Abstract
Up to 60% of cases of the autosomal recessive immunodeficiency hemophagocytic lymphohistiocytosis (HLH) are associated with mutations in the perforin (PRF1) gene. In this study, we expressed wild-type and mutated perforin in rat basophil leukemia cells to study the effect on lytic function of the substitutions A91V and N252S (commonly considered to be neutral polymorphisms) and 22 perforin missense substitutions first identified in HLH patients. Surprisingly, we found that A91V perforin was expressed at reduced levels compared with wild-type perforin, resulting in partial loss of lytic capacity. In contrast, expression and function of N252S-substituted perforin were normal. Most HLH-associated mutations resulted in protein degradation (probably due to misfolding) and complete loss of perforin activity, the exception being R232H, which retained approximately 30% wild-type activity. Several other mutated proteins (H222Q, C73R, F157V, and D313V) had no detectable lytic activity but were expressed at normal levels, suggesting that their functional defect might map downstream at the level of the target cell membrane. One further perforin substitution identified in an HLH patient (V183G) was normally expressed and displayed normal lysis. This report represents the first systematic functional analysis of HLH-associated missense mutations and the 2 most common perforin polymorphisms. (Blood. 2005;105:4700-4706)
Introduction
Perforin, a membrane-disruptive protein secreted by cytotoxic T lymphocytes (CTLs) and natural killer (NK) cells, is essential for killing virus-infected or transformed cells targeted for destruction through the granule exocytosis pathway.1-6 A syndrome of perforin deficiency has recently been described in humans, in that up to 58% of children (typically younger than 2 years) presenting with the rare autosomal recessive disorder familial hemophagocytic lymphohistiocytosis (HLH) carry mutations in both of their perforin (PRF1) alleles.7-9 CTL and NK cell activities are consistently low or absent in HLH patients, and mutations in PRF1 coincide with a marked reduction of perforin staining that has generally been attributed to protein instability.7-9 The cytolytic lymphocytes (CLs) of children diagnosed with HLH are unable to impart a lethal hit to target cells and fail to clear antigen-presenting cells. This causes an uncontrolled activation and expansion of CD4+ and CD8+ T cells and overproduction of inflammatory cytokines, manifested clinically as fever and liver and spleen enlargement and pathologically by hemophagocytosis in the bone marrow and lymphoid organs.10 The clinical and pathologic findings in HLH are reminiscent of the increased expansion of virus-specific T cells and antigen-presenting cells and the inability to down-regulate the immune response seen in perforin-deficient mice infected with a pathogen such as lymphocytic choriomeningitis virus.11,12
A catalog of putative disabling perforin mutations has been compiled as a result of genetic analysis of HLH patients.7-9,13-18 Some of the mutations result in major truncations in perforin (such as the common 50delT mutation, resulting in truncation after amino acid residue 17) that would be expected to inactivate perforin. However, a significant proportion of the changes in PRF1 are missense mutations whose functional significance is not understood. Also, genetic analysis has revealed at least 2 loci other than PRF1 that are HLH associated.19 It is therefore important to exercise caution in ascribing the disease phenotype to an alteration of the perforin sequence, especially when a number of apparently innocent perforin polymorphisms have also been identified. Establishing a causal relationship between a perforin missense mutation and disease should at a minimum require a demonstration of reduced or absent perforin activity in a setting removed from the patient's own CLs. Given the paucity of information currently available on the structure/function relationships of perforin, ascertaining the factors that underpin the loss of function associated with a given perforin mutation may also prove to be an invaluable tool for understanding normal perforin function.
One of the most controversial issues related to mutations in PRF1 is the high frequency of the A91V substitution, found to affect between 3% and 17% of various healthy study populations.18,20-22 The high incidence of the A91V allele suggests that it may represent a neutral polymorphism of perforin.22 As HLH is a rare disease, it is clear that homozygosity for A91V does not of itself result in HLH in the great majority of cases. Nonetheless, some HLH patients with late or atypical presentations were found to be A91V heterozygotes with a potentially inactivating mutation (such as a truncation) encoded on their second perforin allele.20,21 These patients showed reduced CL cytotoxicity as well as significantly reduced levels of perforin expression during the acute stage of the disease. Significantly, it has also been pointed out that atypical presentations of HLH may be misdiagnosed, potentially resulting in underestimation of the disease frequency.18 It might be possible to definitively determine whether the A91V substitution has a pathogenic role in HLH or other immune disorders if the product could be studied by recombinant means.
To date, functional studies of perforin have been greatly hindered by a lack of appropriate methodologies for expressing recombinant perforin, particularly in cells capable of granule-mediated cytotoxicity. Rat basophilic leukemia (RBL) cells were previously shown to be capable of expressing, packaging, and exocytosis of wild-type (WT) perforin, and the secretory pathway of basophils closely mimics that of CTLs and NK cells.23,24 Recently we adapted the RBL cell-based cytotoxicity assay to show that 2 perforin missense mutations, R225W and G429E, are causally associated with HLH.25 Whilst the R225W mutation triggered perforin degradation in the effector cell, G429E perforin was normally synthesized and secreted but significantly reduced the lytic activity of perforin by reducing its Ca2+-dependent binding to the target cell membrane. By contrast, we also found that the putative HLH-causing mutation T435M had no negative effect on perforin function, suggesting that HLH in the affected individual was probably caused by an alternative mechanism.26
In the current study, we have examined the function of 22 further HLH-associated perforin substitutions. Our study employed recombinant technologies to identify 2 categories of perforin mutation: those resulting in protein instability and others that were normally expressed but led to postsynaptic defects resulting in reduced or absent lytic function without a significant effect on protein stability. In addition, we evaluated 2 suspected perforin polymorphisms, A91V and N252S, for their capacity to induce target cell death, as each substitution has been identified in at least one HLH-affected individual.
Materials and methods
Mouse perforin cDNA cloned in pKS(+) Bluescript was mutated using the Transformer or QuickChange kits according to manufacturer's instructions (Stratagene, La Jolla, CA; oligonucleotide primer sequences provided on request). As we described earlier, human perforin cannot be expressed efficiently in RBL cells,25 therefore mouse perforin was used here as an alternative. Human and murine perforin share 68% identity, and we indicated (Tables 1, 2, 3) the conservation of mutated residues between the human, rodent, and Japanese flounder perforins. Human and mouse perforins differ in length by one amino acid, due to an additional amino acid in the leader peptide of human perforin. To avoid confusion in comparison to clinical cases, we have used the amino acid numbering of human perforin throughout this study. The relative positions of mutated residues are identical in the human and the mouse forms of the protein. The WT or mutated perforin P39H, G45E, V50M, D70Y, C73R, A91V, W95R, G149S, F157V, V183G, G220S, T221I, H222R, H222Q, I223D, R232C, R232H, N252S, E261K, C279Y, R299C, D313V, R361W, and Q481P (referred to as such in the text) were cloned into the pIRES2-EGFP (plasmid internal ribosomal entry site 2-enhanced green fluorescent protein) expression plasmid (BD Biosciences Clontech, Palo Alto, CA).25 Two allelic substitutions found in the flounder, R232S and Q481E, were similarly expressed. Each perforin cDNA was sequenced in full on both strands to check the fidelity of site-directed mutagenesis. The resultant expression plasmids were purified using the Qiagen Maxi-kit (Qiagen, Hilden, Germany). Fcϵ-expressing RBL cells were cultured and transiently transfected as described previously.25 GFP-expressing cells were collected 18 to 20 hours later using flow cytometry (FACStar; BD Biosciences). Numerous reports indicated the lack of perforin expression in NK cells of HLH patients, suggesting inherent instability of the mutated proteins. To address the issue, and given the large number of samples analyzed, we had to be able to reliably compare the levels of expression of perforin variants. Therefore, prior to sorting transfected cells, the FACStar flow cytometer was calibrated by using CalibRITE fluorescein isothiocyanate-labeled fluorescent beads (BD Biosciences). We found that this approach provided us with reproducible levels of WT perforin expression and comparable cytotoxicity on a day-to-day basis. The cytotoxicity of RBL cells was analyzed using Jurkat T cells as targets in a 4-hour 51Cr-release assay as described.25 Unless indicated otherwise, each mutated perforin was tested in RBL-based assay at least 3 times at E/T ratios of 30:1, 10:1, and 2:1.
Missense mutations identified in homozygous patients
PFP genotype ID . | Amino acid substitution . | Amino acid . | . | Predicted domain . | Lytic activity, % of WT PFP . | Age at HLH diagnosis, mo . | |
|---|---|---|---|---|---|---|---|
| . | . | Rodent . | Flounder . | . | . | . | |
| A9,13 | Val50Met | Val | Ile | N-terminal | 0 | 4, 84 | |
| B14 | Trp95Arg | Trp | Trp | ? | 0 | 3 | |
| C8,14 | Gly220Ser | Gly | Gly | α-helix | 0 | 2, 1.5 | |
| D9 | His222Arg | His | His | α-helix | 0 | 1 | |
| E13 | Ile224Asp | Ile | Ile | α-helix | 0 | 58 | |
| F7,9* | Arg225Trp | Thr | Thr | α-helix | 025 | NR, 60 | |
| G27* | Thr453Met | Thr | Thr | C2 | 10026 | 3 | |
PFP genotype ID . | Amino acid substitution . | Amino acid . | . | Predicted domain . | Lytic activity, % of WT PFP . | Age at HLH diagnosis, mo . | |
|---|---|---|---|---|---|---|---|
| . | . | Rodent . | Flounder . | . | . | . | |
| A9,13 | Val50Met | Val | Ile | N-terminal | 0 | 4, 84 | |
| B14 | Trp95Arg | Trp | Trp | ? | 0 | 3 | |
| C8,14 | Gly220Ser | Gly | Gly | α-helix | 0 | 2, 1.5 | |
| D9 | His222Arg | His | His | α-helix | 0 | 1 | |
| E13 | Ile224Asp | Ile | Ile | α-helix | 0 | 58 | |
| F7,9* | Arg225Trp | Thr | Thr | α-helix | 025 | NR, 60 | |
| G27* | Thr453Met | Thr | Thr | C2 | 10026 | 3 | |
The original reference for each patient PFP genotype is shown in the first column as a superscript. Amino acid conservation is derived from the amino acid sequence alignment of mammalian and flounder perforins, as in PredictProtein28 (EMBL-Heidelberg). PFP indicates perforin; ?, unknown domain; and NR, not reported.
Missense mutations identified in compound heterozygotes with nonsense or frameshift mutations encoded in the second allele of PRF1
PFP genotype ID . | Amino acid substitution . | Amino acid . | . | Predicted domain . | Lytic activity, % of WT PFP . | Age at HLH diagnosis, mo . | |
|---|---|---|---|---|---|---|---|
| . | . | Rodent . | Flounder . | . | . | . | |
| H9 | Gly45Glu | Gly | Gly | N-terminal | 0 | 8 | |
| I9 | Asp70Tyr | Asp | Asp | ? | 0 | 96 | |
| J9 | Cys73Arg | Cys | Cys | ? | 0 | 4 | |
| K9 | Phe157Val | Phe | Tyr | ? | 0 | 2 | |
| L9 | His222Gln | His | His | α-helix | 0 | 3 | |
| M14 | Arg232Cys* | Arg | Ser | α-helix | 0 | 72 | |
| N8 | Arg232His*† | Arg | Ser | α-helix | 15-20 | 27, 144 | |
| O7 | Asn252Ser‡ | Asp | Glu | ? | 100 | 1, 1 | |
| P8 | Glu261Lys | Glu | Ser | ? | 0 | NR | |
| Q9 | Asp313Val | Asp | Asp | ? | 15 | NR | |
| R9 | Gln481Pro | Gln | Glu | C2 | < 7 | 2 | |
PFP genotype ID . | Amino acid substitution . | Amino acid . | . | Predicted domain . | Lytic activity, % of WT PFP . | Age at HLH diagnosis, mo . | |
|---|---|---|---|---|---|---|---|
| . | . | Rodent . | Flounder . | . | . | . | |
| H9 | Gly45Glu | Gly | Gly | N-terminal | 0 | 8 | |
| I9 | Asp70Tyr | Asp | Asp | ? | 0 | 96 | |
| J9 | Cys73Arg | Cys | Cys | ? | 0 | 4 | |
| K9 | Phe157Val | Phe | Tyr | ? | 0 | 2 | |
| L9 | His222Gln | His | His | α-helix | 0 | 3 | |
| M14 | Arg232Cys* | Arg | Ser | α-helix | 0 | 72 | |
| N8 | Arg232His*† | Arg | Ser | α-helix | 15-20 | 27, 144 | |
| O7 | Asn252Ser‡ | Asp | Glu | ? | 100 | 1, 1 | |
| P8 | Glu261Lys | Glu | Ser | ? | 0 | NR | |
| Q9 | Asp313Val | Asp | Asp | ? | 15 | NR | |
| R9 | Gln481Pro | Gln | Glu | C2 | < 7 | 2 | |
The original reference for each patient PFP genotype is shown in the first column as a superscript. Amino acid conservation is derived from the amino acid sequence alignment of mammalian and flounder perforins, as in PredictProtein28 (EMBL-Heidelberg). PFP indicates perforin; ?, unknown domain; and NR, not reported.
Shown in Figure 6.
Shown in Figure 2.
Shown in Figure 3.
Mutations identified in compound heterozygotes with missense mutations in both alleles of PRF1
PFP genotype ID (age at HLH diagnosis, mo) and amino acid substitution . | Amino acid . | . | Predicted domain . | Lytic activity, % of WT PFP . | |
|---|---|---|---|---|---|
| . | Rodent . | Flounder . | . | . | |
| S15 (120) | |||||
| Pro39His | Pro | Pro | N-terminal | 0 | |
| Gly149Ser | Gly | Gly | ? | 0 | |
| T9 (24) | |||||
| Gly149Ser | Gly | Gly | ? | 0 | |
| Arg299Cys | Arg | Arg | ? | 0 | |
| U9 (48) | |||||
| Gly149Ser | Gly | Gly | ? | 0 | |
| Arg361Trp | Arg | Arg | ? | 0 | |
| V7 (NR) | |||||
| Val183Gly | Ala | Ser | ? | 100 | |
| Cys279Tyr* | Cys | Cys | ? | 0 | |
| W14 (6) | |||||
| Thr221Ile | Thr | Thr | α-helix | 0 | |
| Arg225Trp† | Thr | Thr | α-helix | 025 | |
| X7 (14, 17, 28) | |||||
| Arg225Trp† | Thr | Thr | α-helix | 025 | |
| Gly429Glu† | Gly | Gly | C2 | 3025 | |
PFP genotype ID (age at HLH diagnosis, mo) and amino acid substitution . | Amino acid . | . | Predicted domain . | Lytic activity, % of WT PFP . | |
|---|---|---|---|---|---|
| . | Rodent . | Flounder . | . | . | |
| S15 (120) | |||||
| Pro39His | Pro | Pro | N-terminal | 0 | |
| Gly149Ser | Gly | Gly | ? | 0 | |
| T9 (24) | |||||
| Gly149Ser | Gly | Gly | ? | 0 | |
| Arg299Cys | Arg | Arg | ? | 0 | |
| U9 (48) | |||||
| Gly149Ser | Gly | Gly | ? | 0 | |
| Arg361Trp | Arg | Arg | ? | 0 | |
| V7 (NR) | |||||
| Val183Gly | Ala | Ser | ? | 100 | |
| Cys279Tyr* | Cys | Cys | ? | 0 | |
| W14 (6) | |||||
| Thr221Ile | Thr | Thr | α-helix | 0 | |
| Arg225Trp† | Thr | Thr | α-helix | 025 | |
| X7 (14, 17, 28) | |||||
| Arg225Trp† | Thr | Thr | α-helix | 025 | |
| Gly429Glu† | Gly | Gly | C2 | 3025 | |
The original reference for each patient PFP genotype is shown in the first column as a superscript. Amino acid conservation is derived from the amino acid sequence alignment of mammalian and flounder perforins, as in PredictProtein28 (EMBL-Heidelberg). PFP indicates perforin; ?, unknown domain; and NR, not reported.
Shown in Figure 5.
Cell lysates from transiently transfected RBL cells were resolved on a 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE; Tris-Glycine) gel, which was then analyzed for perforin or tubulin expression by immunoblotting with P1-8 antiperforin or antitubulin antibodies, followed by the secondary horseradish peroxidase-linked anti-rat or anti-mouse immunoglobulin.26 The signal was detected by chemiluminescence (Amersham Biosciences, Little Chalfont, England).
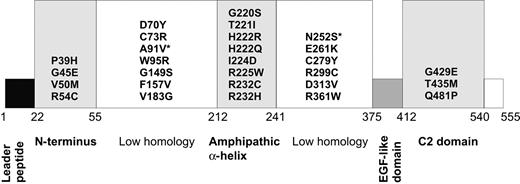
The location of 2 common perforin polymorphisms and missense mutations identified in HLH. The putative domains of perforin are indicated as boxes, and the numerals indicate the approximate amino acid boundaries for each domain, designating the first residue of the leader as residue 1. The N-terminus is predicted to have lytic properties; 2 low-homology regions have no significant similarity to other mammalian protein domains; amphipathic α-helix is homologous to regions of the complement membrane attack complex components C5b to C9; the EGF-like domain is structurally similar to ubiquitous EGF domains, primarily due to highly conserved cysteine residues; the C2 domain is the calcium-binding region responsible for membrane binding of perforin. The asterisked residues A91V and N252S refer to suspected PRF1 polymorphisms.
The location of 2 common perforin polymorphisms and missense mutations identified in HLH. The putative domains of perforin are indicated as boxes, and the numerals indicate the approximate amino acid boundaries for each domain, designating the first residue of the leader as residue 1. The N-terminus is predicted to have lytic properties; 2 low-homology regions have no significant similarity to other mammalian protein domains; amphipathic α-helix is homologous to regions of the complement membrane attack complex components C5b to C9; the EGF-like domain is structurally similar to ubiquitous EGF domains, primarily due to highly conserved cysteine residues; the C2 domain is the calcium-binding region responsible for membrane binding of perforin. The asterisked residues A91V and N252S refer to suspected PRF1 polymorphisms.
Results
In the past we have investigated the basis of the defective function associated with the G429E, R225W, and T435M perforin substitutions.25,26 In the present study we undertook a functional analysis of 22 further suspected HLH-causing missense mutations of PRF1 that map to various perforin domains (Figure 1). To analyze the impact of the mutations on perforin function in isolation from potential defects in other loci, we used a previously validated model for examining the lytic function of perforin. We expressed WT or mutated perforin in RBL cells, then ascertained their ability to lyse Jurkat target cells to which they were conjugated.25 Using this approach we were able to discriminate likely presynaptic and postsynaptic dysfunctions of the various perforin mutants. We also performed a detailed analysis of 2 alterations of the perforin sequence that have thus far been considered to be PRF1 polymorphisms, A91V and N252S. The role of each substitution in disease pathogenesis remains unclear and is the subject of continuing debate.18,21,22
A functional analysis of the suspected perforin polymorphism A91V
A91V is the most common amino acid substitution identified in perforin, with an allele frequency ranging between 3% and 17%.9,22 Assuming normal Mendelian inheritance, this indicates that up to 4% of some populations are likely to be homozygous for the A91V allele. Clearly, if A91V resulted in significantly reduced perforin function, it would be expected to cause many more cases of HLH than are currently reported: approximately 1:50 000 live births in toto or 1:90 000 cases related to perforin dysfunction.29 Therefore, the A91V substitution has generally been regarded as a neutral polymorphism of the PRF1 gene. Despite this, several HLH patients with late and/or atypical presentation have been shown to carry A91V. In one case study, 2 siblings were A91V/W374ter heterozygotes and each developed atypical HLH at 25 and 27 years.20 Both patients had either undetectable or severely reduced NK cell activity and perforin expression in their peripheral blood during active disease.20 No other trigger for the disease could be identified in either individual. In another report, 1 of 2 fraternal twins, both of whom were homozygous for A91V and heterozygous for R232H (1 allele was doubly mutated), developed HLH by the age of 13 years whereas the other twin remained healthy.21 Once again, no trigger was identified prior to disease onset. These reports have pointed out that A91V may play a role in the pathogenesis of HLH and also indicated that the disease might have a late, atypical, or sporadic presentation in this group of individuals.
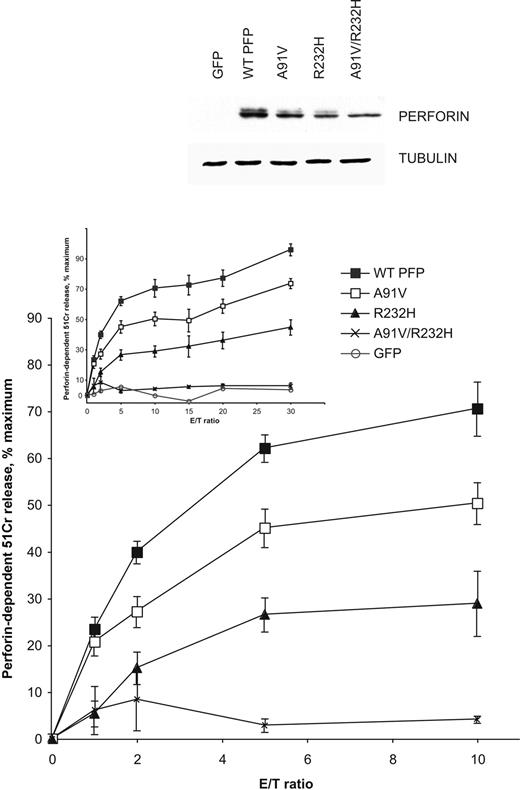
To resolve the uncertainty surrounding a possible pathogenic role for A91V, we expressed recombinant A91V perforin in RBL cells to compare its lytic capacity to WT perforin. We attempted to model the phenotype of patients who have developed HLH in association with A91V. We therefore generated and tested A91V, R232H, and doubly mutated A91V/R232H perforin in our assays. The truncated perforin W374ter (inherited together with A91V by another HLH patient) was considered likely to be inactive, as it lacks the entire carboxyl C2 domain, which is essential for Ca2+-dependent target cell membrane binding by perforin.26 First, we performed 51Cr-release cytotoxicity assays using A91V-, R232H-, or A91V/R232H-perforin-transfected RBL cells (Figure 2). On the basis that it took about twice as many RBL cells to achieve a given level of target cell death, the cytotoxic activity of A91V perforin was consistently reduced by approximately 50% compared with WT perforin. By the same criterion, R232H perforin was slightly less active than A91V and generated approximately 30% of WT perforin activity. Importantly, the doubly mutated A91V/R232H protein was completely inactive (Figure 2). Analysis of protein expression levels by Western blot revealed reduced expression of A91V, R232H, and, to a greater extent, A91V/R232H perforin compared with the WT protein. These observations suggested that both mutations affected the folding and stability of perforin and were likely to impact negatively on its cytotoxicity in the RBL assay.
We also produced recombinant human A91V and WT perforin using the baculovirus expression system.25 We found that the lytic activity of A91V was reduced to less than 10% that of WT perforin (I.V., A. Ciccone, C. House, J.A.T., “The molecular basis for A91V perforin polymorphism dysfunction,” manuscript in preparation). In addition, purified A91V was functionally unstable, in that its lytic activity rapidly diminished to undetectable levels after 48 hours of storage at 4°C. By comparison, WT perforin was stable under these storage conditions for several months. On this basis, we propose that the A91V substitution results in protein misfolding that is most likely responsible for its reduced stability in RBL cells. This instability was augmented in the case of perforin purified from baculovirus-infected insect cells, possibly due to the absence of appropriate intracellular chaperone(s) in insect cells and/or the altered redox environment.
Reduced expression and partial loss of function of A91V and the coinherited substitution R232S. The effect of PRF1 mutations identified in fraternal twins inheriting A91V, R232H, and doubly mutated A91V/R232H perforin (PFP).21 The top panel shows a Western blot of whole-cell extracts from RBL cells expressing the respective mutated perforin and sorted as described in “Materials and methods.” The graphs show 4-hour cytotoxicity assays using transiently transfected and sorted RBL cells as effector cells and 51Cr-labeled Jurkat cells as targets at the effector-target (E/T) ratios indicated. The data shown are the means ± SE of 4 to 9 independent experiments. For clarity, a subset of the data (the lower E/T ratios) is shown again in the larger plot.
Reduced expression and partial loss of function of A91V and the coinherited substitution R232S. The effect of PRF1 mutations identified in fraternal twins inheriting A91V, R232H, and doubly mutated A91V/R232H perforin (PFP).21 The top panel shows a Western blot of whole-cell extracts from RBL cells expressing the respective mutated perforin and sorted as described in “Materials and methods.” The graphs show 4-hour cytotoxicity assays using transiently transfected and sorted RBL cells as effector cells and 51Cr-labeled Jurkat cells as targets at the effector-target (E/T) ratios indicated. The data shown are the means ± SE of 4 to 9 independent experiments. For clarity, a subset of the data (the lower E/T ratios) is shown again in the larger plot.
As a whole, the functional assays indicated that the A91V substitution is an unusual type of PRF1 polymorphism, in that it has a high allele frequency but clearly results in reduced stability and, consequently, partial loss of perforin lytic activity. This conclusion is consistent with clinical studies, in that some individuals possessing A91V appear to be prone to a late or atypical presentation of HLH. We propose that the level of cytotoxic activity of A91V may generally be substantial enough to prevent HLH provided the second allele is WT or even when the mutation is inherited in the homozygous state, as is the case in 1% to 4% of healthy populations. It may still be possible that a coinherited genetic abnormality, epigenetic factors, or the nature of the environmental stimulus (such as a specific pathogen) may predispose an A91V homozygote to HLH, but these factors may be difficult to predict. Even perforin-deficient mice can clear many viral pathogens; however, infection with lymphocytic choriomeningitis virus results in an HLH-like syndrome.12 The genetic analysis of HLH-affected individuals suggests that A91V may predispose to HLH in cases where the second allele has an inactivating mutation, for instance where one allele is completely inactive because of an additional mutation at residue 232 (Figure 2). Given that inactivating mutations of PRF1 occur in approximately 1 in 150 individuals (based on 1:90 000 incidence of PRF1 mutation-related HLH and random segregation of alleles)29 and A91V homozygosity is predicted to occur in 1% to 4% of populations, the children of A91V parents (particularly homozygous parents) may be an “at risk” group for developing atypical HLH or a related immuno-insufficiency, as their chance of inheriting the A91V/null phenotype is predicted to be 1 in 150 (0.67%). One of the challenges in studying perforin experimentally is to determine the minimum level of perforin activity sufficient to maintain adequate CTL and NK cell function. Clinical experience suggests that an individual whose sole functional perforin allele is A91V still has sufficient perforin activity to maintain CL homeostasis for a prolonged period of time, up to 27 years.20 If identified early in life, such individuals may warrant close clinical monitoring.
A functional analysis of the suspected perforin polymorphism N252S
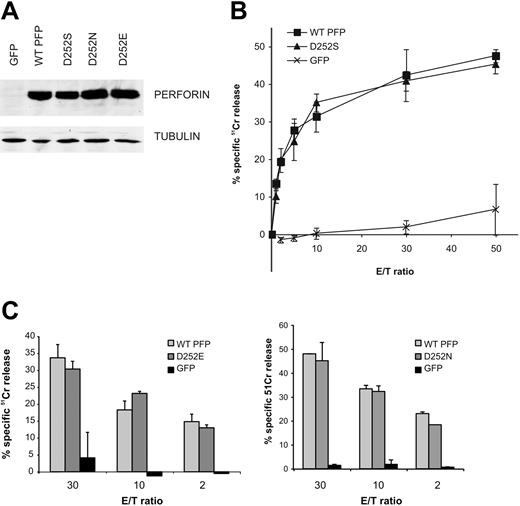
Another suspected PRF1 polymorphism leading to the N252S substitution has been reported in an HLH patient who developed the disease by the age of 1 month.7,8 Despite the early onset of disease, the CTL activity of the patient was reportedly reduced by only 50% compared with aged-matched controls. It was not clear whether the patient had an inactivating mutation in the second allele of PRF17,8 but our results (Figure 3) suggested that this is likely to be the case. In fact, a more recent report from the same group indicated that the described patient had 2 detrimental mutations, which the authors could not originally identify (one of these is reported in the present paper, G149S).30 Another report suggested that N252S coinherited with a detrimental Fas mutation was responsible for autoimmune lymphoproliferative syndrome and lymphoma.27 In contrast to these observations, a 2% allele frequency for the N252S substitution has recently been found in the general population, suggesting that it represents a polymorphism of PRF1.9 Consistent with that notion, N252 is encoded only in the human PFNI gene, whereas rodent and flounder perforin have aspartic acid or glutamic acid, respectively (PredictProtein28 ; EMBL-Heidelberg, Heidelberg, Germany). This suggests that perforin might withstand at least semiconservative amino acid substitutions at residue 252. To elucidate the effect of the N252S substitution on perforin function, we generated several perforin mutations, D252N, D252E and D252S, and analyzed their activity in the RBL cytotoxicity assay (Figure 3). We found that all of these substitutions retained WT perforin activity. Assuming codominant expression, these observations suggested that an individual carrying the N252S allele and an inactivating mutation in their other PRF1 allele30 would have approximately 50% of normal perforin activity, consistent with the level of CTL activity observed in the HLH patients in the original report.8 Taken together, our data and epidemiologic studies9 indicate that the N252S substitution alone could not have been causative of disease but, rather, that an additional genetic defect(s) might have been responsible. We therefore concluded that N252S probably represents a neutral PRF1 polymorphism.
Functional analysis of missense mutations associated with HLH
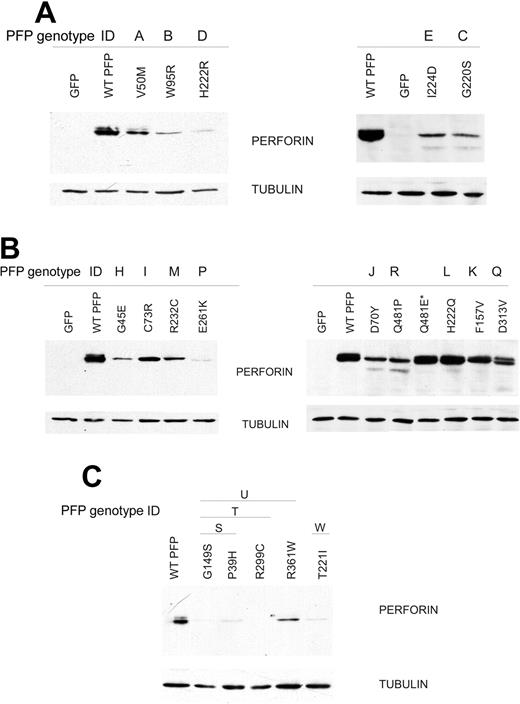
The majority of HLH patients with suspected detrimental missense mutations in PRF1 have undetectable levels of perforin expression in their CLs.9 Traditionally, this has been attributed to the instability of the mutated perforin, in keeping with a presynaptic defect of function. However, an alternative reason for the absence of perforin activity might be related to immune dysregulation; since HLH is characterized by hyperactivation of CLs, it is plausible that the diminished perforin levels are a result of the cells constitutively secreting their granule contents in an attempt to clear persistent antigen or to down-regulate the number of antigen-presenting cells. This notion is supported by our earlier observation that G429E perforin is expressed at normal levels in RBL cells but displayed significantly reduced cytotoxic activity.25 In the current study, we grouped perforin mutations according to the combinations of alleles reported in various HLH patients. Figure 4A and Table 1 show a summary of the results for alleles found in homozygous patients; Figure 4B and Table 2 show the corresponding data for alleles coexpressed with a nonsense of frame-shift mutation of perforin; whereas Figure 4C and Table 3 refer to alleles identified only in compound heterozygous patients with missense mutations in both alleles. This approach was chosen so that wherever possible, our findings might be usefully applied to the interpretation of corresponding clinical reports.
Normal expression and function of perforin with a serine substitution at residue 252. (A) The Western immunoblot shows the relative expression of D252S, D252N (as in human perforin), and D252E (as in flounder perforin) in transiently transfected RBL cells. (B) The line graph shows the lytic activity of D252S perforin (equivalent to N252S in humans) in the 51Cr-release cytotoxicity assay. (C) The bar charts compare the lytic capacity of perforin variants at position 252 grafted onto mouse perforin: D252E found in flounder (left) and D252N in human perforin (right). The data shown are mean ± SD and are representative of 3 independent experiments.
Normal expression and function of perforin with a serine substitution at residue 252. (A) The Western immunoblot shows the relative expression of D252S, D252N (as in human perforin), and D252E (as in flounder perforin) in transiently transfected RBL cells. (B) The line graph shows the lytic activity of D252S perforin (equivalent to N252S in humans) in the 51Cr-release cytotoxicity assay. (C) The bar charts compare the lytic capacity of perforin variants at position 252 grafted onto mouse perforin: D252E found in flounder (left) and D252N in human perforin (right). The data shown are mean ± SD and are representative of 3 independent experiments.
Analysis of missense mutations of PRF1 on the expression of perforin. RBL cells were transfected to express perforin bearing each of the missense mutations listed, then sorted with a fluorescence-activated cell sorter (FACS). The mutations were classified according to the HLH patient's genotype: (A) those identified in homozygous patients; (B) mutations identified in compound heterozygotes, where the second allele encoded a frame-shift or premature termination of the protein; and (C) mutations identified in compound heterozygotes with missense mutations in both alleles of PRF1. The Western immunoblots show the relative level of expression of mutated perforin in equivalent numbers of FACS-sorted RBL cells (see “Materials and methods”).
Analysis of missense mutations of PRF1 on the expression of perforin. RBL cells were transfected to express perforin bearing each of the missense mutations listed, then sorted with a fluorescence-activated cell sorter (FACS). The mutations were classified according to the HLH patient's genotype: (A) those identified in homozygous patients; (B) mutations identified in compound heterozygotes, where the second allele encoded a frame-shift or premature termination of the protein; and (C) mutations identified in compound heterozygotes with missense mutations in both alleles of PRF1. The Western immunoblots show the relative level of expression of mutated perforin in equivalent numbers of FACS-sorted RBL cells (see “Materials and methods”).
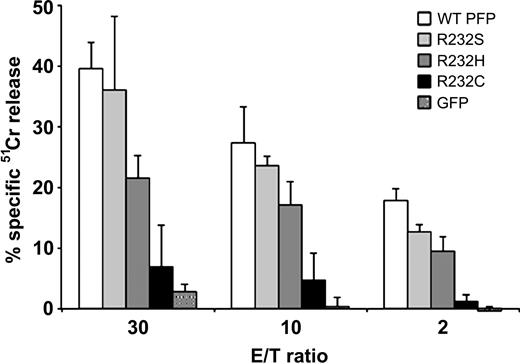
Effects of various substitutions at residue 232 of perforin on RBL-mediated cytotoxicity.51Cr-release cytotoxicity assays using transfected RBL cells and Jurkat target cells at the E/T ratios indicated, comparing the cytotoxic function of R232C and R232H (substitutions identified in HLH patients) with WT and R232S (flounder) perforins. Data shown are means ± SD.
Effects of various substitutions at residue 232 of perforin on RBL-mediated cytotoxicity.51Cr-release cytotoxicity assays using transfected RBL cells and Jurkat target cells at the E/T ratios indicated, comparing the cytotoxic function of R232C and R232H (substitutions identified in HLH patients) with WT and R232S (flounder) perforins. Data shown are means ± SD.
We began our analysis of missense mutations by investigating whether a given mutation resulted in a presynaptic or postsynaptic dysfunction. The analysis of expression levels in RBL cells revealed that the majority of perforin mutations result in unstable/unfolded protein. Thus, according to Western blot analysis, perforin with the mutation P39H, G45E, G45R, V50M, D70Y, W95R, G149S, G220S, T221I, H222R, R232C, R232H, E261K, C279Y, R299C, R361W, or Q481P was undetectable or greatly reduced in RBL cells compared with WT perforin. This was not surprising, as the mutated residues were uniformly highly conserved in the flounder and had undergone nonconservative substitution. It is likely that presynaptic defects of the mutated proteins were related to their misfolding or abnormal trafficking, leading to degradation. Predictably, all of the unstable perforin variants had minimal detectable cytotoxic activity in the RBL cell-based 51Cr-release assay (Figure 4A-C; Tables 1, 2, 3). This was consistent with clinical reports, in that the corresponding HLH patients had undetectable NK/CTL perforin and absent NK activity. Interestingly, the age of HLH presentation varied widely, even among patients with the same mutated allele, and was independent of whether the mutated perforin displayed some minor residual activity (Tables 1, 2, 3). To support our findings, we also engineered amino acid substitutions R232S (Figure 5) and Q481E (Figure 4B; Table 2) to mirror residues found in the corresponding position in flounder perforin. Unlike R232H (Figures 2 and 5), R232S had normal activity, whereas R232C (reported in one HLH patient)14 also had severely diminished function (Figures 4A, 5; Table 2). Flounder Q481E perforin also had WT expression levels (Figure 4B; Table 2) and activity (data not shown).
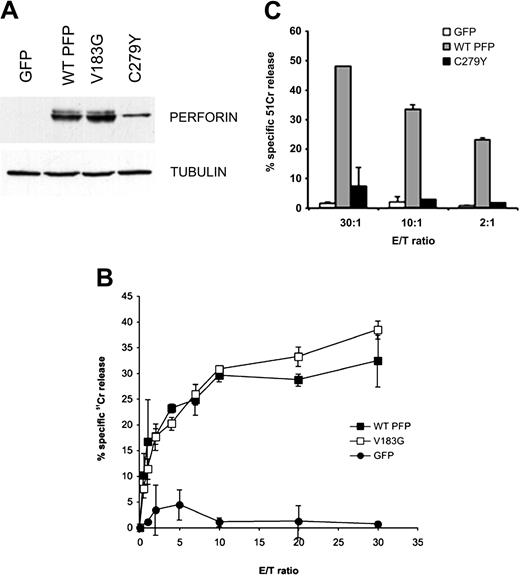
V183G perforin has normal function but the C279Y substitution results in loss of perforin function. (A) Western immunoblot shows the relative expression of the wild-type, V183G, and C279Y perforins in transiently transfected FACS-sorted RBL cells. (B-C) 51Cr-release cytotoxicity assays using transfected RBL cells and Jurkat target cells at the E/T ratios indicated and comparing the putative perforin mutation V183G (B) and C279Y perforin (C) with WT perforin. Data shown are mean ± SD.
V183G perforin has normal function but the C279Y substitution results in loss of perforin function. (A) Western immunoblot shows the relative expression of the wild-type, V183G, and C279Y perforins in transiently transfected FACS-sorted RBL cells. (B-C) 51Cr-release cytotoxicity assays using transfected RBL cells and Jurkat target cells at the E/T ratios indicated and comparing the putative perforin mutation V183G (B) and C279Y perforin (C) with WT perforin. Data shown are mean ± SD.
Another grouping of perforin mutations analyzed here was expressed quite differently from those described above. Contrary to clinical reports showing poor expression, V183G (Figure 6) and H222Q perforin were expressed in RBL cells at a level equivalent to WT perforin (Figure 4B-C), and the expression levels of C73R, F157V, and D313V perforin were only marginally reduced (Figure 4B). Unlike the mutations that led to protein instability, these mutations are likely to be defective at the postsynaptic level and, as in the case of G429E,25 may be of use in defining the normal functions of perforin on the target cell membrane.
Subsequently we used the 51Cr-release cytotoxicity assay to analyze the cytotoxic properties of these mutated perforins. We were surprised to find that the lytic activity of V183G perforin, which has been implicated in HLH, was indistinguishable from that of the WT protein (Figure 6). Interestingly, V183 is not conserved among rodent and flounder perforin, with alanine and serine found in rat and flounder perforins, respectively. We concluded that the V183G mutation was unlikely to play a causative role in HLH for perforin genotype V (Figure 6; Table 3), even though the second allele had an inactivating C279Y substitution. Given our experimental observations and the lack of amino acid conservation, we propose that the V183G substitution may be a neutral polymorphism of PRF1, and HLH in the corresponding patient was likely to be caused by some other mechanism independent of perforin. In addition, perforin mutations did not appear to have an appreciable “dominant-negative” effect on the function of WT perforin, as this property would be expected to affect perforin function in the patients' parents. Mutation of the conserved histidine H222Q resulted in normal expression of perforin in RBL cells, but the transfected RBL cells had no detectable cytotoxic activity (data not shown). Similar results were observed with nonconservatively substituted residues C73R, F157V, and D313V mutations, whose expression levels in RBL cells were only slightly reduced compared with the WT perforin. As discussed, the apparent loss of perforin from CLs of these HLH patients might be related to constitutive CL activation, especially during the acute stage of disease.
Discussion
In conclusion, we have presented a comprehensive functional analysis of the missense mutations and polymorphisms of PRF1 thus far reported in association with HLH. Our data indicate that the instability of mutated perforin is a more common cause of perforin-related HLH than postsynaptic dysfunction. We established that the A91V mutation is an unusual case of “polymorphism” in that it significantly affects the stability and cytolytic activity of perforin, most likely due to incorrect folding of the protein. The fact that A91V is carried by a significant proportion of the healthy population in the homozygous state is in keeping with our experimental findings that this substitution nonetheless retains a significant proportion of WT function.
Prepublished online as Blood First Edition Paper, March 8, 2005; DOI 10.1182/blood-2004-12-4935.
J.A.T. is a Senior Principal Research Fellow of the National Health and Medical Research Council (NHMRC) of Australia and is supported by Program grants from the NHMRC and the Juvenile Diabetes Research Foundation.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr M. J. Smyth for helpful discussions. The authors thank Ralph Rossi and Andrew Fryga from the Cell Sorting Facility for their helpful suggestions.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal