMCs from patient 1 and from a healthy donor were resuspended (2 × 106 cells per mL) in RPMI 1640 medium supplemented with 10% fetal calf serum, 1% l-glutamine, and 50 IU/mL human rIL-2 and then stimulated for 3 days with 1 µg/mL phytohemagglutinin. The cells were then resuspended (2 × 106 cells/mL) in 96-well plates in RPMI 1640 medium containing 50 IU/mL human rIL-2; the wells were then topped up with HVS supernatant (30% total volume). Thereafter, the culture medium was replaced every 3 to 4 days. Immortalized CD8+ lymphocytes were used as HVS-transformed CTL lines.
Lysosomal degranulation assays
To evaluate lysosomal exocytosis of CTL lines, cells (2 × 105 in 96-well round-bottom plates) were mixed with an equal number of P815 cells in the presence of 0.5 µg/mL anti-CD3 mAb (OKT3) for 2 hours. To determine the effect of an UNC13D variant on degranulation, alloantigen-specific CTLs (2 × 106 cells per well) or HVS-transformed CTLs (5 × 106 cells per well) were transfected with plasmids (10 μg per well) by electroporation and cultured with human rIL-2 (100 IU/mL) in 24-well plates. Forty-eight hours later, cells were harvested and stimulated for 2 hours with P815 cells and 0.5 µg/mL OKT3. Cells were then resuspended in phosphate-buffered saline supplemented with 0.5% bovine serum albumin and 2 mM EDTA, stained with anti-CD3, anti-CD8, and anti-CD107a mAbs, and analyzed by flow cytometry.
Cytolytic assay
Cytolytic activity was determined in a lactate dehydrogenase release assay (Non-Radioactive Cytotoxicity Assay, CytoTox 96 Assay; Promega, Madison, WI), according to the manufacturer’s instructions. Alloantigen-specific CTLs or HVS-transformed CTLs were cocultured for 6 hours with KI-LCL cells alone or with P815 cells and 0.5 µg/mL OKT3, respectively. For each experiment, 1 × 104 KI-LCL cells or 5 × 103 P815 cells were plated in 96-well microtiter plates as targets. Background spontaneous and maximal lactate dehydrogenase release were determined by culturing target cells in medium alone or in medium containing Triton X-100. To determine the functional effects of an UNC13D variant, HVS-transformed CTLs were transfected with FLAG-tagged cDNA carrying wild-type or missense mutated UNC13D linked to T2A-EGFP. Then, 24 hours later, EGFP+ cells were sorted using a FACSAria III (BD Biosciences, San Jose, CA) cytometer, incubated for another 24 hours with human rIL-2 (100 IU/mL), and used for the assay.
Antibodies
Rabbit polyclonal Abs raised against human munc13-4 protein have been described previously.15,35 Rabbit polyclonal anti-integrin αIIb (Santa Cruz Biotechnology, Dallas, TX), mouse polyclonal anti-β-actin (Sigma-Aldrich, St. Louis, MO), and mouse monoclonal anti-FLAG M2 (Sigma-Aldrich) Abs served as primary Abs for western blotting. The Abs used for the flow cytometric analyses were mouse monoclonal phycoerythrin (PE)-Cy7–conjugated anti-CD3 (Beckman Coulter, Brea, CA), Brilliant Violet 421–conjugated anti-CD8 (BioLegend, San Diego, CA), PE-conjugated anti-CD41a (BD Biosciences), PE-conjugated anti-CD107a (eBioscience, San Diego, CA), allophycocyanin-conjugated anti-DYKDDDK (FLAG) (Miltenyi Biotec), and fluorescein isothiocyanate–conjugated donkey anti-rabbit immunoglobulin G (Jackson ImmunoResearch Laboratories, West Grove, PA).
Statistical analysis
One-way analysis of variance, followed by the Tukey post hoc test, was used to compare multiple groups. P < .05 was considered significant. All statistical analyses were performed using GraphPad Prism (version 7.02) software (GraphPad Software, Inc., La Jolla, CA).
Results
The Munc13-4 expression assay effectively identifies FHL3 patients
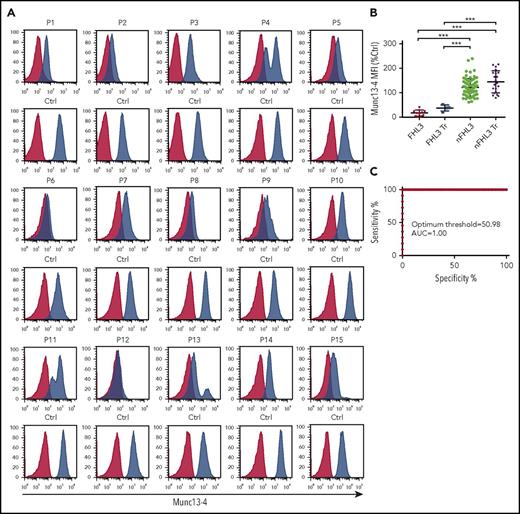
Between 2011 and 2016, we screened 143 HLH patients for FHL3 by measuring expression of intraplatelet munc13-4 by flow cytometry. Of these, 108 underwent UNC13D genetic analysis and 15 patients were found to carry biallelic pathogenic UNC13D variants (Table 2). Platelets from all 15 patients showed a marked reduction in munc13-4 expression, regardless of the UNC13D mutation type (Figure 1A). Bimodal populations comprising normal and munc13-4−deficient platelets were evident in patients receiving platelet transfusions (P4, P11, P13, and P15; Figure 1A). No other patients showed reduced expression or bimodal distribution of munc13-4 protein. When the munc13-4 protein in patient samples was measured in terms of a relative expression ratio (ie, to that in control samples), the ratios determined by the flow cytometric intraplatelet munc13-4 expression assay effectively discriminated FHL3 patients from those with other forms of HLH (optimum threshold, 50.98%; area under the curve, 1.00) (Figure 1B-C).
UNC13D mutations found in the 15 patients diagnosed with FHL3
| Patient . | First allele . | Second allele . | ||
|---|---|---|---|---|
| Mutation . | Predicted effect . | Mutation . | Predicted effect . | |
| P1 | c.1721_1722insA | FS | c.1763G>A | NS |
| P2 | c.754-1G>C | SE | c.2759A>G | MS |
| P3 | c.118-308C>T | TD | Duplication of exon 7-12 | ED |
| P4 | c.817C>T | NS | c.534-538delGACA | FS |
| P5 | c.754-1G>C | SE | c.118-308C>T | TD |
| P6 | c.1992+1G>A | SE | c.767G>A | MS |
| c.1240C>T | MS | |||
| P7 | c.118-308C>T | TD | c.118-308C>T | TD |
| P8 | c.2381delT | FS | c.322-1G>A | SE |
| P9 | c.754-1G>C | SE | c.1596+1G>C | SE |
| P10 | c.118-308C>T | TD | c.1596+1G>C | SE |
| P11 | c.118-308C>T | TD | c.1596+1G>C | SE |
| P12 | c.247C>T | NS | c.1596+1G>C | SE |
| P13 | c.28C>T | NS | c.723delG | FS |
| c.754-1G>C | SE | |||
| P14 | c.118-308C>T | TD | c.1596+1G>C | SE |
| P15 | c.1596+1G>C | SE | c.3081delG | FS |
| Patient . | First allele . | Second allele . | ||
|---|---|---|---|---|
| Mutation . | Predicted effect . | Mutation . | Predicted effect . | |
| P1 | c.1721_1722insA | FS | c.1763G>A | NS |
| P2 | c.754-1G>C | SE | c.2759A>G | MS |
| P3 | c.118-308C>T | TD | Duplication of exon 7-12 | ED |
| P4 | c.817C>T | NS | c.534-538delGACA | FS |
| P5 | c.754-1G>C | SE | c.118-308C>T | TD |
| P6 | c.1992+1G>A | SE | c.767G>A | MS |
| c.1240C>T | MS | |||
| P7 | c.118-308C>T | TD | c.118-308C>T | TD |
| P8 | c.2381delT | FS | c.322-1G>A | SE |
| P9 | c.754-1G>C | SE | c.1596+1G>C | SE |
| P10 | c.118-308C>T | TD | c.1596+1G>C | SE |
| P11 | c.118-308C>T | TD | c.1596+1G>C | SE |
| P12 | c.247C>T | NS | c.1596+1G>C | SE |
| P13 | c.28C>T | NS | c.723delG | FS |
| c.754-1G>C | SE | |||
| P14 | c.118-308C>T | TD | c.1596+1G>C | SE |
| P15 | c.1596+1G>C | SE | c.3081delG | FS |
ED, exon duplication; FS, frameshift; MS, missense; NS, nonsense; SE, splice error; TD, transcriptional dysregulation.
Flow cytometric detection of intraplatelet munc13-4 protein expression effectively identifies FHL3 patients. (A) Flow cytometric analysis of intraplatelet munc13-4 expression in FHL3 patients and healthy controls (Ctrl). Blue histograms represent staining with the anti-munc13-4 antibody; red histograms represent staining with isotype control Abs. P4, P11, P13, and P15 received platelet transfusions (Tr) several days before blood samples were taken. (B) Munc13-4 expression levels in platelets from enrolled patients relative to that of controls. Each dot shows the relative mean fluorescence intensity (MFI) of munc13-4 protein expression in a patient sample compared with that in a control sample: (MFI of munc13-4 staining − MFI of isotype staining) in a patient/(MFI of munc13-4 staining − MFI of isotype control staining) in a control. Five patients with FHL2, 1 patient with X-linked lymphoproliferative syndrome 1 (XLP1), and 1 patient with XLP2 were included in the non-FHL3 (nFHL3) group. Data from 4 FHL3 and 21 nFHL3 patients who received a transfusion within several days of sampling are shown separately. (C) Receiver operating characteristics curve analysis of relative munc13-4 MFI to discriminate FHL3 patients from those with other types of HLH. Patients who received a platelet transfusion were excluded from the analysis. Error bars indicate the mean ± standard deviation (SD). *P < .05; **P < .01; and ***P < .001. AUC, area under the curve.
Flow cytometric detection of intraplatelet munc13-4 protein expression effectively identifies FHL3 patients. (A) Flow cytometric analysis of intraplatelet munc13-4 expression in FHL3 patients and healthy controls (Ctrl). Blue histograms represent staining with the anti-munc13-4 antibody; red histograms represent staining with isotype control Abs. P4, P11, P13, and P15 received platelet transfusions (Tr) several days before blood samples were taken. (B) Munc13-4 expression levels in platelets from enrolled patients relative to that of controls. Each dot shows the relative mean fluorescence intensity (MFI) of munc13-4 protein expression in a patient sample compared with that in a control sample: (MFI of munc13-4 staining − MFI of isotype staining) in a patient/(MFI of munc13-4 staining − MFI of isotype control staining) in a control. Five patients with FHL2, 1 patient with X-linked lymphoproliferative syndrome 1 (XLP1), and 1 patient with XLP2 were included in the non-FHL3 (nFHL3) group. Data from 4 FHL3 and 21 nFHL3 patients who received a transfusion within several days of sampling are shown separately. (C) Receiver operating characteristics curve analysis of relative munc13-4 MFI to discriminate FHL3 patients from those with other types of HLH. Patients who received a platelet transfusion were excluded from the analysis. Error bars indicate the mean ± standard deviation (SD). *P < .05; **P < .01; and ***P < .001. AUC, area under the curve.
Reported pathogenic missense UNC13D variants cause reduced munc13-4 expression
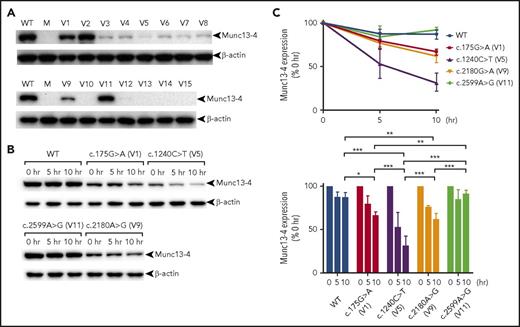
We next determined whether the reported UNC13D missense variants cause reduced munc13-4 expression and can therefore be detected rapidly using our screening method. Thus, we analyzed 11 variants identified in the literature (see “Materials and methods” for the criteria), together with 3 missense variants found in our patients plus a benign variant (Table 1). We found that most UNC13D missense variants caused significant reduction in munc13-4 expression when transfected into HEK293T cells (Figure 2A). Because the c.175G>A and c.2180G>A variants caused reduced, but residual, munc13-4 protein expression, we next evaluated the stability of the respective translated proteins. Cycloheximide chase analysis revealed that wild-type protein and a benign p.Phe857Cys (c.2599A>G) variant were highly stable, whereas the p.Arg414Cys (c.1240C>T) variant, which proved to be pathogenic in P6 (who was diagnosed at our laboratory),13 was degraded rapidly. Munc13-4 proteins harboring p.Ala59Thr (c.175G>A) or p.Arg727Gln (c.2180G>A) substitutions showed intermediate stability (Figure 2B-C).
Pathogenic UNC13D missense variants cause reduced munc13-4 expression. HEK293T cells were transfected with FLAG-tagged cDNA carrying wild-type or missense mutated UNC13D sequences. Cells were harvested after overnight culture (A) or after additional incubation in the presence of cycloheximide for 0, 5, and 10 hours (B), and munc13-4 protein expression levels were analyzed by western blotting. (C) Relative expression of variant munc13-4 proteins at the indicated time points after treatment with cycloheximide. V1 to V15 are the UNC13D variants listed in Table 1. Results in panels A and B are representative of >3 independent experiments. Error bars indicate the mean ± SD. *P < .05; **P < .01; ***P < .001. M, mock; WT, wild type.
Pathogenic UNC13D missense variants cause reduced munc13-4 expression. HEK293T cells were transfected with FLAG-tagged cDNA carrying wild-type or missense mutated UNC13D sequences. Cells were harvested after overnight culture (A) or after additional incubation in the presence of cycloheximide for 0, 5, and 10 hours (B), and munc13-4 protein expression levels were analyzed by western blotting. (C) Relative expression of variant munc13-4 proteins at the indicated time points after treatment with cycloheximide. V1 to V15 are the UNC13D variants listed in Table 1. Results in panels A and B are representative of >3 independent experiments. Error bars indicate the mean ± SD. *P < .05; **P < .01; ***P < .001. M, mock; WT, wild type.
Establishment of FHL3 model cell lines for functional analysis of UNC13D variants
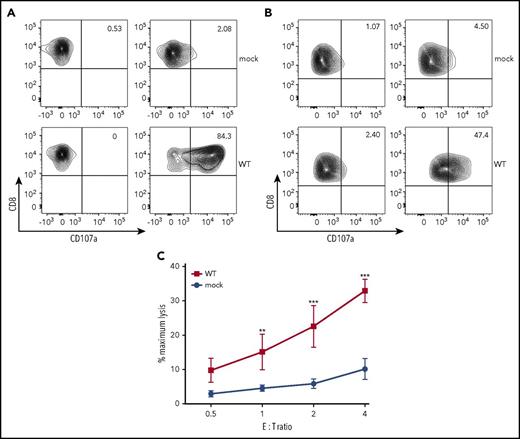
To analyze the pathogenicity of UNC13D variants, we established human FHL3 model CTL lines that could be used for degranulation and cytolytic assays. We first established an alloantigen-specific CTL line from P13 (Allo13). Allo13 cells showed defective degranulation when compared with a CTL line established from a healthy control (supplemental Figure 1A); degranulation was restored upon transient expression of wild-type UNC13D cDNA (Figure 3A). Allo13 cells showed defective cytolytic activity when compared with cells from healthy individuals (supplemental Figure 1B), but could not be used in the assay due to low transfection efficacy and poor viability after electroporation. We therefore transformed CD8+ T cells from P1 with HVS and established an immortalized CTL line (HVS-T1). Compared with the control CTL line established from a healthy individual (HVS-TH), HVS-T1 cells were defective in both degranulation (supplemental Figure 2A) and cytolytic activity (supplemental Figure 2B). They showed higher transfection efficacy and better viability than Allo13 cells after electroporation and, again, expression of wild-type UNC13D cDNA restored degranulation and cytolytic ability to levels comparable to those of HVS-TH (Figure 3B-C).
FHL3 model CTL lines for functional analysis of UNC13D variants. Allo13 (A) or HVS-T1 (B) cells transfected with FLAG-tagged cDNA carrying wild-type UNC13D sequences linked to the T2A peptide and EGFP were stimulated with P815 cells and an anti-CD3 mAb (OKT3). CD3+CD8+EGFP+ cells were gated, and degranulation was assessed by flow cytometry. Results are representative of 3 independent experiments. (C) HVS-T1 cells were transfected with FLAG-tagged cDNA carrying wild-type UNC13D sequences linked to the T2A peptide and EGFP. EGFP+ cells were sorted and incubated with P815 (target cells). Data represent the mean ± SD of 3 independent experiments. *P < .05; **P < .01; ***P < .001. E:T ratio, effector-to-target ratio.
FHL3 model CTL lines for functional analysis of UNC13D variants. Allo13 (A) or HVS-T1 (B) cells transfected with FLAG-tagged cDNA carrying wild-type UNC13D sequences linked to the T2A peptide and EGFP were stimulated with P815 cells and an anti-CD3 mAb (OKT3). CD3+CD8+EGFP+ cells were gated, and degranulation was assessed by flow cytometry. Results are representative of 3 independent experiments. (C) HVS-T1 cells were transfected with FLAG-tagged cDNA carrying wild-type UNC13D sequences linked to the T2A peptide and EGFP. EGFP+ cells were sorted and incubated with P815 (target cells). Data represent the mean ± SD of 3 independent experiments. *P < .05; **P < .01; ***P < .001. E:T ratio, effector-to-target ratio.
Expression level of transcribed munc13-4 protein determines the pathogenicity of an UNC13D variant
To further test whether munc13-4 protein expression analysis can identify FHL3 patients reliably, we measured expression and function of munc13-4 proteins encoded by the missense UNC13D variants in the established CTL cell lines.
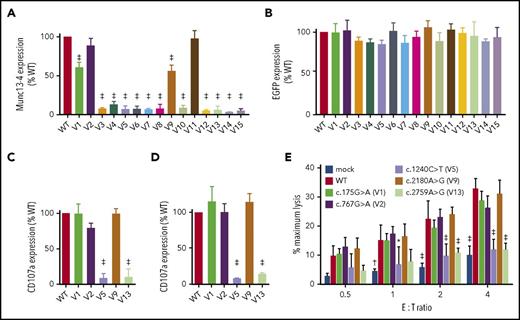
Transfection of UNC13D missense variants into HVS-T1 cells reproduced munc13-4 protein expression observed in HEK293T cells (Figure 4A), whereas EGFP expression by each variant was comparable (Figure 4B). Transfection of the c.767G>A variant found in P6, which retained near normal levels of munc13-4 expression, restored degranulation of Allo13 and HVS-T1 cells (Figure 4C-D; supplemental Figure 3). It also restored cytolytic activity in HVS-T1 cells to levels equivalent to that observed for wild-type UNC13D (Figure 4D). By contrast, transfection of the c.1240C>T variant found in P6 and the c.2759A>G variant found in P2, which resulted in almost complete loss of munc13-4 expression, did not (Figure 4C-E; supplemental Figure 3). These findings mirrored the functional CTL defects observed in patients. Interestingly, cells transfected with UNC13D variants that induced residual protein expression (c.175G>A and c.2180G>A) showed degranulation and cytolytic activity comparable with that of cells transfected with the wild-type UNC13D construct (Figure 4C-E; supplemental Figure 4). These findings indicate that munc13-4 proteins encoded by these 2 UNC13D variants can function to a certain extent, although they are less stable than the wild-type protein.
Expression level of the transcribed munc13-4 protein determines the pathogenicity of an UNC13D variant. HVS-T1 cells were transfected with FLAG-tagged cDNA carrying wild-type or missense UNC13D sequences linked to the T2A peptide and EGFP. CD3+CD8+EGFP+ cells were gated, and expression of munc13-4 (A) and EGFP (B) in cells transfected with missense UNC13D variants relative to that in cells transfected with the wild-type UNC13D sequence was analyzed by flow cytometry. Relative expression of munc13-4 protein (A) was calculated as follows: MFI of anti-FLAG–stained cells transfected with UNC13D variants − MFI of anti-FLAG–stained mock-transfected cells/MFI of anti-FLAG−stained cells transfected with wild-type UNC13D − MFI of anti-FLAG–stained mock-transfected cells. Relative expression of EGFP (B) was calculated as follows: MFI of EGFP in cells transfected with UNC13D variants/MFI of EGFP in cells transfected with wild-type UNC13D. Values represent the mean ± SD of 3 independent experiments. (C) Allo13 or (D) HVS-T1 cells were transfected with FLAG-tagged cDNA carrying wild-type or missense UNC13D sequences linked to the T2A peptide and EGFP, followed by incubation with P815 and an anti-CD3 mAb (OKT3). CD3+CD8+EGFP+ cells were gated, and degranulation was assessed by flow cytometry. Relative degranulation was calculated as follows: MFI of CD107a expression by cells transfected with UNC13D variants − MFI of CD107a expression by mock-transfected cells/MFI of CD107a expression by cells transfected with wild-type UNC13D − MFI of CD107a expression by mock-transfected cells. Data represent the mean ± SD of 3 independents experiments. (E) HVS-T1 cells were transfected with FLAG-tagged cDNA carrying wild-type or missense UNC13D sequences linked to the T2A peptide and EGFP. EGFP+ cells were sorted and incubated with P815 cells and an anti-CD3 mAb (OKT3) at the indicated effector-to-target (E:T) ratios. Specific lysis was then calculated. Data represent the mean ± SD of 3 independent experiments. *P < .05, †P < .01, ‡P < .001, compared with the wild-type UNC13D construct.
Expression level of the transcribed munc13-4 protein determines the pathogenicity of an UNC13D variant. HVS-T1 cells were transfected with FLAG-tagged cDNA carrying wild-type or missense UNC13D sequences linked to the T2A peptide and EGFP. CD3+CD8+EGFP+ cells were gated, and expression of munc13-4 (A) and EGFP (B) in cells transfected with missense UNC13D variants relative to that in cells transfected with the wild-type UNC13D sequence was analyzed by flow cytometry. Relative expression of munc13-4 protein (A) was calculated as follows: MFI of anti-FLAG–stained cells transfected with UNC13D variants − MFI of anti-FLAG–stained mock-transfected cells/MFI of anti-FLAG−stained cells transfected with wild-type UNC13D − MFI of anti-FLAG–stained mock-transfected cells. Relative expression of EGFP (B) was calculated as follows: MFI of EGFP in cells transfected with UNC13D variants/MFI of EGFP in cells transfected with wild-type UNC13D. Values represent the mean ± SD of 3 independent experiments. (C) Allo13 or (D) HVS-T1 cells were transfected with FLAG-tagged cDNA carrying wild-type or missense UNC13D sequences linked to the T2A peptide and EGFP, followed by incubation with P815 and an anti-CD3 mAb (OKT3). CD3+CD8+EGFP+ cells were gated, and degranulation was assessed by flow cytometry. Relative degranulation was calculated as follows: MFI of CD107a expression by cells transfected with UNC13D variants − MFI of CD107a expression by mock-transfected cells/MFI of CD107a expression by cells transfected with wild-type UNC13D − MFI of CD107a expression by mock-transfected cells. Data represent the mean ± SD of 3 independents experiments. (E) HVS-T1 cells were transfected with FLAG-tagged cDNA carrying wild-type or missense UNC13D sequences linked to the T2A peptide and EGFP. EGFP+ cells were sorted and incubated with P815 cells and an anti-CD3 mAb (OKT3) at the indicated effector-to-target (E:T) ratios. Specific lysis was then calculated. Data represent the mean ± SD of 3 independent experiments. *P < .05, †P < .01, ‡P < .001, compared with the wild-type UNC13D construct.
Discussion
Functional analysis of NK-cell and CTL degranulation enables early identification of patients needing molecular evaluation for FHL3.11-13 However, these evaluations are often hampered by limited availability of patient material; therefore, genetic analysis remains the main method of diagnosis. However, the conventional sequencing technique may fail to detect some UNC13D mutations.23 In addition, the clinical significance of a rare variant, particularly a missense variant, is often difficult to interpret. In the case of FHL2, a method based on a comprehensive functional analysis of perforin variants in the human NK cell line has been established.36 For FHL3, a platform based on the use of rat basophilic leukemia cells for functional characterization of UNC13D variants has been proposed; however, functional analysis is limited to degranulation, and the cytolytic activity cannot be evaluated.32
Here, we developed model human FHL3 CTL lines defective in both degranulation and cytolytic activity (supplemental Figures 1 and 2). Complementation with wild-type munc13-4 restored full function to these cells (Figure 3). These cell lines can be used for comprehensive functional analysis of a given UNC13D variant.
We previously reported a rapid FHL3 screening method based on detection of munc13-4 expression in platelets.15 Here, we evaluated the reliability of this munc13-4 protein expression assay. We found that expression of munc13-4 by platelets from all FHL3 patients diagnosed at our laboratory was significantly lower than that by platelets from non-FHL3 patients (Figure 1A-B and Murata et al15 ). The munc13-4 expression assay showed high specificity and sensitivity, regardless of the particular mutations harbored by the patients (Figure 1C). Patients harboring the deep-intronic c.118-308C>T mutation showed slightly higher levels of protein expression than those harboring other types of mutation (Figure 1A); however, the patient with homozygotic deep-intronic mutations (P7) showed markedly reduced munc13-4 expression, which was readily identifiable using our method. Furthermore, our method identified P3, who harbored intragenic duplication of UNC13D, which conventional genetic analysis failed to detect.25
To date, all reported pathogenic UNC13D mutations evaluated for protein expression cause a marked reduction in munc13-4 protein expression. However, UNC13D missense mutations are responsible for a considerable number of FHL3 cases worldwide, and munc13-4 expression levels have not been evaluated in most of these (supplemental Table 1). To further confirm the reliability of the munc13-4 protein expression assay as a FHL3 screening method, we transiently transfected HEK293T cells with variant UNC13D cDNA sequences to find out if the reported missense mutations caused reduced munc13-4 protein expression. Indeed, all tested UNC13D missense mutations reduced munc13-4 expression to some extent (Figure 2A). Two UNC13D variants, c.175G>A and c.2180G>A, caused only a slight reduction in protein expression (Figure 2A). Although proteins encoded by these variants showed marked instability when compared with that of wild-type munc13-4 (Figure 2B-C), the functional characteristics of these variants could not be evaluated. Therefore, we developed human CTL-based FHL3 model cell lines (Allo13 and HVS-T1) to determine the functional characteristics of UNC13D variants (Figure 3; supplemental Figures 1 and 2). Transfection of HEK293T cells and HVS-T1 cells with UNC13D variants induced the same munc13-4 protein expression profile (Figures 2A and 4A). Surprisingly, the c.175G>A and c.2180G>A variants in Allo13 and HVS-T1 cells restored degranulation and cytolytic activity to levels comparable with those in cells transfected with wild-type constructs (Figure 4C-E; supplemental Figure 4). These 2 variants were identified in the same patient (UPN483), who presented with HLH symptoms at 0.7 months of age22 ; however, the patient lacked splenomegaly, a finding that is almost always present in FHL3 patients. CD107a expression by the patient NK cells was reduced, although the specificity of the assay is low.12,13 In addition, the homozygous c.175G>A variant was reported in a 5-year-old healthy child with normal degranulation,20 and the allele frequency of c.175G>A was relatively high (0.01591 by ExAC Browser: http://exac.broadinstitute.org/variant/17-73839326-C-T). These observations suggest that the c.175G>A variant is unlikely to cause HLH. Because the c.2180G>A variant showed very similar functional characteristics to c.175G>A, we think that this variant is also very unlikely to trigger HLH. However, it is possible that these variants predispose their carrier individuals to HLH under extreme conditions because their transcribed munc13-4 proteins are less stable than the wild-type protein.
This study raises an important question: why do all reported pathogenic missense UNC13D variants cause a marked reduction in munc13-4 protein expression? The precise mechanism lies beyond the scope of the current study, but we believe that protein instability is the main cause. Transfection of UNC13D variants into HEK293T cells and HSV-T1 cells induced the same munc13-4 protein expression profile (Figures 2A and 4A). Because these 2 cell lines are ontogenically remote, the result likely reflects the intrinsic characteristics of the munc13-4 protein. We used the T2A peptide to coexpress UNC13D and EGFP sequences in HVS-T1 cells.37-39 Expression of munc13-4 and EGFP proteins in HVS-T1 cells correlated well (supplemental Figure 5), and EGFP expression levels were comparable between each construct (Figure 4B). Therefore, it is likely that each UNC13D variant is translated to a similar extent and that the pathogenicity of a reported missense UNC13D variant is mostly determined by the stability of its translated munc13-4 protein.
It is noteworthy that a variant UNC13D cannot be simply judged as “pathogenic” or “nonpathogenic”; this is because the stability of munc13-4 protein encoded by a particular UNC13D variant falls along a continuous spectrum (Figure 4A). Whereas some munc13-4 variants that are mildly unstable rarely trigger HLH under physiological conditions, others that are slightly more unstable may easily trigger HLH.
We acknowledge that the study has several limitations. First, we analyzed only mutations associated with a documented reduction in lysosomal degranulation and/or cytolytic function in patients so as not to include rare but benign UNC13D variants. It is possible that there are some pathogenic UNC13D variants that maintain normal munc13-4 expression levels. However, the novel analysis system used in the current study can be readily applied to other UNC13D variants to evaluate their functional significance. Comparing the expression and function of munc13-4 proteins encoded by various UNC13D variants in vitro and ex vivo would help to unravel the structure and functional regulation of the munc13-4 protein. Second, we transfected UNC13D cDNA plasmids into HEK293T cells or CTL cell lines, which would result in higher protein expression than that observed under physiological conditions. However, this means that the missense UNC13D variants evaluated in the current study would likely cause even lower munc13-4 expression when patient samples are used. Because all pathogenic missense UNC13D variants analyzed in the current study caused a marked reduction in munc13-4 expression, even under conditions in which their expression is artificially forced, it is likely that most pathogenic UNC13D mutations are amenable to rapid detection by the munc13-4 expression assay.
As reported by Bergsten et al, early recognition of FHL patients, along with risk-group stratification, is likely to improve the outcome of HLH therapy.40 Although screening for FHL has improved over recent years there is no standardized diagnostic approach. Impaired NK-cell and CTL cytotoxicity is central to the pathogenesis of FHL8 ; however, many cases of secondary HLH show reduced NK-cell activity,41-43 and CTL cytotoxicity assays are not easily performed. Flow cytometry analysis of perforin expression by NK cells is both a sensitive and reliable method for identifying FHL2 patients; in addition, the NK-cell degranulation assay is a comprehensive method for identifying patients with a degranulation defect.11,12,17,18 However, impaired NK-cell degranulation is often observed during the acute phase of secondary HLH.12,13 CTLs are an alternative, sensitive tool for use in lysosomal exocytosis assays, but they cannot differentiate FHL3-5. Here, we show that analyzing expression of munc13-4 enables reliable identification of FHL3 patients. Because FHL2 and FHL3 account for the majority of FHL cases,9,40 combined analysis of munc13-4 and perforin enables rapid diagnosis, thereby increasing the chance of early stem cell transplantation for most FHL patients and improving the prognosis.
In conclusion, we developed novel human CTL-based FHL3 model cell lines that can be used to evaluate the functional effects of a UNC13D variant. By using these cell lines, we demonstrated that the munc13-4 expression level determines the pathogenicity of a reported UNC13D missense variant. Together with data obtained from FHL3 patients diagnosed at our laboratory and from the literature, we conclude that most FHL3 patients are likely amenable to rapid screening/detection by the munc13-4 expression assay.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors are grateful to all of the participating patients, their families, and the referring physicians.
This work was supported by Japan Society for the Promotion of Science Grants-in-Aid for Scientific Research (JSPS KAKENHI; grant number 26461582), by the Cooperative Research Project Program of Joint Usage/Research Center at the Institute of Development, Aging and Cancer, Tohoku University, and by grants from the “Research on Measures for Intractable Diseases” project, which matched funding subsidy from the Japanese Ministry of Health, Labor, and Welfare.
Authorship
Contribution: T.Y., R.N., and T.H. designed the research; H.S., S.S., and E.H. performed patient screening; H.S. performed functional analysis of UNC13D variants; T.W. prepared the HVS-T1 and HVS-TH cells; O.O. performed genetic analyses; R.S. and H.H. prepared the anti-munc13-4 Abs; H.S., T.Y., K.I., T.K., T.W., R.N., H.H., E.I., and T.H. analyzed the data and discussed the results; and H.S. and T.Y. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Takahiro Yasumi, Department of Pediatrics, Graduate School of Medicine, Kyoto University, 54 Kawahara-cho, Shogoin, Sakyo-ku, Kyoto 606-8507, Japan; e-mail: yasumi@kuhp.kyoto-u.ac.jp.