Abstract
Mutations in the perforin gene (PRF1) are a common cause of the fatal immune dysregulation disorder, familial hemophagocytic lymphohistiocytosis (type 2 FHL, FHL2). Here we report a female infant born with biallelic PRF1 mutations: a novel substitution, D49N, and a previously identified in-frame deletion, K285del. We assessed the effects of each mutation on the cytotoxicity of human NK cells in which the expression of endogenous perforin was ablated with miR30-based short hairpin (sh) RNAs. Both mutations were detrimental for function, thereby explaining the clinically severe presentation and rapidly fatal outcome. We demonstrate that D49N exerts its deleterious effect by generating an additional (third) N-linked glycosylation site, resulting in protein misfolding and degradation in the killer cell. Our data provide a rationale for treating some cases of type 2 familial hemophagocytic lymphohistiocytosis, based on the pharmacologic inhibition or modification of glycosylation.
Introduction
Familial hemophagocytic lymphohistiocytosis (FHL) is a fatal autosomal recessive disorder of immune regulation.1-5 Perforin (PRF1) mutations are a common cause, accounting for at least 30% of FHL cases.6-10 Perforin is expressed exclusively by cytotoxic lymphocytes, and its pore-forming activity is essential for the granule-mediated apoptosis of virus-infected and cancerous cells.11 Although biallelic loss-of-function PRF1 mutations result in aggressive FHL, which develops within weeks of birth, mutations that retain some activity are generally protective through infancy and early childhood but result in atypical late-onset FHL, hematologic malignancy, and/or life-threatening viral infections.12
In this study, we report a novel missense mutation, D49N, which was coinherited with a previously identified, yet uncharacterized, common deletion, K285del. We generated a novel functional assay in human NK cells to demonstrate that both mutations are detrimental to perforin-mediated cytotoxicity. Furthermore, we show that D49N introduces a novel N-linked glycosylation site that leads to protein misfolding and instability.
Methods
Perforin knockdown KHYG1 human NK cells
Perforin knockdown cell lines were created by overexpressing miR30-based shRNAs targeting the 3′-untranslated region of PRF1. The sequence of the 21mer constructs cloned into the retroviral pLMP-cherry vector was 5′-AUCCCGAUUCACCCUGUCCAA-3′ and 5′-UCGGCUAUCGUUAGUGCUAGU-3′. HEK293T cells were used to package the amphotropic retrovirus and the viral supernatant collected to transduce KHYG1 cells. Cherry red-positive cells were FACS sorted for selection of KHYG1 shPRF1 cells.
KHYG1 perforin complementation assays
For KHYG1 complementation assays, the coding sequence of human wild-type (WT) or mutated perforin was cloned into the retroviral murine stem cell virus green fluorescent protein (GFP) vector. HEK293T cells were used to package the amphotropic retrovirus and the viral supernatant collected to transduce KHYG1 shPRF1 cells. For selection of perforin-expressing KHYG1 shPRF1 cells, GFP+ cells were FACS sorted based on identical mean-relative fluorescence.
DNA sequencing, cell culture, and Western immunoblotting have been described previously12 (for details see supplemental Methods, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Results and discussion
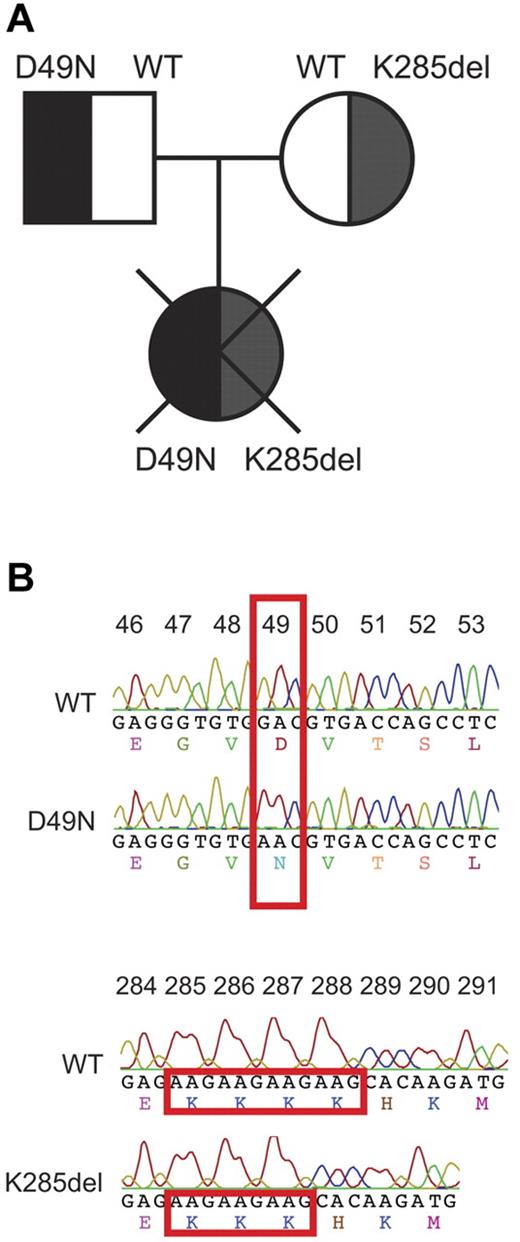
We obtained peripheral blood from a female infant, the first-born of healthy, unrelated parents. She presented with fever, thrombocytopenia, and elevated bilirubin one day after birth, then rapidly developed neutropenia, hepatosplenomegaly, raised liver enzymes, ascites, and coagulopathy. At day 9, hemophagocytic lymphohistiocytosis (HLH) was diagnosed from a bone marrow aspirate showing pronounced hemophagocytosis, in addition to elevated ferritin, hypofibrinogenemia, and persistent pancytopenia. Therapy was commenced as per the HLH-2004 protocol, using etoposide, cyclosporine, and dexamethasone, but the infant did not respond and died on day 14 of progressive multiorgan failure. The PRF1 gene was sequenced in the patient (and parents), revealing biallelic PRF1 mutations: c.145G > A (exon 2) resulted in the D49N substitution, and an in-frame deletion c.[853_855delAAG] (exon 3) resulted in deletion of lysine 285 (K285del; Figure 1).
Identification of biallelic perforin mutations inherited by a female patient presenting with HLH. (A) Family tree indicating the perforin phenotype in each member of the patient's family. (B) Comparison of the perforin gene sequence of the patient and a normal WT control. One mutation was G145 > A, mapping to exon 2, and resulting in the substitution of aspartate 49 by asparagine (D49N), whereas the other was a deletion of nucleotides 853 to 855 in exon 3, resulting in the in-frame deletion of lysine 285 (K285del).
Identification of biallelic perforin mutations inherited by a female patient presenting with HLH. (A) Family tree indicating the perforin phenotype in each member of the patient's family. (B) Comparison of the perforin gene sequence of the patient and a normal WT control. One mutation was G145 > A, mapping to exon 2, and resulting in the substitution of aspartate 49 by asparagine (D49N), whereas the other was a deletion of nucleotides 853 to 855 in exon 3, resulting in the in-frame deletion of lysine 285 (K285del).
Because of limited patient material, cytotoxic activity of the patient's cytotoxic lymphocytes could not be tested. Instead, we designed a surrogate assay using the human NK cell line KHYG-1 to assess the effects of each PRF1 mutation; previous studies of PRF1 mutants have been limited to rodent cells.12,13 We created a perforin “knockdown” cell line using miR30-based shRNA constructs targeting the 3′-untranslated region of PRF1 (shPRF1). We found that reduced perforin protein expression correlated well with decreased killing of K562 target cells (not shown). Reintroducing human WT perforin into the cells (shPRF1+ WT) restored perforin levels and cytotoxic activity (Figure 2A).
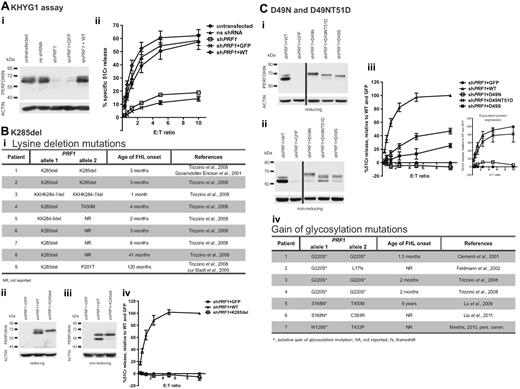
Patient's perforin mutations, D49N and K285del, are detrimental to NK cell cytotoxicity. (A) KHYG1 cells were virally transduced with nonsilencing (ns) shRNA or shPRF1 constructs to down-regulate perforin expression. Perforin expression was then restored in KHYG1 shPRF1 cells by virally transducing cells with WT-perforin construct (KHGY1 shPRF1+ WT). KHYG1 shPRF1 cells were also transduced with the empty vector for control (KHYG1 shPRF1+ GFP). (i) Western immunoblot shows the relative amounts of perforin expression in each of the stable KHYG1 cell populations. (ii) Four-hour 51Cr release assays, against target K562 cells at the effector/target (E:T) ratios indicated, show a 90% reduction in function in KHYG1 shPRF1 cells and full restoration in KHYG1 shPRF1+ WT cells. Data are mean ± SE of 8 independent experiments. (Bi) Table shows patients 1 to 9 identified in the literature who inherited deletion mutations in the intensely basic (282KKKKHK) region of PRF1. (ii-iv) KHYG1 shPRF1 cells were virally transduced with K285del-perforin, sorted on the basis of identical mean GFP fluorescence (compared with KHYG1 shPRF1+ GFP and KHYG1 shPRF1+ WT cells), and then analyzed for perforin expression and cytotoxicity. For clarity, background levels seen for KHYG1 shPRF1+ GFP cells were subtracted from total 51Cr release levels (to reflect the activity of reintroduced recombinant perforin) and standardized against WT-perforin at a 10:1 E/T ratio. Data are mean ± SE of 3 independent experiments. (Ci-iii) KHYG1 shPRF1 cells were virally transduced with D49N- and D49NT51D-perforin, sorted on the basis of identical mean GFP fluorescence (compared with KHYG1 shPRF1+ GFP and KHYG1 shPRF1+ WT cells), and then analyzed for perforin expression and cytotoxicity. The values plotted represent standardized 51Cr release levels, as described in subpanels ii through iv. The data shown are mean ± SE of 7 independent experiments. (iii inset) Perforin-expressing KHYG1 shPRF1 cells were sorted to achieve identical protein expression and analyzed for perforin cytotoxicity. The values plotted represent standardized 51Cr release levels (as described in subpanels ii-iv). Data are mean ± SE of 3 to 5 independent experiments for each cell line. Corresponding Western blots are shown in supplemental Figure 4. (iv) Table lists FHL patients who inherited putative gain of glycosylation mutations in perforin.
Patient's perforin mutations, D49N and K285del, are detrimental to NK cell cytotoxicity. (A) KHYG1 cells were virally transduced with nonsilencing (ns) shRNA or shPRF1 constructs to down-regulate perforin expression. Perforin expression was then restored in KHYG1 shPRF1 cells by virally transducing cells with WT-perforin construct (KHGY1 shPRF1+ WT). KHYG1 shPRF1 cells were also transduced with the empty vector for control (KHYG1 shPRF1+ GFP). (i) Western immunoblot shows the relative amounts of perforin expression in each of the stable KHYG1 cell populations. (ii) Four-hour 51Cr release assays, against target K562 cells at the effector/target (E:T) ratios indicated, show a 90% reduction in function in KHYG1 shPRF1 cells and full restoration in KHYG1 shPRF1+ WT cells. Data are mean ± SE of 8 independent experiments. (Bi) Table shows patients 1 to 9 identified in the literature who inherited deletion mutations in the intensely basic (282KKKKHK) region of PRF1. (ii-iv) KHYG1 shPRF1 cells were virally transduced with K285del-perforin, sorted on the basis of identical mean GFP fluorescence (compared with KHYG1 shPRF1+ GFP and KHYG1 shPRF1+ WT cells), and then analyzed for perforin expression and cytotoxicity. For clarity, background levels seen for KHYG1 shPRF1+ GFP cells were subtracted from total 51Cr release levels (to reflect the activity of reintroduced recombinant perforin) and standardized against WT-perforin at a 10:1 E/T ratio. Data are mean ± SE of 3 independent experiments. (Ci-iii) KHYG1 shPRF1 cells were virally transduced with D49N- and D49NT51D-perforin, sorted on the basis of identical mean GFP fluorescence (compared with KHYG1 shPRF1+ GFP and KHYG1 shPRF1+ WT cells), and then analyzed for perforin expression and cytotoxicity. The values plotted represent standardized 51Cr release levels, as described in subpanels ii through iv. The data shown are mean ± SE of 7 independent experiments. (iii inset) Perforin-expressing KHYG1 shPRF1 cells were sorted to achieve identical protein expression and analyzed for perforin cytotoxicity. The values plotted represent standardized 51Cr release levels (as described in subpanels ii-iv). Data are mean ± SE of 3 to 5 independent experiments for each cell line. Corresponding Western blots are shown in supplemental Figure 4. (iv) Table lists FHL patients who inherited putative gain of glycosylation mutations in perforin.
We then examined the effects of the mutations on perforin-mediated cytotoxicity. All the lysines in the conserved sequence 282KKKKHK in perforin are encoded by AAG, making this region prone to replication slippage. Indeed, in-frame deletions have been reported in 9 unrelated patients (Figure 2Bi),4,7,14 but these mutations have never been studied functionally. We expressed K285del-perforin in KHYG1 shPRF1 cells and found that the deletion was detrimental to cytotoxicity (Figure 2Bii-iv). These results are consistent with clinical observations, as patients homozygous for deletions in this region invariably develop FHL within weeks to months of birth. The only K285del patient presenting with late-onset FHL inherited a second PRF1 mutation with partial activity (patient 9, Figure 2Bi).12
The second mutation, D49N, is novel. Because of a threonine (T51) 2 residues downstream of the mutation site, the asparagine-49 fulfilled the requirement for a consensus N-linked glycosylation site (N-x-S/T), which, by contrast with 2 conserved glycosylation sites (positions 205 and 548), is located centrally in the structurally labile MACPF domain.15,16 Similar to K285del, D49N did not rescue cytotoxicity in shPRF1 KHYG1 cells (Figure 2C). Consistent with glycan addition, D49N-perforin migrated slower on SDS-PAGE, and its expression level was lower than WT (Figure 2Ci-ii). The D49N-perforin was also undetectable by intracellular FACS staining using a different antibody (δG9; supplemental Figure 1). To further corroborate abnormal folding, D49N-perforin was far more susceptible to trypsin digestion than WT-perforin (supplemental Figure 3).
To determine whether glycosylation per se contributed to the loss of cytotoxicity, a second mutation, T51D, was introduced with D49N to disrupt the glycosylation consensus sequence. As expected, this second mutation prevented the addition of the glycan and restored the protein's migration on SDS-PAGE (Figure 2Ci-ii; supplemental Figure 2). However, D49NT51D-perforin was still expressed at lower levels than WT-perforin (Figure 2Cii; supplemental Figure 1) and remained more susceptible to trypsin digestion (supplemental Figure 3), indicating that protein folding remained defective. Accordingly, although the ability to kill target K562 was significantly improved, it was still reduced compared with WT-perforin (Figure 2Ciii). This was not entirely surprising, as D49 is highly conserved across species. Indeed, the activity of an experimental mutant D49S was reduced compared with WT-perforin but was still significantly higher than that of D49N (Figure 2Ci-iii; supplemental Figure 2). To determine the intrinsic activity of D49NT51D-perforin, cells expressing identical levels of the D49NT51D and WT-perforin were sorted (supplemental Figure 4). Cytotoxicity of resultant KHYG1 cells was essentially identical, confirming gain of glycosylation as a pathogenic mechanism of D49N (Figure 2Ciii inset).
Overall, these data demonstrate that the D49N substitution has a marked adverse effect on perforin cytotoxicity because of the introduction of a new N-linked glycan, which in turn results in protein misfolding and degradation. The ability of both D49N and K285del to completely abolish perforin cytotoxicity was consistent with the clinical presentation at birth and rapid lethality in the affected patient.
Given the simplicity of the consensus sequence for N-glycosylation (N-x-S/T), we expected to identify a number of other disease-causing PRF1 mutations leading to gain of glycosylation. Indeed, our retrospective analysis revealed 7 such examples (Figure 2Civ). Patients inheriting such mutations in the homozygous state died of FHL at an early age.14,17-20 All of these mutations (including D49N) map near the center of the MACPF domain, immediately adjacent to the evolutionary conserved “bent” β-sheet,16 which is known to undergo marked conformational rearrangement when perforin inserts into the plasma membrane. The severe derangement of perforin structure predicted from this change was consistent with the early onset of FHL. Although less than 1.5% of known disease-causing missense mutations overall give rise to gains of glycosylation,21 this phenomenon appears somewhat more common in FHL2, affecting 4 of the approximately 50 PRF1 missense mutations to date. This difference may represent a statistical anomaly because of a relatively small number of catalogued perforin mutations, but may also reflect the inherent instability of a monomeric perforin, which does not permit a structural challenge as significant as gain of glycosylation.16
Gain of glycosylation mutations are associated with many other human diseases, including dysfibrinogenemia, anti-thrombin deficiency, and primary immunodeficiencies.22 For patients bearing these mutations, pharmaceuticals that inhibit glycosidases (eg, derivatives of castanospermine and NB-DNJ [N-butyl-deoxynojirimycin] or kifunensin)23 have been shown to offer promising therapeutic benefits by inhibiting glycosylation and/or regulating the protein folding environment. Indeed, kifunensin significantly increased the activity of all NK cell lines. In contrast, castanospermine and NB-DNJ were detrimental for KHYG1 effector function (supplemental Figure 5).
Where appropriate, therapeutic approaches based on modifiers of glycosylation may therefore be explored as a temporary therapeutic intervention preceding potentially curative HSCT.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Prof J.-L. Cassanova for providing the modifiers of glycosylation.
This study was supported in part by the Cancer Council Victoria (J.C.) and National Health and Medical Research Council of Australia (A.J.B., J.A.L., J.A.T., and I.V.).
Authorship
Contribution: J.C. designed and carried out the experiments, analyzed the data, and drafted the manuscript; K.T. performed genetic analyses; A.J.B. and J.A.L. helped design and conducted the experiments; M.L. and B.W. conducted the clinical studies; and J.A.T. and I.V. designed the study and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ilia Voskoboinik, Peter MacCallum Cancer Centre, Cancer Immunology Program, St Andrews Place, East Melbourne, VIC 3002, Australia; e-mail: ilia.voskoboinik@petermac.org.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal