Abstract
B-cell chronic lymphocytic leukemia (B-CLL) is characterized by the clonal expansion of CD5-expressing B lymphocytes that produce mAbs often reactive with microbial or autoantigens. Long-term culture of B-CLL clones would permit the collection and characterization of B-CLL mAbs to study antigen specificity and of B-CLL DNA to investigate molecular mechanisms promoting the disease. However, the derivation of long-term cell lines (eg, by EBV), has not been efficient. We have improved the efficiency of EBV B-CLL transformation of CpG oligonucleotide-stimulated cells by incubating patient peripheral blood mononuclear cells in the presence of an irradiated mouse macrophage cell line, J774A.1. Using this approach, peripheral blood mononuclear cells isolated from 13 of 21 B-CLL patients were transformed as documented by IGHV-D-J sequencing. Four clones grew and retained CD5 expression in culture for 2 to 4 months. However, despite documentation of EBV infection by expression of EBNA2 and LMP1, B-CLL cells died after removal of macrophage feeder cells. Nevertheless, using electrofusion technology, we generated 6 stable hetero-hybridoma cell lines from EBV-transformed B-CLL cells, and these hetero-hybridomas produced immunoglobulin. Thus, we have established enhanced methods of B-CLL culture that will enable broader interrogation of B-CLL cells at the genetic and protein levels.
Introduction
B-cell chronic lymphocytic leukemia (B-CLL) is characterized by the clonal expansion of CD5-expressing B lymphocytes in blood, bone marrow, and lymphoid tissues in vivo.1 Patients with B-CLL can be divided into 2 subgroups based on the presence or absence of immunoglobulin (Ig) heavy variable (IGHV) gene mutations.2 Patients with unmutated IGHVs (U-CLL) have worse clinical outcomes than patients with mutated IGHVs (M-CLL).3,4 In addition, the use of specific IGHVs and IGK/LVs differs between B-CLL cells and normal B lymphocytes and between the U-CLL and M-CLL subgroups.2,5,6 For example, IGHV1-69 is most often found in U-CLL cases and IGHV4-34 most often in M-CLL.2 Furthermore, U-CLL clones frequently display stereotyped B-cell antigen receptors (BCRs) with very similar heavy chain complementarity determining region 3 (HCDR3s) because of common IGHV-D-J rearrangements.7-12 Finally, most U-CLL cells and certain M-CLL cells express autoreactive BCRs.13-15 Collectively, these data indicate that the structure and probably the antigen reactivity of the BCRs of B-CLL cells are intimately linked to the development and evolution of the disease.1,16
For this reason, characterization of the antigen specificity of B-CLL clones has become a topic of great interest. In line with the frequent autoreactivity of B-CLL cells, recent studies have defined the products of cell death and molecular catabolism as major targets of these BCRs/mAbs.17-20 These analyses have been carried out using mAbs expressed as recombinant Igs17-20 or collected from the supernatants of B-CLL cells stimulated to differentiate in vitro13,14,17 or from EBV-transformed B-CLL cells.17
Although the use of native Igs secreted by B-CLL cells has certain advantages, the latter approach has been limited by the low EBV transformation efficiency of primary B-CLL cells and the difficulty in producing stable EBV-transformed B-cell lines. The refractoriness of B-CLL cells to transformation by EBV, an oncogenic herpesvirus that transforms normal human B cells efficiently in vitro,21,22 is in part the result of an unusual response to EBV infection, in which infected B-CLL cells do not express EBV latent membrane protein 1 (LMP1), which is required for transformation of B cells.23,24
In this study, we have improved the efficiency of primary B-CLL cell transformation after EBV infection by coculturing patient peripheral blood mononuclear cells (PBMCs) with irradiated mouse feeder cells (J774A.1 cells) in the presence of Toll-like receptor 9 (TLR9) ligands (CpG oligonucleotides). Under these conditions, a majority of B-cell clones derived by EBV transformation were of leukemic origin as documented by IGHV-D-J DNA sequencing. Some of these cells were maintained in culture for up to 4 months, expressed surface membrane CD5, and synthesized EBNA2 and LMP1. When these clones were hybridized by electrofusion with an appropriate partner, stable hetero-hybridoma B-CLL cell lines of defined specificity were generated. This more reproducible and efficient system of EBV-induced growth transformation should help define the antigen reactivities of B-CLL clones as well as providing a replenishable source of B-CLL cells and DNA for genetic analyses.
Methods
Cell lines
J774A.1 (TIB-67) and K6H6/B5 (CRL-1823) cell lines were purchased from ATCC. Culture medium was RPMI 1640 supplemented with 15% FBS, 2mM l-glutamine, 1mM sodium pyruvate, 1% nonessential amino acids, 15mM HEPES, 100 U/mL penicillin G, and 100 μg/mL streptomycin (Invitrogen).
Isolation of CLL PBMC and EBV transformation
After obtaining informed consent in accordance with the Declaration of Helsinki as part of an institutional review board-approved protocol of the Feinstein Institute for Medical Research, North Shore–Long Island Jewish Health System (Manhasset, NY), peripheral blood samples were collected from 66 B-CLL patients (47 U-CLL and 19 M-CLL cases; Tables 1 and 2). PBMCs were isolated by density-gradient centrifugation (Ficoll-Paque; Pharmacia LKB Biotechnology) and cryopreserved with a programmable cell-freezing machine (CryoMed). The IGHV-D-J rearrangements of these cases were amplified and sequenced as described.6
Characteristics of B-CLL PBMCs used to test EBV-transformation conditions
| CLL ID . | Mutation status* . | IGHV mutation, %† . | IGHV . | IGHD . | IGHJ . | HCDR3 length‡ . | IgM+ wells, %§ . | Mean IgM level, μg/mL . |
|---|---|---|---|---|---|---|---|---|
| CLL246 | U | 0.3 | 1-69 | 3-3 | 6 | 21 | 100 | 3.3 |
| CLL487 | M | 4.8 | 2-05 | 1-1 | 4 | 17 | 68.8 | 0.36 |
| CLL493 | U | 0 | 1-69 | 5-5 | 4 | 21 | 96.3 | 0.24 |
| CLL526 | U | 0 | 1-69 | 3-3 | 6 | 23 | 100 | 2.12 |
| CLL698 | U | 0 | 1-69 | 3-3 | 6 | 21 | 100 | 5.81 |
| CLL816 | U | 0 | 1-69 | 3-3 | 6 | 25 | 95.8 | 0.17 |
| CLL821 | U | 0 | 1-69 | 3-3 | 4 | 22 | 100 | 0.29 |
| CLL822 | U | 0 | 2-05 | 6-13 | 6 | 25 | 100 | 0.30 |
| CLL939 | M | 6.6 | 3-66 | 3-10 | 3 | 11 | 50.4 | 0.05 |
| CLL1073 | U | 0 | 1-69 | 1-26 | 6 | 20 | 85.0 | 2.04 |
| CLL1075 | M | 5.9 | 1-69 | 5-12 | 4 | 16 | 100 | 1.63 |
| CLL1080 | M | 2.8 | 3-09 | 3-3 | 4 | 15 | 50.0 | 1.03 |
| CLL1223 | M | 6.2 | 2-05 | 3-9 | 4 | 17 | 85.0 | 2.03 |
| CLL1296 | M | 7.6 | 3-07 | 3-22 | 5 | 13 | 93.8 | 2.31 |
| CLL1324 | U | 0 | 1-69 | 3-10 | 6 | 23 | 100 | 2.53 |
| CLL ID . | Mutation status* . | IGHV mutation, %† . | IGHV . | IGHD . | IGHJ . | HCDR3 length‡ . | IgM+ wells, %§ . | Mean IgM level, μg/mL . |
|---|---|---|---|---|---|---|---|---|
| CLL246 | U | 0.3 | 1-69 | 3-3 | 6 | 21 | 100 | 3.3 |
| CLL487 | M | 4.8 | 2-05 | 1-1 | 4 | 17 | 68.8 | 0.36 |
| CLL493 | U | 0 | 1-69 | 5-5 | 4 | 21 | 96.3 | 0.24 |
| CLL526 | U | 0 | 1-69 | 3-3 | 6 | 23 | 100 | 2.12 |
| CLL698 | U | 0 | 1-69 | 3-3 | 6 | 21 | 100 | 5.81 |
| CLL816 | U | 0 | 1-69 | 3-3 | 6 | 25 | 95.8 | 0.17 |
| CLL821 | U | 0 | 1-69 | 3-3 | 4 | 22 | 100 | 0.29 |
| CLL822 | U | 0 | 2-05 | 6-13 | 6 | 25 | 100 | 0.30 |
| CLL939 | M | 6.6 | 3-66 | 3-10 | 3 | 11 | 50.4 | 0.05 |
| CLL1073 | U | 0 | 1-69 | 1-26 | 6 | 20 | 85.0 | 2.04 |
| CLL1075 | M | 5.9 | 1-69 | 5-12 | 4 | 16 | 100 | 1.63 |
| CLL1080 | M | 2.8 | 3-09 | 3-3 | 4 | 15 | 50.0 | 1.03 |
| CLL1223 | M | 6.2 | 2-05 | 3-9 | 4 | 17 | 85.0 | 2.03 |
| CLL1296 | M | 7.6 | 3-07 | 3-22 | 5 | 13 | 93.8 | 2.31 |
| CLL1324 | U | 0 | 1-69 | 3-10 | 6 | 23 | 100 | 2.53 |
U indicates unmutated type; and M, > 2% mutated compared with germline according to IMGT.27
IGHV mutation (%) was compared with germline according to IMGT.27
HCDR3 length indicates number of amino acid residues in the HCDR3.
To determine frequencies of IgM+ wells, PBMCs from B-CLL patients were plated at 5000 cells per well in the presence of irradiated J774A.1 cells (50 000 cells per well) and ODN2006 (12.5 μg/mL). The culture supernatants were analyzed for IgM production. For comparison, we used EBV-transformed B-cell cultures under the same condition from 20 healthy control PBMCs and found IgM production in 100% of the wells tested with an average IgM level of 6.7 μg/mL.
Overall statistics
| Procedures . | IGHV1-69 (U/M) . | IGHV2 or IGHV3 (U/M) . | All (U/M) . |
|---|---|---|---|
| Total no. of samples stimulated with EBV | 37 (35/2) | 29 (12/17) | 66 (47/19) |
| No. of samples with at least 50% of wells with IgM | 25 (23/2) | 16 (8/8) | 41 (31/10) |
| % samples producing total IgM* | 67.6% | 55.2% | 62.1% |
| No. of EBV-B-cell lines sequenced | 11 (10/1) | 10 (5/5) | 21 (15/6) |
| No. of EBV-B-cell lines with matched Ig sequences† | 9 (9/0) | 4 (3/1) | 13 (12/1) |
| % EBV-B-cell lines from B-CLL* | 81.8% | 40.0% | 61.9% |
| No. of EBV-B-cell lines electrofused | 8 (8/0) | 2 (1/1) | 10 (9/1) |
| No. of hetero-hybridoma cloned and sequenced | 6 (6/0) | 1 (0/1) | 7 (6/1) |
| No. of matched hetero-hybridoma clones | 5 (5/0) | 1 (0/1) | 6 (5/1) |
| Procedures . | IGHV1-69 (U/M) . | IGHV2 or IGHV3 (U/M) . | All (U/M) . |
|---|---|---|---|
| Total no. of samples stimulated with EBV | 37 (35/2) | 29 (12/17) | 66 (47/19) |
| No. of samples with at least 50% of wells with IgM | 25 (23/2) | 16 (8/8) | 41 (31/10) |
| % samples producing total IgM* | 67.6% | 55.2% | 62.1% |
| No. of EBV-B-cell lines sequenced | 11 (10/1) | 10 (5/5) | 21 (15/6) |
| No. of EBV-B-cell lines with matched Ig sequences† | 9 (9/0) | 4 (3/1) | 13 (12/1) |
| % EBV-B-cell lines from B-CLL* | 81.8% | 40.0% | 61.9% |
| No. of EBV-B-cell lines electrofused | 8 (8/0) | 2 (1/1) | 10 (9/1) |
| No. of hetero-hybridoma cloned and sequenced | 6 (6/0) | 1 (0/1) | 7 (6/1) |
| No. of matched hetero-hybridoma clones | 5 (5/0) | 1 (0/1) | 6 (5/1) |
Numbers in parentheses represent unmutated (U) type/mutated (M) type, respectively.
The overall efficiency of B-CLL outgrowth was based on the following equation: % samples producing IgM (62.1%) × % B-cell lines with the matched Ig sequences (61.9%) = 38.4%.
The EBV-transformed B (EBV-B) cells with matched Ig sequences had IGHV sequences that matched those of the previously identified B-CLL IgM sequences from the same patient samples.30
PBMCs thawed at the time of use were incubated with EBV-containing supernatant from the marmoset cell line B95-8 for 4 hours at 37°C in a 5% CO2 incubator. A TLR9 agonist, ODN2006 (12.5 μg/mL; Invivogen), was used to activate B cells and boost transformation efficiency as described by Traggiai et al.25 Cyclosporine A (0.5 μg/mL) was added to suppress EBV-specific cytotoxic T-cell activity. After infection with EBV, PBMCs were resuspended in culture medium containing ODN2006, distributed at 2500 to 10 000 cells per well in 96-well u-bottom plates, and cultured in the presence of feeder cells, J774A.1 (50 000 cells per well) that had been exposed to γ-irradiation (40 Gy) from a Shepherd irradiator.
ELISA
Three weeks after EBV infection, culture supernatant was collected from each well, and levels of total IgM were measured using an IgM-specific ELISA to define IgM-producing B-cell lines. Briefly, ELISA plates (Corning Life Sciences) were coated with 5 μg/mL of goat anti–human Ig (reactive with all isotypes; BioSource International) in 0.1M sodium bicarbonate buffer. After incubating overnight at 4°C, plates were blocked with PBS containing 15% goat serum, 4% whey protein, 0.5% Tween-20, and 0.05% NaN3. Then test supernatants diluted in the blocking buffer were distributed to wells and incubated for 1.5 hours at room temperature. After washing with PBS-0.5% Tween-20, bound human IgM was detected with HRP-conjugated goat anti–human IgM (μ-chain specific; Jackson ImmunoResearch Laboratories) and peroxidase substrate tetramethylbenzidine (Kirkegaard and Perry Laboratories) using a SpectraMax Plus384 plate reader (Molecular Devices). The detection limit of IgM in each well was 60 ng/mL; negative wells with undetectable levels of IgM were assigned 10 ng/mL to permit logarithmic transformation of the data.
Isolation and sequencing of Ig transcripts
Live EBV-transformed B cells from selected wells were sorted as single cells into 96-well PCR plates containing 20 μL/well of a reverse transcription (RT) reaction buffer (Invitrogen) that included 5 μL of 5× first-strand cDNA buffer, 0.5 μL of RNAseOut, 1.25 μL of DTT, 0.0625 μL of Igepal and 13.25 μL of dH2O. Plates were stored at −80°C until use. Cell sorting was performed on a BD FACSAria (BD Biosciences). RNA was also extracted from hetero-hybridoma cell lines in bulk culture using an RNA extraction kit (QIAGEN). The genes encoding IGHV and IGKV/IGLV chains were amplified by RT and nested PCR using a previously reported method.26 All PCR products were purified using PCR purification kit (QIAGEN) and sequenced in forward and reverse directions using an ABI3700 instrument and BigDye sequencing kit (Applied Biosystems). Sequences were analyzed using the ImMunoGeneTics information system (http://imgt.org/) to identify immunoglobulin variable region gene segments and somatic mutations.27
Quantitative real-time RT-PCR of EBNA2 and LMP1 mRNAs.
RNA was extracted from PBMCs or EBV-infected PBMCs using QIAGEN RNeasy. RT by random cDNA priming was performed according to the manufacturer's protocol (Applied Biosystems High Capacity cDNA RT kit). Real-time PCR was performed either using Quanta SYBR Green (LMP1) or specific TaqMan-based probes (EBNA2) in an Applied Biosystems Step One Plus instrument. LMP1 mRNAs were detected using primers within exons 2 and 3, and these reactions were normalized with the SETDB1 gene, which we have found to remain consistent from PBMC through EBV transformation. Forward primer for LMP1 (exon 2) was: 5′-AATTTGCACGGACAGGCATT-3′, whereas the reverse primer was LMP1 (exon 3): 5′-AAGGCCAAAAGCTGCCAGAT-3′. Forward primer for SETDB1 was: 5′-TCCATGGCATGCTGGAGCGG-3′ and reverse SETDB1 was: 5′-CAGAGGGTTCTTGCCCCGGT-3′. EBNA2 mRNAs were detected by TaqMan using a forward primer spanning the Y2/YH exon junction (5′-GCTTAGCCAGTAACCCAGCACT-3′) and a reverse primer within YH (5′-TGCTTAGAAGGTTGTTGGCATG) with a probe contained within the YH exon (5′-CCCAACCACAGGTTCAGGCAAAACTTT-3′) that was labeled with the 6-carboxyfluorescein phosphoramidite (FAM) reporter dye at the 5′-end and 6-carboxytetramethylrhodamine (TAMRA) at the 3′ end. These transcripts were also normalized to SETDB1.
Western blotting
Cells were lysed in a 1% Triton X-100 containing buffer (20mM Tris, pH 7.5, 100mM NaCl, 10% glycerol, 1% Triton X-100, 1mM EDTA, 1mM DTT, 20mM NaF, 10mM sodium pyrophosphate, and complete protease inhibitors without EDTA) and normalized to total protein content by a Bradford assay (Bio-Rad). All samples were run on Novex 4%-12% Bis-Tris gels and blotted using standard procedures. LMP1 was detected using the S12 antibody (1:10 dilution of hybridoma supernatant, kind gift of E. Kieff, Harvard Medical School, Boston, MA).
Electrofusion
K6H6/B5 myeloma partner cells and EBV-transformed B cells were washed twice with an electrofusion medium (Cyto Pulse Sciences) before fusion. A 1:2 B cell to myeloma cell ratio was used in fusion. Electrofusion was achieved using a PA-4000/PA-101 apparatus with FE-20/800 electrode fusion chamber (Cyto Pulse Sciences). Instrument settings were used according to the methods described previously with minor modifications.28 Pre-fusion dielectrophoresis was performed with an alternating current voltage of 75 V at 0.8 MHz for 15 seconds. Cells were fused with a single square-wave direct current voltage of 300 V for 0.04 ms. Postfusion dielectrophoresis was performed with an alternating current voltage of 20 V at 0.2 MHz for 30 seconds. After fusion, cells were harvested and distributed into 96-well flat-bottom plates at 4000 B cells per well and incubated in culture medium supplemented with 100μM hypoxanthine, 0.4μM aminopterin, 16μM thymidine, and 0.5μM ouabain.
Results
Use of J774A.1 as feeder cells
To establish optimal culture conditions for EBV transformation of B-CLL cells, we determined IgM production from infected cells cultured in the presence and absence of irradiated mouse macrophage feeder cell line, J774A.1. A type B CpG oligonucleotide, ODN2006, was used in all cultures.25 Growth transformation was judged to have occurred when B cells grew for 2 to 3 weeks in culture with 2 distinct morphologic features of EBV-transformed B cells, enlarged cell size, and clump formation of proliferating lymphoblast cells with Ig production.25,29 Thereafter, we measured IgM concentrations in culture supernatants as a proxy for cell growth and function.
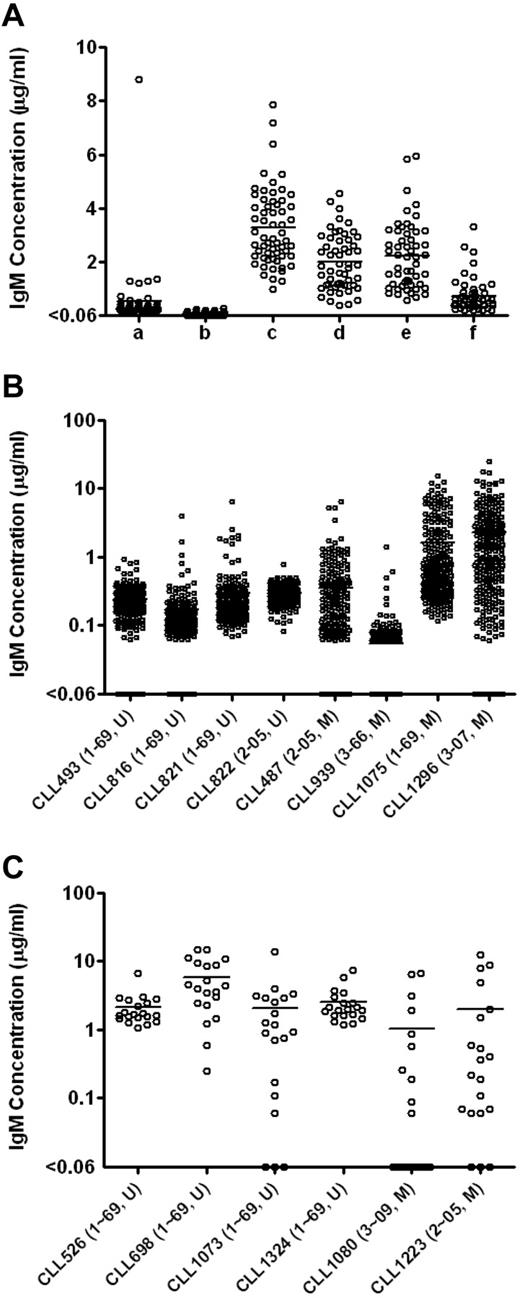
The use of J774A.1 feeder cells significantly increased levels of IgM detected (Figure 1A). For example, 5000 PBMCs stimulated by ODN2006 and cultured with 50 000 J774A.1 cells produced a mean of 3.3 μg/mL IgM, whereas culturing the same number of PBMCs with ODN2006 in the absence of feeder cells produced minimal levels of IgM (mean = 0.1 μg/mL, P < .0001, Student t test).
Total IgM levels in the EBV-transformed B-cell cultures enhanced by feeder cells. (A) Total IgM levels in the EBV-transformed B-cell cultures derived from CLL246 PBMCs under different culture conditions. After EBV infection, PBMCs were plated as follows: lane a indicates 10 000 PBMCs per well without feeder cells; lane b, 5000 PBMCs per well without feeder cells; lane c, 5000 PBMCs per well with 50 000 cells per well of J774A.1; lane d, 2500 PBMCs per well with 50 000 cells per well of J774A.1; lane e, 2500 PBMCs per well with 25 000 cells per well of J774A.1; and lane f, 2500 PBMCs per well with 12 500 cells per well of J774A.1. The J774A.1 feeder cells were γ-irradiated before use. Levels of IgM in the culture supernatants were measured by ELISA. Each data point represents the level of IgM in each well (54-60 wells per culture condition). U indicates unmutated type; and M, > 2% mutated compared with germline according to ImMunoGeneTics.27 Total IgM levels in the EBV-transformed B-cell cultures derived from additional B-CLL samples were measured in 240 wells per sample (B) or in 20 wells per sample (C). After EBV infection, 5000 PBMCs were incubated in the presence of 50 000 cells per well of irradiated J774A.1 cells. Each data point represents the level of IgM in each well. To determine the minimal number of PBMCs necessary for B-CLL cell activation as defined by IgM production, total of 100 000 PBMCs were plated in total of 20 wells (5000 PBMCs per well) in panel C.
Total IgM levels in the EBV-transformed B-cell cultures enhanced by feeder cells. (A) Total IgM levels in the EBV-transformed B-cell cultures derived from CLL246 PBMCs under different culture conditions. After EBV infection, PBMCs were plated as follows: lane a indicates 10 000 PBMCs per well without feeder cells; lane b, 5000 PBMCs per well without feeder cells; lane c, 5000 PBMCs per well with 50 000 cells per well of J774A.1; lane d, 2500 PBMCs per well with 50 000 cells per well of J774A.1; lane e, 2500 PBMCs per well with 25 000 cells per well of J774A.1; and lane f, 2500 PBMCs per well with 12 500 cells per well of J774A.1. The J774A.1 feeder cells were γ-irradiated before use. Levels of IgM in the culture supernatants were measured by ELISA. Each data point represents the level of IgM in each well (54-60 wells per culture condition). U indicates unmutated type; and M, > 2% mutated compared with germline according to ImMunoGeneTics.27 Total IgM levels in the EBV-transformed B-cell cultures derived from additional B-CLL samples were measured in 240 wells per sample (B) or in 20 wells per sample (C). After EBV infection, 5000 PBMCs were incubated in the presence of 50 000 cells per well of irradiated J774A.1 cells. Each data point represents the level of IgM in each well. To determine the minimal number of PBMCs necessary for B-CLL cell activation as defined by IgM production, total of 100 000 PBMCs were plated in total of 20 wells (5000 PBMCs per well) in panel C.
To determine the optimal number of irradiated J774A.1 cells needed for B-CLL transformation, we cocultured EBV-stimulated PBMCs (2500 cells per well) from CLL246 (U-CLL case expressing IGHV1-69) in the presence of 12 500 to 50 000 irradiated J774A.1 cells per well. EBV-stimulated PBMCs cocultured with 25 000 and 50 000 J774A.1 cells produced similar levels of IgM (averages of 2.2 and 2.0 μg/mL, respectively) that were significantly higher than that produced by PBMCs cocultured with 12 500 J774A.1 cells (average of 0.7 μg/mL; P < .0001, Bonferroni multiple comparison test).
Production of IgM by PBMCs from a large cohort of B-CLL cases
We next used 5000 PBMCs per well in the presence of 50 000 J774A.1 cells and 12.5 μg/mL ODN2006 to test EBV transformation efficiency of PBMCs from 14 additional B-CLL patients. Because of our interest in B cells using IGHVs that can be used in antiviral antibodies (K.-K.H., D.M.K., X.C. et al, unpublished data, July 2011), 15 patients (9 U-CLL cases, 8 expressing IGHV1-69 and 1 IGHV2-05; and 6 M-CLL cases, 1 using IGHV1-69, 2 IGHV2-05, 1 IGHV3-07, 1 IGHV3-09, and 1 IGHV3-66 gene segments) were used. As shown in Figure 1B-C in “Use of J774A.1 as feeder cells,” PBMCs from all patients produced detectable levels of IgM in more than or equal to 50% of the wells tested (Table 1), indicating that CpG-activated B cells from most B-CLL patients, regardless of their IGHV gene mutation status, were induced to secrete IgM by EBV in the presence of J774A.1 cells.
Finally, using these conditions, we infected 100 000 PBMCs from each of 51 additional B-CLL patients with EBV and plated 5000 cells each into 20 wells to generate leukemic B-cell transformants. Combined with the 15 samples described in the preceding paragraph, a total of 66 PBMC samples (47 U-CLL and 19 M-CLL cases) were infected with EBV. Of these, 41 samples (62.1%; 31 U-CLL and 10 M-CLL cases) secreted IgM in more than or equal to 50% of the wells tested, indicating efficient outgrowth of infected cells (data summarized in Table 2). When the 41 samples were divided into subgroups based on IGHV gene segment use, 25 IGHV1-69 (67.6%), 4 IGHV2 (36.4%), and 12 IGHV3 (66.7%) samples were transformed into IgM-producing B cells in more than or equal to 50% of the wells tested. Higher transformation frequencies were obtained from cases expressing IGHV3-11 (3 of 3, 3 U-CLL), IGHV3-30 (2 of 2, 1 U-CLL and 1 M-CLL), IGHV3-09 (2 of 3, 1 U-CLL and 1 M-CLL), and IGHV2-05 (4 of 7, 1 U-CLL and 3 M-CLL).
Molecular confirmation of B-CLL cell transformation
To determine whether B-CLL cells, rather than contaminating normal B cells, were transformed in our cultures after 5 to 6 weeks of continuous culture, we collected B cells from IgM-producing wells of 21 EBV-stimulated patient PBMC samples, sorted them as single cells into 96-well PCR plates, and performed IGV sequencing; these findings were then compared with the IGV sequences previously identified as originating from B-CLL cells of the same subjects.30 In 13 of 21 patient samples (61.9%), we identified the correct B-CLL IGHV sequences (Table 3).
Matched Ig sequences in single-cell sorted EBV-infected cell lines
| CLL ID . | IGHV . | IGHV mutation, % . | No. of cell lines sorted* . | Total no. of sequences . | No. of matched IGHV genes† . | Matched IGHV genes, % . | CD5+ CD19+ cells in PBMCs, % . |
|---|---|---|---|---|---|---|---|
| CLL246‡ | 1-69 | 0.3 | 3 | 32 | 32 | 100 | ND |
| CLL493‡ | 1-69 | 0 | 1 | 7 | 7 | 100 | 99.8 |
| CLL526‡ | 1-69 | 0 | 2 | 5 | 4 | 80 | ND |
| CLL675 | 3-23 | 0 | 1 | 1 | 0 | 0 | 99.8 |
| CLL698 | 1-69 | 0 | 5 | 17 | 2 | 11.8 | ND |
| CLL701 | 1-69 | 0 | 2 | 17 | 8 | 47.1 | 75.7 |
| CLL753 | 3-74 | 0 | 2 | 12 | 7 | 58.3 | 95.5 |
| CLL815‡ | 1-69 | 0 | 1 | 9 | 9 | 100 | 99.5 |
| CLL861‡ | 1-69 | 0 | 1 | 13 | 13 | 100 | 99.6 |
| CLL1073 | 1-69 | 0 | 2 | 14 | 0 | 0 | 94.7 |
| CLL1121‡ | 3-11 | 0 | 3 | 18 | 17 | 94.4 | 99.5 |
| CLL1153‡ | 1-69 | 0 | 2 | 11 | 10 | 90.9 | 80.7 |
| CLL1191 | 3-23 | 0 | 1 | 16 | 1 | 6.3 | 99.9 |
| CLL1288 | 3-11 | 0 | 1 | 10 | 0 | 0 | ND |
| CLL1324‡ | 1-69 | 0 | 4 | 18 | 16 | 88.9 | 98.7 |
| CLL1011 | 1-69 | 3.8 | 1 | 2 | 0 | 0 | 99.4 |
| CLL1080 | 3-09 | 2.8 | 1 | 17 | 0 | 0 | 99.4 |
| CLL1193‡ | 3-13 | 6.3 | 1 | 9 | 9 | 100 | ND |
| CLL1200 | 2-05 | 3.8 | 2 | 25 | 0 | 0 | 96.6 |
| CLL1223 | 2-05 | 6.2 | 1 | 9 | 0 | 0 | 97.1 |
| CLL1296 | 3-07 | 7.6 | 1 | 3 | 0 | 0 | 99.6 |
| CLL ID . | IGHV . | IGHV mutation, % . | No. of cell lines sorted* . | Total no. of sequences . | No. of matched IGHV genes† . | Matched IGHV genes, % . | CD5+ CD19+ cells in PBMCs, % . |
|---|---|---|---|---|---|---|---|
| CLL246‡ | 1-69 | 0.3 | 3 | 32 | 32 | 100 | ND |
| CLL493‡ | 1-69 | 0 | 1 | 7 | 7 | 100 | 99.8 |
| CLL526‡ | 1-69 | 0 | 2 | 5 | 4 | 80 | ND |
| CLL675 | 3-23 | 0 | 1 | 1 | 0 | 0 | 99.8 |
| CLL698 | 1-69 | 0 | 5 | 17 | 2 | 11.8 | ND |
| CLL701 | 1-69 | 0 | 2 | 17 | 8 | 47.1 | 75.7 |
| CLL753 | 3-74 | 0 | 2 | 12 | 7 | 58.3 | 95.5 |
| CLL815‡ | 1-69 | 0 | 1 | 9 | 9 | 100 | 99.5 |
| CLL861‡ | 1-69 | 0 | 1 | 13 | 13 | 100 | 99.6 |
| CLL1073 | 1-69 | 0 | 2 | 14 | 0 | 0 | 94.7 |
| CLL1121‡ | 3-11 | 0 | 3 | 18 | 17 | 94.4 | 99.5 |
| CLL1153‡ | 1-69 | 0 | 2 | 11 | 10 | 90.9 | 80.7 |
| CLL1191 | 3-23 | 0 | 1 | 16 | 1 | 6.3 | 99.9 |
| CLL1288 | 3-11 | 0 | 1 | 10 | 0 | 0 | ND |
| CLL1324‡ | 1-69 | 0 | 4 | 18 | 16 | 88.9 | 98.7 |
| CLL1011 | 1-69 | 3.8 | 1 | 2 | 0 | 0 | 99.4 |
| CLL1080 | 3-09 | 2.8 | 1 | 17 | 0 | 0 | 99.4 |
| CLL1193‡ | 3-13 | 6.3 | 1 | 9 | 9 | 100 | ND |
| CLL1200 | 2-05 | 3.8 | 2 | 25 | 0 | 0 | 96.6 |
| CLL1223 | 2-05 | 6.2 | 1 | 9 | 0 | 0 | 97.1 |
| CLL1296 | 3-07 | 7.6 | 1 | 3 | 0 | 0 | 99.6 |
ND indicates not determined.
After 5-6 weeks of continuous culture, selected IgM-producing wells from 21 EBV-infected B-CLL samples were sorted as single cells for Ig sequencing.
The cells with matched Ig sequences had IGHV sequences that matched those of the previously identified B-CLL IgM sequences from the same patient samples.30
Each PBMC sample produced at least 1 cell line with all matched IGHV sequences.
When we divided the 21 B-CLL samples into subgroups based on expressed IGHVs, 9 of 11 IGHV1-69 (81.8%), none of 2 IGHV2, and 4 of 8 IGHV3 (50.0%; IGHV3-11, 3-13, 3-23, and 3-74) samples had been transformed by EBV. Of note, 12 of 15 U-CLL cases (80.0%) produced B-cell lines with matching IGHV sequences, in contrast to only 1 of 6 M-CLL cases (16.7%; Table 3). Of these 13 transformed B-CLL cell lines that matched the leukemic Ig sequence, 9 were clonal as the single-sorted B cells had only matching IGHV sequences (Table 4), whereas the remainder contained low frequencies of cells with non–B-CLL IGV sequences. Transformation frequency was not associated with percentage of CD5+ CD19+ cells in PBMC samples used (Table 3). These data suggest that, under the conditions used, B-CLL with IGHV1-69 or other clones expressing unmutated IGHVs are more easily transformed by EBV than M-CLL cells.
Summary of Ig sequence data from 9 EBV-transformed B-CLL cell lines
| IGHV . | IGKV/IGLV* . | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CLL ID . | Cell line . | Source . | Family . | Mutation, % . | CDR1 . | CDR2 . | CDR3 . | Family . | Mutation, % . | CDR1 . | CDR2 . | CDR3 . |
| CLL246 | B-CLL | PBMCs | 1-69 | 0.3 | GGTFSSYA | IIPILGIA | ARSDQNYDFWSGYFRYYGMDV | ND | ||||
| CLL246-H3 | EBV-B cells | 1-69 | 0.7 | GGTFSSYA | IIPILGIA | ARSDQNYNFWSGYFRYYGMDV | κ3-20 | 0.0 | QSVSSSY | GAS | QQYGSSPET | |
| CLL493 | B-CLL | PBMCs | 1-69 | 0.0 | GGTFSSYA | IIPIFGTA | ARDGTNLRGWIQLWPGAYFDY | κ1-33 | 0.0 | QDISNY | DAS | QQYDNLPYT |
| CLL493-1 | EBV-B cells | 1-69 | 0.0 | GGTFSSYA | IIPIFGTA | ARDGTNLRGWIQLWPGAYFDY | κ1-33 | 0.0 | QDISNY | DAS | QQYDNLPYT | |
| CLL526 | B-CLL | PBMCs | 1-69 | 0.0 | GGTFSSYA | IIPIFGTA | ARVGYYDFWSGYYPNYYYYGMDV | λ3-9 | 0.0 | NIGSKN | RDS | QVWDSSTV |
| CLL526-G10 | EBV-B cells | 1-69 | 0.0 | GGTFSSYA | IIPIFGTA | ARVGYYDFWSGYYPNYYYYGMDV | λ3-9 | 0.0 | NIGSKN | RDS | QVWDSSTV | |
| CLL815 | B-CLL | PBMCs | 1-69 | 0.0 | GGTFSSYA | IIPIFGTA | ARGYCSSTSCYLWAIGVWYFDL | ND | ||||
| CLL815-C6 | EBV-B cells | 1-69 | 0.0 | GGTFSSYA | IIPIFGTA | ARGYCSSTSCYLWAIGVWYFDL | κ3-11 | 0.0 | QSVSSY | DAS | QQRSNWPYT | |
| CLL861 | B-CLL | PBMCs | 1-69 | 0.0 | GGTFSSYA | IIPIFGTA | ARGGEYDYIWGSYRPNDAFDI | κ3-20 | 0.0 | QSVSSSY | GAS | QQYGSSPGT |
| CLL861-D2 | EBV-B cells | 1-69 | 0.0 | GGTFSSYA | IIPIFGTA | ARGGEYDYIWGSYRPNDAFDI | κ3-20 | 0.0 | QSVSSSY | GAS | QQYGSSPGT | |
| CLL1153 | B-CLL | PBMCs | 1-69 | 0.0 | GGTFSSYA | IIPIFGTA | ARDDSYYDFWSGWYY | κ3-15 | 0.0 | QSVSSN | GAS | QQYNNWPLYT |
| CLL1153-E5 | EBV-B cells | 1-69 | 0.0 | GGTFSSYA | IIPIFGTA | ARDDSYYDFWSGWYY | κ3-15 | 0.0 | QSVSSN | GAS | QQYNNWPLYT | |
| CLL1324 | B-CLL | PBMCs | 1-69 | 0.0 | GGTFSSYA | IIPIFGTA | AREASYGSGSYYQQYYYYYYMDV | λ3-19 | 0.0 | SLRSYY | GKN | NSRDSSGNPYV |
| CLL1324-C9 | EBV-B cells | 1-69 | 0.0 | GGTFSSYA | IIPIFGTA | AREASYGSGSYYQQYYYYYYMDV | λ3-19 | 0.0 | SLRSYY | GKN | NSRDSSGNPYV | |
| CLL1121 | B-CLL | PBMCs | 3-11 | 0.0 | GFTFSDYY | ISSSGSTI | ARDRRGYDFWSGYLGPTPQGDY | ND | ||||
| CLL1121-E11 | EBV-B cells | 3-11 | 0.0 | GFTFSDYY | ISSSGSTI | ARDRRGYDFWSGYLGPTPQGDY | κ2-30 | 0.0 | QSLVYSDGNTY | KVS | MQGTHWPPL | |
| CLL1193 | B-CLL | PBMCs | 3-13 | 6.3 | GFTFSNYD | IGTAGDT | VRGPRGYCSGDGCFSDSFDY | ND | ||||
| CLL1193-C9 | EBV-B cells | 3-13 | 6.3 | GFTFSNYD | IGTAGDT | VRGPRGYCSGDGCFSDSFDY | κ1-16 | 3.7 | QGINHH | AAS | QQYNNYPLT | |
| IGHV . | IGKV/IGLV* . | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CLL ID . | Cell line . | Source . | Family . | Mutation, % . | CDR1 . | CDR2 . | CDR3 . | Family . | Mutation, % . | CDR1 . | CDR2 . | CDR3 . |
| CLL246 | B-CLL | PBMCs | 1-69 | 0.3 | GGTFSSYA | IIPILGIA | ARSDQNYDFWSGYFRYYGMDV | ND | ||||
| CLL246-H3 | EBV-B cells | 1-69 | 0.7 | GGTFSSYA | IIPILGIA | ARSDQNYNFWSGYFRYYGMDV | κ3-20 | 0.0 | QSVSSSY | GAS | QQYGSSPET | |
| CLL493 | B-CLL | PBMCs | 1-69 | 0.0 | GGTFSSYA | IIPIFGTA | ARDGTNLRGWIQLWPGAYFDY | κ1-33 | 0.0 | QDISNY | DAS | QQYDNLPYT |
| CLL493-1 | EBV-B cells | 1-69 | 0.0 | GGTFSSYA | IIPIFGTA | ARDGTNLRGWIQLWPGAYFDY | κ1-33 | 0.0 | QDISNY | DAS | QQYDNLPYT | |
| CLL526 | B-CLL | PBMCs | 1-69 | 0.0 | GGTFSSYA | IIPIFGTA | ARVGYYDFWSGYYPNYYYYGMDV | λ3-9 | 0.0 | NIGSKN | RDS | QVWDSSTV |
| CLL526-G10 | EBV-B cells | 1-69 | 0.0 | GGTFSSYA | IIPIFGTA | ARVGYYDFWSGYYPNYYYYGMDV | λ3-9 | 0.0 | NIGSKN | RDS | QVWDSSTV | |
| CLL815 | B-CLL | PBMCs | 1-69 | 0.0 | GGTFSSYA | IIPIFGTA | ARGYCSSTSCYLWAIGVWYFDL | ND | ||||
| CLL815-C6 | EBV-B cells | 1-69 | 0.0 | GGTFSSYA | IIPIFGTA | ARGYCSSTSCYLWAIGVWYFDL | κ3-11 | 0.0 | QSVSSY | DAS | QQRSNWPYT | |
| CLL861 | B-CLL | PBMCs | 1-69 | 0.0 | GGTFSSYA | IIPIFGTA | ARGGEYDYIWGSYRPNDAFDI | κ3-20 | 0.0 | QSVSSSY | GAS | QQYGSSPGT |
| CLL861-D2 | EBV-B cells | 1-69 | 0.0 | GGTFSSYA | IIPIFGTA | ARGGEYDYIWGSYRPNDAFDI | κ3-20 | 0.0 | QSVSSSY | GAS | QQYGSSPGT | |
| CLL1153 | B-CLL | PBMCs | 1-69 | 0.0 | GGTFSSYA | IIPIFGTA | ARDDSYYDFWSGWYY | κ3-15 | 0.0 | QSVSSN | GAS | QQYNNWPLYT |
| CLL1153-E5 | EBV-B cells | 1-69 | 0.0 | GGTFSSYA | IIPIFGTA | ARDDSYYDFWSGWYY | κ3-15 | 0.0 | QSVSSN | GAS | QQYNNWPLYT | |
| CLL1324 | B-CLL | PBMCs | 1-69 | 0.0 | GGTFSSYA | IIPIFGTA | AREASYGSGSYYQQYYYYYYMDV | λ3-19 | 0.0 | SLRSYY | GKN | NSRDSSGNPYV |
| CLL1324-C9 | EBV-B cells | 1-69 | 0.0 | GGTFSSYA | IIPIFGTA | AREASYGSGSYYQQYYYYYYMDV | λ3-19 | 0.0 | SLRSYY | GKN | NSRDSSGNPYV | |
| CLL1121 | B-CLL | PBMCs | 3-11 | 0.0 | GFTFSDYY | ISSSGSTI | ARDRRGYDFWSGYLGPTPQGDY | ND | ||||
| CLL1121-E11 | EBV-B cells | 3-11 | 0.0 | GFTFSDYY | ISSSGSTI | ARDRRGYDFWSGYLGPTPQGDY | κ2-30 | 0.0 | QSLVYSDGNTY | KVS | MQGTHWPPL | |
| CLL1193 | B-CLL | PBMCs | 3-13 | 6.3 | GFTFSNYD | IGTAGDT | VRGPRGYCSGDGCFSDSFDY | ND | ||||
| CLL1193-C9 | EBV-B cells | 3-13 | 6.3 | GFTFSNYD | IGTAGDT | VRGPRGYCSGDGCFSDSFDY | κ1-16 | 3.7 | QGINHH | AAS | QQYNNYPLT | |
The B-CLL IGHV and IGKV/IGLV sequences were obtained from PBMCs as described previously.30 The IGHV and IGKV/IGLV sequences were obtained from sorted B cells of 9 cultured EBV-transformed B (EBV-B) cell lines. The IGHV and IGKV/IGLV mutations (%) were compared with germline according to IMGT.27
ND indicates not determined.
For clarity, only the dominant IGKV/IGLV sequences are shown for each cell line.
Characterization of EBV-transformed B-CLL cell lines
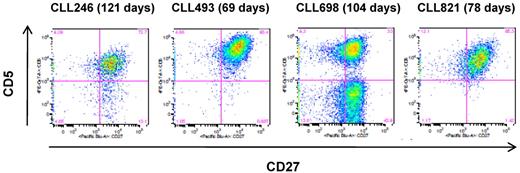
To characterize the longevity of the EBV-transformed B-CLL cells, 10 cell lines that contained the leukemic Ig sequences (9 IGHV1-69 and 1 IGHV 3-11; all U-CLL) were selected and expanded for 2 to 4 months (range, 69-130 days). Of the 10 cultures, 6 retained viable cells in the presence of irradiated J774A.1 feeder cells, and 4 of these (CLL246, CLL493, CLL698, and CLL821; all IGHV1-69, U-CLL) proliferated and produced IgM at low to moderate rates in the presence of the feeder cells (data not shown). These cell lines also showed surface expression of CD5 (42.2%-97.4% of cell population; Figure 2) and exhibited the leukemic IGV sequences. CLL698 also showed a subset of CD5-negative cells (Figure 2C) and a normal B cell Ig sequence. In contrast, the remaining 2 cell lines (CLL701, IGHV1-69; and CLL1121, IGHV3-11) did not express CD5 and were normal B cells (data not shown).
Surface expression of CD5 on the EBV-transformed B-CLL cells. The cells were cultured in the presence of irradiated J774A.1 cells for the indicated period of time. The flow cytometric analysis was performed on the CD19+ cell population. The number in parentheses indicates the total number of days in culture in the presence of irradiated J774A.1 cells.
Surface expression of CD5 on the EBV-transformed B-CLL cells. The cells were cultured in the presence of irradiated J774A.1 cells for the indicated period of time. The flow cytometric analysis was performed on the CD19+ cell population. The number in parentheses indicates the total number of days in culture in the presence of irradiated J774A.1 cells.
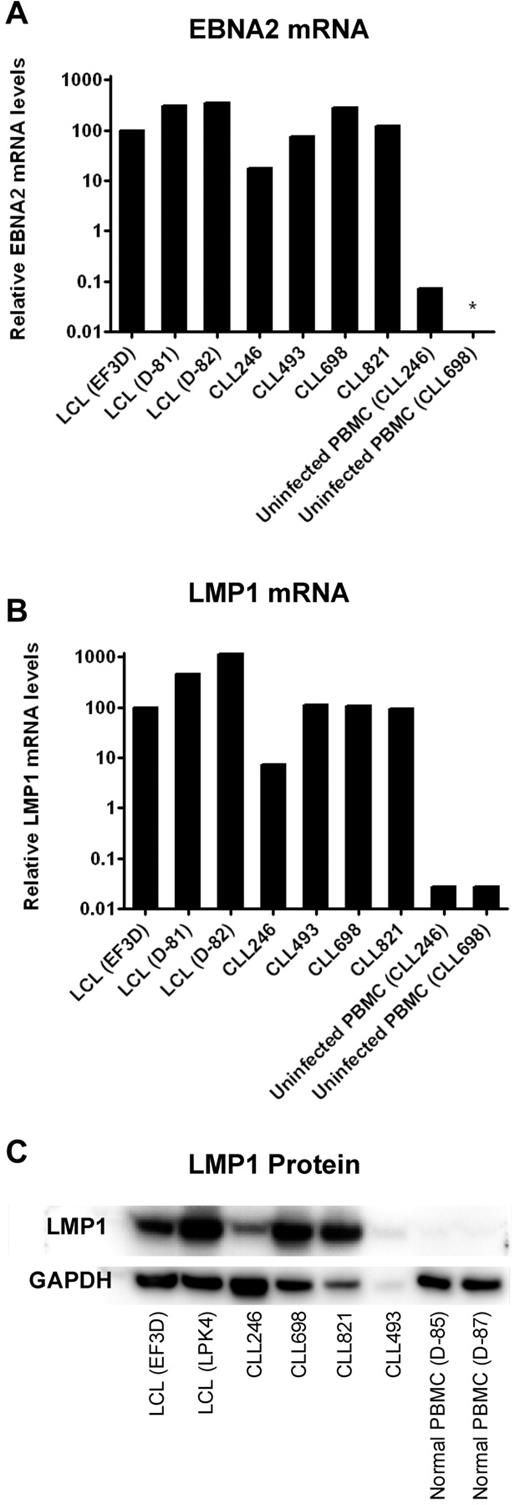
LMP1 is required for successful immortalization of B cells and B-CLL cells as well.23,24 Therefore, we determined the levels of this and another EBV-encoded gene product, EBNA2, in the 4 cell lines that survived prolonged in vitro culture. CLL246-, CLL493-, CLL698-, and CLL821-derived cell lines expressed EBNA2 and LMP1 mRNA at levels comparable with that of the positive controls, EF3D, D-81, and D-82, lymphoblastoid cell lines (LCLs) established from EBV infection and outgrowth of PBMCs from a normal donor (Figure 3). In comparison, uninfected PBMCs from patients CLL246 and CLL698 expressed undetectable or more than 100-fold lower levels of LMP1 or EBNA2. Moreover, the 4 cell lines expressed LMP1 protein at levels consistent with observed mRNA levels. For example, CLL246 expressed less LMP1 mRNA and protein than CLL698 and CLL821. CLL493 also expressed LMP1 protein; however, the low level of expression might be the result of the lower amount of protein available for extraction from this sample (1.5 μg/mL vs 5.0 μg/mL for the other samples).
Expression of EBNA2 and LMP1 in EBV-transformed B-CLL cells after prolonged culture in vitro. B-CLL cells were cultured in the presence of irradiated J774A.1 cells for the period of time described in Figure 2. Four lymphoblastoid cell lines (LCLs EF3D, D-81, D-82, and LPK4) were generated by EBV infection of PBMCs from normal donors and were used as positive controls. Uninfected PBMCs from 2 B-CLL patients (CLL246 and CLL698) or 2 normal donors served as negative controls. For comparisons of mRNA levels of EBNA2 (A) and LMP1 (B), the cycle threshold (Ct) values of viral RNA (LMP1 or EBNA2) were subtracted by the Ct values of SETDB1 RNA. The 2^ − (viral RNA Ct − SETDB1 Ct) values were then normalized such that EF3D (an LCL) was 100% (because there were no detectable copies of EBNA2 or LMP1 in uninfected PBMCs). *Not detected. (C) For Western blots of LMP1, 5 μg each of the proteins extracted from cell lysates was loaded in each lane except for CLL493 (1.5 μg because of limited availability).
Expression of EBNA2 and LMP1 in EBV-transformed B-CLL cells after prolonged culture in vitro. B-CLL cells were cultured in the presence of irradiated J774A.1 cells for the period of time described in Figure 2. Four lymphoblastoid cell lines (LCLs EF3D, D-81, D-82, and LPK4) were generated by EBV infection of PBMCs from normal donors and were used as positive controls. Uninfected PBMCs from 2 B-CLL patients (CLL246 and CLL698) or 2 normal donors served as negative controls. For comparisons of mRNA levels of EBNA2 (A) and LMP1 (B), the cycle threshold (Ct) values of viral RNA (LMP1 or EBNA2) were subtracted by the Ct values of SETDB1 RNA. The 2^ − (viral RNA Ct − SETDB1 Ct) values were then normalized such that EF3D (an LCL) was 100% (because there were no detectable copies of EBNA2 or LMP1 in uninfected PBMCs). *Not detected. (C) For Western blots of LMP1, 5 μg each of the proteins extracted from cell lysates was loaded in each lane except for CLL493 (1.5 μg because of limited availability).
Combined, these data indicate that relatively long-term cultures of EBV-transformed B-CLL cell lines can be established in the presence of J774A.1 feeder cells. However, none of these cell lines was immortalized (ie, grew continuously in the absence of feeder cells).
Electrofusion and generation of B-CLL hetero-hybridoma cell lines
Finally, because the EBV-transformed B-CLL cells required the presence of J774A.1 feeder cells, we sought to generate B-CLL hetero-hybridoma cell lines by electrofusion. We selected 10 EBV-transformed B-cell cultures that contained IGHV-D-J sequences (in either single cell sorted or bulk cultures) matching those of the B-CLL cells from the same patient. These cultures were generated from 9 U-CLL cases (8 IGHV1-69 and 1 IGHV3-11) and one M-CLL case (IGHV3-07) and were expanded to 0.5 to 1 million cells independent of J774 feeder cells. Cells were then electrofused with the parental HGPRTase-negative K6H6/B5 cell line, creating 7 hetero-hybridoma cell lines. Ig DNA sequencing demonstrated that 6 cell lines (5 IGHV1-69 and 1 IGHV3-07) had IGHV sequences that matched those of the known B-CLL IgM sequences from the same patient samples (Table 5),30 whereas the remaining hetero-hybridoma cell line (IGHV1-69) did not have the matching B-CLL IGHV sequence. These B-CLL cell lines produced IgM mAbs at concentrations ranging from 5 to 150 μg/mL. Thus, stable hetero-hybridomas were generated from 60% of B-CLL EBV-transformed B-cell cultures.
Summary of Ig sequence data from 6 B-CLL hetero-hybridoma cell lines
| IGHV . | IGKV/IGLV . | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CLL ID . | Cell line . | Source . | Family . | Mutation, % . | CDR1 . | CDR2 . | CDR3 . | Family . | Mutation, % . | CDR1 . | CDR2 . | CDR3 . |
| CLL246 | B-CLL | PBMCs | 1-69 | 0.3 | GGTFSSYA | IIPILGIA | ARSDQNYDFWSGYFRYYGMDV | ND | ||||
| CLL246-M1 | Hybridoma | 1-69 | 0.7 | GGTFSSYA | IIPILGIA | ARSDQNYDFWSGYFRYYGMDV | κ3-20 | 0.0 | QSVSSSY | GAS | QQYGSSPET | |
| CLL526 | B-CLL | PBMCs | 1-69 | 0.0 | GGTFSSYA | IIPIFGTA | ARVGYYDFWSGYYPNYYYYGMDV | λ3-9 | 0.0 | NIGSKN | RDS | QVWDSSTV |
| CLL526-H10 | Hybridoma | 1-69 | 0.0 | GGTFSSYA | IIPIFGTA | ARVGYYDFWSGYYPNYYYYGMDV | λ3-9 | 0.0 | NIGSKN | RDS | QVWDSSTV | |
| CLL698 | B-CLL | PBMCs | 1-69 | 0.0 | GGTFSSYA | IIPIFGTA | ARGDLPYYDFWSGYHYYGMDV | κ1-5 | 0.4 | QSISSW | KAS | QQYNSYST |
| CLL698-A11 | Hybridoma | 1-69 | 0.3 | GGTFSSYA | IIPIFGTA | ARGDLPYYDFWSGYHYYGMDV | κ1-5 | 0.4 | QSISSW | KAS | QQYNSYST | |
| CLL821 | B-CLL | PBMCs | 1-69 | 0.0 | GGTFSSYA | IIPIFGTA | ASNAAPLRFLEWSYYTEMGGDY | ND | ||||
| CLL821–1 | Hybridoma | 1-69 | 0.0 | GGTFSSYA | IIPIFGTA | ASNAAPLRFLEWSYYTEMGGDY | κ1-17 | 0.0 | QGIRND | AAS | LQHNSYPYT | |
| CLL1296 | B-CLL | PBMCs | 3-07 | 7.6 | GFTFSSYW | INQGGSDM | ARDSSGYQGRFDP | ND | ||||
| CLL1296-1 | Hybridoma | 3-07 | 7.6 | GFTFSSYW | INQGGSDM | ARDSSGYQGRFDP | κ1-8 | 4.3 | QDITSY | AAS | QQYYSYPRT | |
| CLL1324 | B-CLL | PBMCs | 1-69 | 0.0 | GGTFSSYA | IIPIFGTA | AREASYGSGSYYQQYYYYYYMDV | λ3-19 | 0.0 | SLRSYY | GKN | NSRDSSGNPYV |
| CLL1324-E9 | Hybridoma | 1-69 | 0.0 | GGTFSSYA | IIPIFGTA | AREASYGSGSYYQQYYYYYYMDV | λ3-19 | 0.0 | SLRSYY | GKN | NSRDSSGNPYV | |
| IGHV . | IGKV/IGLV . | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CLL ID . | Cell line . | Source . | Family . | Mutation, % . | CDR1 . | CDR2 . | CDR3 . | Family . | Mutation, % . | CDR1 . | CDR2 . | CDR3 . |
| CLL246 | B-CLL | PBMCs | 1-69 | 0.3 | GGTFSSYA | IIPILGIA | ARSDQNYDFWSGYFRYYGMDV | ND | ||||
| CLL246-M1 | Hybridoma | 1-69 | 0.7 | GGTFSSYA | IIPILGIA | ARSDQNYDFWSGYFRYYGMDV | κ3-20 | 0.0 | QSVSSSY | GAS | QQYGSSPET | |
| CLL526 | B-CLL | PBMCs | 1-69 | 0.0 | GGTFSSYA | IIPIFGTA | ARVGYYDFWSGYYPNYYYYGMDV | λ3-9 | 0.0 | NIGSKN | RDS | QVWDSSTV |
| CLL526-H10 | Hybridoma | 1-69 | 0.0 | GGTFSSYA | IIPIFGTA | ARVGYYDFWSGYYPNYYYYGMDV | λ3-9 | 0.0 | NIGSKN | RDS | QVWDSSTV | |
| CLL698 | B-CLL | PBMCs | 1-69 | 0.0 | GGTFSSYA | IIPIFGTA | ARGDLPYYDFWSGYHYYGMDV | κ1-5 | 0.4 | QSISSW | KAS | QQYNSYST |
| CLL698-A11 | Hybridoma | 1-69 | 0.3 | GGTFSSYA | IIPIFGTA | ARGDLPYYDFWSGYHYYGMDV | κ1-5 | 0.4 | QSISSW | KAS | QQYNSYST | |
| CLL821 | B-CLL | PBMCs | 1-69 | 0.0 | GGTFSSYA | IIPIFGTA | ASNAAPLRFLEWSYYTEMGGDY | ND | ||||
| CLL821–1 | Hybridoma | 1-69 | 0.0 | GGTFSSYA | IIPIFGTA | ASNAAPLRFLEWSYYTEMGGDY | κ1-17 | 0.0 | QGIRND | AAS | LQHNSYPYT | |
| CLL1296 | B-CLL | PBMCs | 3-07 | 7.6 | GFTFSSYW | INQGGSDM | ARDSSGYQGRFDP | ND | ||||
| CLL1296-1 | Hybridoma | 3-07 | 7.6 | GFTFSSYW | INQGGSDM | ARDSSGYQGRFDP | κ1-8 | 4.3 | QDITSY | AAS | QQYYSYPRT | |
| CLL1324 | B-CLL | PBMCs | 1-69 | 0.0 | GGTFSSYA | IIPIFGTA | AREASYGSGSYYQQYYYYYYMDV | λ3-19 | 0.0 | SLRSYY | GKN | NSRDSSGNPYV |
| CLL1324-E9 | Hybridoma | 1-69 | 0.0 | GGTFSSYA | IIPIFGTA | AREASYGSGSYYQQYYYYYYMDV | λ3-19 | 0.0 | SLRSYY | GKN | NSRDSSGNPYV | |
Discussion
In this study, we have shown that the mouse macrophage cell line, J774A.1, enhances the outgrowth of EBV-transformed B cells from CpG oligonucleotide-stimulated B-CLL cells by more than 30-fold (Figure 1A), yielding an overall efficiency of B-CLL cell outgrowth of 38% (Table 2). The leukemic origin of the cell lines was documented by IGHV-D-J sequencing and transformation by EBV verified by the expression of the EBV-related LMP1 mRNA and protein in each case tested. Although single-cell analyses demonstrated growth of both U-CLL and M-CLL clones, M-CLL cells were less responsive to transformation. Furthermore, none of the EBV-transformants became feeder cell independent. Finally, using electrofusion to an HGPRTase-deficient myeloma cell line, the secretory and long-term growth capacities of EBV transformants were further enhanced.
The mechanism(s) by which the J774A.1 macrophage cell line promotes survival and outgrowth of B-CLL cells after CpG stimulation and EBV infection has not been defined. J774A.1 murine cells produce numerous cytokines and chemokines31,32 ; however, the biologic effects of these murine agonists on survival and growth of B-CLL cells is not clear. J774A.1 cells also produce growth factors that facilitate hybridoma growth and cloning,33 and conditioned medium from J774A.1 cells enhances the viability of EBV-transformed normal human B cells.34 It is also noteworthy that coculture of B-CLL cells with “nurse-like cells,” derivatives of the myelomonocytic lineage, or stromal cells prevent spontaneous apoptosis of B-CLL cells in vitro, by both cell contact and soluble mediators.35-38
J774A.1 cells also help clear apoptotic cells in vitro.39 In this study, recognition by scavenger receptors of oxidized phosphatidylserine may be a mechanism of apoptotic cell clearance by J774A.1 cells, and such uptake could culminate in the presentation of apoptotic antigens to B-CLL cells. In addition, because in our cell culture system transformation was most successful for U-CLL cells, clones that often bind and presumably respond to apoptotic cells via surface Ig BCR-signaling after binding unprocessed apoptotic antigens could be an additional component for successful transformation.17-20 Proliferation of J774A.1 cells was not necessary for their trophic effect in this system as they were exposed to 40 Gy of γ-irradiation, which was determined experimentally to completely inhibit proliferation and to prevent overgrowth of the feeder cell line in vitro. Irradiation may also generate apoptotic cell debris, leading to additional stimulation of predominantly U-CLL clones.
Unlike normal human B lymphocytes that lose CD5 surface membrane expression after EBV immortalization,40,41 several B-CLL cell lines that were maintained in culture for a number of months continued to express CD5. This observation suggests that the influence of the macrophage feeder cells extends beyond survival and outgrowth and affects the fundamental biology of the transformed cells or that continuous activation via BCRs by apoptotic cells17-20 maintains CD5 expression.
Even though it is known that LMP1 is required for successful immortalization of B cells23,24 and our B-CLL cells expressed both LMP1 mRNA and protein while cultured on macrophages (Figure 3), when the feeder cell support was eliminated, the B-CLL cells died. This suggests that ongoing input from the macrophages was needed to maintain viability and also implies that extracellular signals produced by the feeder cells induced LMP1 expression and cell proliferation. In this regard, it is noteworthy that several cytokines, including IL-4 and IL-10, induce LMP1 expression in B cells.42,43 Future studies will be needed to determine the genetic differences that exist between these B-CLL cells and those that became completely feeder cell independent.44
Even though our yield of EBV-transformed B-CLL cells was relatively high (∼ 38%; Table 2), in some instances normal B cells from the B-CLL patient's blood outgrew the leukemic cells. For example, at week 5, 94.4% of wells from the EBV-infected CLL1121 cell line contained single B cells exhibiting an IGHV-D-J rearrangement that matched the originating B-CLL clone (Table 3); however, at 137 days, this culture consisted of only normal B cells. Nevertheless, our approach will be helpful for studies of B-CLL cell biology, antibody production, and genetics. Examples of future studies of B-CLL biology are understanding in detail the mechanisms whereby macrophage products support the growth of B-CLL cells and in particular their effects on supporting the outgrowth of EBV transformants, which we assume is via up-regulation of LMP1. Indeed, it is intriguing that the majority of B-CLL clones taken from patients do not exhibit EBNA-1 or LMP1, suggesting an active mechanism whereby these are not infected by EBV or cannot express the necessary molecules to sustain proliferation in vivo.
Stimulation with CpG oligonucleotides facilitates transformation of normal human memory B lymphocytes by EBV,25 and this combined approach has been used to generate potentially therapeutic human mAbs.22,25,45 Because B-CLL cells are antigen-experienced and “memory-like,” we used such oligonucleotides in these studies. Others have shown that mitogenic agents, such as Staphylococcus aureus Cowan strain I, thioredoxin, and IL-2, also aid EBV infection of CLL cells. In the presence of irradiated human embryonic lung fibroblasts, the authors generated 3 immortalized cell lines (1 U-CLL and 2 M-CLL cases),17,44 although IGHV sequence confirmation between the original B-CLL and these 3 cell lines is not available.
Of note, M-CLL cells were more difficult to transform than U-CLL in our study. We found that 80% (12 of 15) of U-CLL samples gave rise to transformed B-CLL cells, but only approximately 17% (1 of 6) M-CLL (CLL1193; Table 2). The reason for this difference is not clear at this juncture. One possibility is the greater likelihood of recognition of apoptotic antigens by U-CLL BCRs as discussed above. Another possibility, based on the requirement of cellular activation by CpGs and other stimulants to promote EBV transformation, is the relative unresponsiveness of M-CLL clones, compared with U-CLL, to TLR9- and BCR-mediated signals.46-49 Furthermore, because TLR-9 is not expressed by all B-CLL clones, other TLR ligands for receptors expressed by more B-CLL clones (eg, TLR-7)50 might enhance EBV transformation.
Finally, the ability of these B-CLL–derived cell lines to secrete Ig and to grow long-term was enhanced by generating hetero-hybridomas by electrofusion. Using modified methods to those described previously by Yu et al,28 we generated hetero-hybridomas at an efficiency of 60% (6 hetero-hybridoma clones of 10 EBV-transformed B cell lines used); this was somewhat lower than more than the 80% efficiency we have previously achieved with EBV-transformed normal human B cells using the same methods (data not shown), suggesting that optimization of fusion methods, including pulse parameters, and of partner cell lines may be necessary.
The availability of DNA and RNA from B-CLL–derived cell lines will enable the determination of specific genetic abnormalities that might represent initiation or progression factors in the disease. In addition, using this approach, access to large amounts of native mAb, especially after hetero-hybridoma formation, will be feasible and practical, thereby facilitating study of the specificity of B-CLL clones to pathogens, tumor antigens, and auto-antigens and structural properties of B-CLL–derived antibodies/BCRs, in particular those with stereotyped structure.2,7,30 The latter feature may be especially valuable because initial studies suggest that B-CLL cells with similar BCR structure and apoptotic autoantigen reactivity may be useful in predicting disease outcome.20
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Laura Guogas for database management, and Adam Brewer, Radharani De, Raul Salinas-Mondragon, Thaddeus Gurley, and Yi Yang for expert technical assistance.
This work was supported by the Bill and Melinda Gates Foundation (Collaboration for AIDS Vaccine Discovery grant, B.F.H.), the Center For HIV/AIDS Vaccine Immunology (grant U19 AI067854), and the National Institutes of Health National Cancer Institute (R01 grant CA81554, N.C.).
National Institutes of Health
Authorship
Contribution: K.-K.H., D.M.K., X.C., M.G., D.J.M., J.F.W., and M.A.L. devised and performed experiments; R.C., C.C.C., and X.-J.Y. collected and analyzed patient samples; H.-X.L., C.C.C., N.C., and B.F.H. interpreted results; and K.-K.H., N.C., and B.F.H. designed the research and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Barton F. Haynes, Duke Human Vaccine Institute, Duke University Medical Center, 2 Genome Court, Durham, NC 27710; e-mail: hayne002@mc.duke.edu; and Nicholas Chiorazzi, Feinstein Institute for Medical Research, 350 Community Dr, Manhasset, NY 11030; e-mail: nchizzi@nshs.edu.