Key Points
Vesicular system causing artemisinin resistance modifies malaria parasites and host red cells.
Abstract
Artemisinin resistance threatens worldwide malaria control and elimination. Elevation of phosphatidylinositol-3-phosphate (PI3P) can induce resistance in blood stages of Plasmodium falciparum. The parasite unfolded protein response (UPR) has also been implicated as a proteostatic mechanism that may diminish artemisinin-induced toxic proteopathy. How PI3P acts and its connection to the UPR remain unknown, although both are conferred by mutation in P falciparum Kelch13 (K13), the marker of artemisinin resistance. Here we used cryoimmunoelectron microscopy to show that K13 concentrates at PI3P tubules/vesicles of the parasite’s endoplasmic reticulum (ER) in infected red cells. K13 colocalizes and copurifies with the major virulence adhesin PfEMP1. The PfEMP1-K13 proteome is comprehensively enriched in multiple proteostasis systems of protein export, quality control, and folding in the ER and cytoplasm and UPR. Synthetic elevation of PI3P that induces resistance in absence of K13 mutation also yields signatures of proteostasis and clinical resistance. These findings imply a key role for PI3P-vesicle amplification as a mechanism of resistance of infected red cells. As validation, the major resistance mutation K13C580Y quantitatively increased PI3P tubules/vesicles, exporting them throughout the parasite and the red cell. Chemical inhibitors and fluorescence microscopy showed that alterations in PfEMP1 export to the red cell and cytoadherence of infected cells to a host endothelial receptor are features of multiple K13 mutants. Together these data suggest that amplified PI3P vesicles disseminate widespread proteostatic capacity that may neutralize artemisinins toxic proteopathy and implicate a role for the host red cell in artemisinin resistance. The mechanistic insights generated will have an impact on malaria drug development.
Introduction
Symptoms and pathologies of Plasmodium falciparum malaria are entirely due to parasite stages that infect and remodel host red blood cells. At least a subset of these stages now show resistance to artemisinins: frontline, antimalarial drugs for which we still have no replacements. Emergence and spread of artemisinin resistance threatens worldwide malaria control and elimination.1-3 PfKelch13 (K13) is a primary marker of artemisinin resistance.4-7 K13 mutations confer resistance in “ring” stage parasites formed immediately after invasion,8-10 as measured by the Ring Stage Survival Assay (RSA), an in vitro correlate of in vivo clinical resistance.11 The major mutation K13C580Y diminishes binding to and ubiquitinylation-dependent proteosomal degradation of phosphatidylinositol-3-kinase (PfPI3K) to increase kinase levels.10 Notably, elevation of PfPI3K’s lipid product phosphatidylinositol-3-phosphate (PI3P) confers artemisinin resistance.10 Population transcriptomics of over 1000 clinical isolates separately revealed that the parasite endoplasmic reticulum (ER) unfolded protein response (UPR) was associated with K13 mutation.12 But where and how PI3P acts and its interaction with UPR (if any)13,14 in infected red cells remains unidentified. In addition, although clinical artemisinin resistance was first identified as delayed clearance of ring-infected red cells from circulation, its consequences for parasite-induced changes in the host cell are unknown. Here we examine localization and dynamics of parasite PI3P, K13, and proteostasis systems that include UPR to delineate a mechanism that explains why hundreds of parasite determinants and multiple parasite organellar systems are implicated in resistance.15-23 Furthermore, we provide evidence that drug resistance affects properties of the host red cell linked to immunity.
Methods
Antibodies
Pan-“PfEMP1” antibodies were raised to recombinant conserved C-terminal acidic-terminal sequence (ATS) domain of PfEMP1 by the commercial vendor Genscript Inc. Anti-PI3P was from Echelon Biosciences. Anti-human Band 3 was a gift from Phillip S. Low. All other antibodies were from Thermo Scientific Inc. (Rockford, IL).
Microscopy
For immunoelectron microscopy, late trophozoite/schizont stages of the Plasmodium falciparum strain PfNF54K13WT or PfNF54K13C580Y were purified and fixed in 4% paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA) in 0.25 M of N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (HEPES) (pH 7.4) for 1 hour at room temperature and processed for immunoelectron microscopy (IEM).24 Mouse anti-PI3P antibody was used at 1/250 to 1/400 dilution in phosphate-buffered saline/1% fish skin gelatin; rat anti-K13 antibody was used at 1/10 dilution, and guinea pig anti-Pf–binding immunoglobulin protein (BiP) antibody at 1/10 dilution. Samples were viewed in a Philips CM120 Electron Microscope (Eindhoven, The Netherlands) under 80 kV. For fluorescence microscopy, live and fixed cells were imaged by DeltaVision Deconvolution microscopy.25
Parasite culture, transfection, cytoadherence, and K13 membrane association
P falciparum laboratory strains and their transgenic counterparts as well as clinical Cambodian isolates (a kind gift from Arjen Dondorp) were prepared, as has been previously described.10 Clinical strain isolation and culture were approved by the Oxford Tropical Medicine Research Ethical Committee, the Ministry of Health in Cambodia (trial registered under NCT00493363), and the University of Notre Dame. For cytoadherence assays, P falciparum CS2 strains were panned by binding to chondroitin sulfate (CSA), as has been described.26 For K13-membrane association, infected red cells were treated as indicated; soluble and insoluble components were analyzed with sodium dodecyl sulfate (SDS)–polyacrylamide gel electrophoresis and Western blots.
Isolation and analyses of the PfEMP1 immunoproteome
P falciparum purified schizonts/segmenters were lysed in 0.05% saponin, followed by 0.5% NP-40 in 20 mM HEPES (pH 7.9), 10 mM KCl, 1 mM EDTA, 1 mM EGTA, and 1 mM dithiothreitol (with protease inhibitor cocktail; Roche Diagnostics) at 4°C. Protein extracts were solubilized in 1% volume-to-volume ratio Triton X-100 and 1% weight-to-volume ratio sodium deoxycholate and were incubated with anti-PfEMP1 ATS antibodies (or mock treated) and then with protein G agarose. After washing, bound proteins were eluted in 50 mM glycine, pH 2.5, digested with trypsin, and subjected to liquid chromatography–tandem mass spectrometry (MS/MS), and peptides were identified using MaxQuant 1.5.2.8.27 MS/MS spectra were searched against a combined PlasmoDb P falciparum 3D7 (version 24) and UniProt human protein database. Hypergeometric analyses were used to identify enrichment in the clinical transcriptome12 or Malaria Parasite Metabolic Pathways 2016 in PlasmoDB (http://plasmodb.org/plasmo/).
Statistical information and data availability
The Mann-Whitney U test or 1-way analysis of variance with a Tukey or Bonferroni post hoc analysis was used to compare the mean values between treatment groups. Statistical analysis was performed with GraphPad Prism (version 6.02). P values were 2 sided, with P < .05 being considered significant. All data in this study are included in this article (and the supplemental Methods, tables, and figures, available on the Blood Web site).
Additional details are presented in the supplemental Methods.
Results
Detection and dynamics of PI3P and K13 in P falciparum
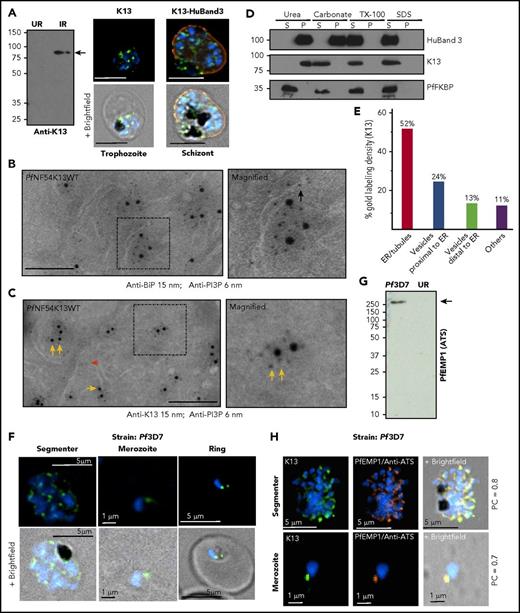
PI3P is indistinguishable from phosphatidylinositol-4-phosphate (PI4P) by mass spectrometry because of identical mass and charge. Conventional indirect immunofluorescence assays (IFAs) use detergents (or methanol) to permeabilize cells, which may result in loss of lipids (including PI3P/PI4P, because they are poorly fixed by formaldehyde-based crosslinking used in IFAs). Rather, tagged reporter proteins that bind selectively to PI3P (and PI4P) are used to visualize their sites of concentration in live (and fixed) cells.28 Two PI3P-binding protein reporters (EEA1 and p40Phox, transgenically expressed), carrying a malaria parasite leader signal sequence, are recruited into and accumulate in perinuclear, ER-vesicular domains in early rings29 (data for SS-EEA1 are shown in Figure 1A). When the ring parasite matures and replicates into multinucleated “schizonts,” each daughter inherits PI3P (Figure 1B), which in emerging extracellular “merozoite” parasites appears as a dynamic cluster of vesicles that are transferred to newly formed rings (Figure 1C; supplemental Movie 1). A reporter for cytoplasmic PI3P failed to reveal distinct, prominent sites of concentration in young rings (supplemental Figure 1A-D), but focal regions were seen in later-stage “trophozoites/schizonts” (as has been previously reported30 ; Figure 1D). We were unable to develop parasites dually expressing both secretory and cytoplasmic (EEA1) reporters (not shown). We therefore undertook cryo-IEM to detect endogenous PI3P in infected red cells (using antibodies to PI3P previously validated in IEM studies in yeast31 ). As is shown in Figure 1E (and supplemental Figure 1E-G), PI3P was detected at the cytoplasmic and luminal faces (yellow and black arrows) of the food vacuole (fv) and apicoplast. In addition, a high density of PI3P was found in tubules characteristic of the ER in both cytoplasmic and luminal orientations (yellow and black arrows), as well as in clusters of fine vesicles cytoplasmic to ER tubules (yellow double arrows).
Dynamics and localization of PI3P in P falciparum–infected red cells. (A-B) In live P falciparum–infected red cells (3D7 strain; Pf3D7), transgenic expression of a secretory form of the PI3P-binding protein EEA1 fused to mCherry (SS-EEA1WT-mCherry; red) reveals secretory PI3P in a perinuclear region in early ring parasites (A).25 Not shown here, but as previously shown, a single-point mutant of EEA1 that fails to bind PI3P is secreted to the parasitophorous vacuole (PV),25 marked by the dotted circles in the middle panels, where the fluorescence image is merged with the bright field (B). Perinuclear localization is also seen in later schizont stages. The boxed regions in the left-hand panels are magnified in the right-hand panels. (C) Time-lapse images of the parasite’s extracellular merozoite stage (arrowhead) invading red cells to become an intracellular ring (arrow). (D) In live Pf3D7, the transgenic cytosolic form of the PI3P-binding protein EEA1 (cEEA1WT–green fluorescent protein) is seen associated with punctate vesicles and organelles of late trophozoites/schizonts (as was previously reported30 ) and distinct from perinuclear foci seen for SS-EEA1WT-mCherry (shown in panels A-B). The boxed region in the left-hand panel is magnified in the right-hand panel. Parasite nucleus stained with Hoechst 33242 (blue). Scale bar, 5 µm. Live cells were imaged using indicator-free RPMI1640 (Gibco) by DeltaVision Deconvolution microscopy25 with a 100×, NA-1.4 objective on an Olympus IX inverted fluorescence microscope on a temperature-controlled stage at 37°C and a Photometrics cooled custom CCD camera (CH350/LCCD) driven by DeltaVision Software from Applied Precision Inc. (Seattle, WA). (E) Cryo-IEM of PfNF54 (wild-type) parasites probed with antibodies to PI3P and secondary antibody 10-nm gold conjugate. Gold particles are detected in the apicoplast, food vacuole, and tubules (suggestive) of ER. Black arrows indicate PI3P in lumen of tubule; yellow arrows indicate cytoplasmic PI3P; double yellow arrows indicate PI3P vesicular clusters cytoplasmic to ER tubules. Experimental replicates, n = 3. Scale bar, 100 nm. Imaged in a Philips CM120 Electron Microscope (Eindhoven, The Netherlands) under 80 kV. BF, bright field; cEEA1, cytosolic form of the PI3P-binding protein EEA1; GFP, green fluorescent protein; P, parasite nucleus; R, red cell.
Dynamics and localization of PI3P in P falciparum–infected red cells. (A-B) In live P falciparum–infected red cells (3D7 strain; Pf3D7), transgenic expression of a secretory form of the PI3P-binding protein EEA1 fused to mCherry (SS-EEA1WT-mCherry; red) reveals secretory PI3P in a perinuclear region in early ring parasites (A).25 Not shown here, but as previously shown, a single-point mutant of EEA1 that fails to bind PI3P is secreted to the parasitophorous vacuole (PV),25 marked by the dotted circles in the middle panels, where the fluorescence image is merged with the bright field (B). Perinuclear localization is also seen in later schizont stages. The boxed regions in the left-hand panels are magnified in the right-hand panels. (C) Time-lapse images of the parasite’s extracellular merozoite stage (arrowhead) invading red cells to become an intracellular ring (arrow). (D) In live Pf3D7, the transgenic cytosolic form of the PI3P-binding protein EEA1 (cEEA1WT–green fluorescent protein) is seen associated with punctate vesicles and organelles of late trophozoites/schizonts (as was previously reported30 ) and distinct from perinuclear foci seen for SS-EEA1WT-mCherry (shown in panels A-B). The boxed region in the left-hand panel is magnified in the right-hand panel. Parasite nucleus stained with Hoechst 33242 (blue). Scale bar, 5 µm. Live cells were imaged using indicator-free RPMI1640 (Gibco) by DeltaVision Deconvolution microscopy25 with a 100×, NA-1.4 objective on an Olympus IX inverted fluorescence microscope on a temperature-controlled stage at 37°C and a Photometrics cooled custom CCD camera (CH350/LCCD) driven by DeltaVision Software from Applied Precision Inc. (Seattle, WA). (E) Cryo-IEM of PfNF54 (wild-type) parasites probed with antibodies to PI3P and secondary antibody 10-nm gold conjugate. Gold particles are detected in the apicoplast, food vacuole, and tubules (suggestive) of ER. Black arrows indicate PI3P in lumen of tubule; yellow arrows indicate cytoplasmic PI3P; double yellow arrows indicate PI3P vesicular clusters cytoplasmic to ER tubules. Experimental replicates, n = 3. Scale bar, 100 nm. Imaged in a Philips CM120 Electron Microscope (Eindhoven, The Netherlands) under 80 kV. BF, bright field; cEEA1, cytosolic form of the PI3P-binding protein EEA1; GFP, green fluorescent protein; P, parasite nucleus; R, red cell.
PI3P levels are affected by K13 mutations,10 but the location of K13 in relation to PI3P remains unknown. IFAs localized endogenous K13 in diffuse and focal distribution in trophozoite and schizonts (Figure 2A; supplemental Figure 2A). Cryo-IEM detected PI3P (6 nm gold) in the ER lumen marked by PfBiP (15 nm gold; Figure 2B, black arrow; supplemental Figure 2B) and enriched with K13 on ER tubules and vesicles (Figure 2C, single and double yellow arrows; supplemental Figure 2C). K13 lacks a secretory signal sequence consistent with its association with cytoplasmic PI3P. The gold particles for PI3P look well aligned, suggesting membrane structures beneath, even when tiny vesicles are difficult to see in cryosections. Biochemical fractionation studies suggested that K13 was peripherally associated with membranes (Figure 2D). Stereological analyses of gold particles indicated that ∼75% of K13 localized to ER tubules, vesicles, or both (Figure 2E). K13 was also found in vesicles distal from the ER (13%; they may also be derived from organelles other than the ER), and low levels (11%) were detected all over the parasite. Only low levels of PI3P vesicles free of K13 were observed (Figure 2C, red arrow), and PI3P with a K13 label (yellow arrows) appeared as well-structured clusters. Along the ER, cytoplasmic PI3P (single and double yellow arrows) consistently exceeded luminal lipid (black arrows). Together, these data suggested K13 as a marker for PI3P, concentrated in the ER. Quantitative IEM could not be undertaken at the ring stage of infection because of the small size of rings (1-2 μm), making them easy to miss in thin sections taken through infected red cells. However, IFAs suggested that K13 was assembled in merozoites within schizonts and transferred to newly formed rings (Figure 2F).
Stage-specific expression and localization of K13 in relation to ER-PI3P, PfBiP, and PfEMP1. (A) Custom antibodies to K13 (supplemental Methods) detect an 83-kDa band in Pf3D7-infected red cells (IRs) but not uninfected red cells (URs) in Western blots (molecular weights in kDa) and localize K13 (green) by IFA in trophozoite and schizont stages (counterstained for host band 3, red), as imaged with a 100×, NA-1.4 objective on an Olympus IX inverted fluorescence microscope using DeltaVision Deconvolution microscopy.25 (B-C) Cryo-IEM of PfNF54K13WT dually probed for PI3P (6 nm gold) and ER marker BiP (15 nm gold) (B) or K13 (15 nm gold) (C). Black arrow indicates PI3P in lumen of ER tubule; yellow arrows indicate cytoplasmic PI3P; double yellow arrows indicate PI3P vesicular clusters outside of ER tubules on the cytoplasmic face; red arrow indicates low level of PI3P vesicles devoid of K13. Scale bar, 100 nm. (D) Membrane association of K13. Lysates of Pf3D7 were treated as indicated, separated by centrifugation (15 000 g for 30 min) into membrane pellet and soluble supernatant fractions and probed in Western blots for parasite and host (human) markers. Adding 6 M urea (a strong chaotropic agent) for 30 min at 23°C to parasite cell lysates failed to release K13 from the pellet (of parasite cell lysates), although the cytosolic parasite protein PfFKBP was quantitatively detected in the soluble fraction, suggesting that K13 was membrane associated. Sodium carbonate 100 mM, pH 11.5, for 30 min on ice released K13 from the pellet, suggesting that it was peripherally (but not integrally) associated with membranes (and consistently, K13 was also released by 1% Tx-100 for 30 min at room temperature or 1% SDS for 30 min at room temperature). Human band 3 was only released by 1% Triton or 1% SDS, confirming that it was integrally membrane associated. Molecular weight standards (in kDa) are as shown. (E) Stereological analyses of K13-gold particle distribution by cryo-IEM. Vesicles close to the ER appear to bud from ER tubules. Vesicles distal to the ER may be derived from other organellar membranes and cannot be ascribed solely to the ER. (F) IFA single optical sections localizing K13 in segmenter, merozoite, and ring stages. Scale bars are as shown. (G) Anti-ATS antibodies recognize a band >250 kDa in Western blots (as was expected for PfEMP1) in IRs but not URs. Molecular weight standards (in kDa) are as shown. (H) IFA showing single optical sections colocalizing K13 and PfEMP1 (labeled by anti-ATS) in Pf3D7 segmenter and merozoites. Pearson’s correlation coefficients are as indicated. Experimental replicates, n = 3. Scale bars are as indicated. Parasite nucleus (blue) is stained with Hoechst 33242. HuBand3, Human band 3; P, pellet; PC, Pearson’s correlation coefficient; S, supernatant.
Stage-specific expression and localization of K13 in relation to ER-PI3P, PfBiP, and PfEMP1. (A) Custom antibodies to K13 (supplemental Methods) detect an 83-kDa band in Pf3D7-infected red cells (IRs) but not uninfected red cells (URs) in Western blots (molecular weights in kDa) and localize K13 (green) by IFA in trophozoite and schizont stages (counterstained for host band 3, red), as imaged with a 100×, NA-1.4 objective on an Olympus IX inverted fluorescence microscope using DeltaVision Deconvolution microscopy.25 (B-C) Cryo-IEM of PfNF54K13WT dually probed for PI3P (6 nm gold) and ER marker BiP (15 nm gold) (B) or K13 (15 nm gold) (C). Black arrow indicates PI3P in lumen of ER tubule; yellow arrows indicate cytoplasmic PI3P; double yellow arrows indicate PI3P vesicular clusters outside of ER tubules on the cytoplasmic face; red arrow indicates low level of PI3P vesicles devoid of K13. Scale bar, 100 nm. (D) Membrane association of K13. Lysates of Pf3D7 were treated as indicated, separated by centrifugation (15 000 g for 30 min) into membrane pellet and soluble supernatant fractions and probed in Western blots for parasite and host (human) markers. Adding 6 M urea (a strong chaotropic agent) for 30 min at 23°C to parasite cell lysates failed to release K13 from the pellet (of parasite cell lysates), although the cytosolic parasite protein PfFKBP was quantitatively detected in the soluble fraction, suggesting that K13 was membrane associated. Sodium carbonate 100 mM, pH 11.5, for 30 min on ice released K13 from the pellet, suggesting that it was peripherally (but not integrally) associated with membranes (and consistently, K13 was also released by 1% Tx-100 for 30 min at room temperature or 1% SDS for 30 min at room temperature). Human band 3 was only released by 1% Triton or 1% SDS, confirming that it was integrally membrane associated. Molecular weight standards (in kDa) are as shown. (E) Stereological analyses of K13-gold particle distribution by cryo-IEM. Vesicles close to the ER appear to bud from ER tubules. Vesicles distal to the ER may be derived from other organellar membranes and cannot be ascribed solely to the ER. (F) IFA single optical sections localizing K13 in segmenter, merozoite, and ring stages. Scale bars are as shown. (G) Anti-ATS antibodies recognize a band >250 kDa in Western blots (as was expected for PfEMP1) in IRs but not URs. Molecular weight standards (in kDa) are as shown. (H) IFA showing single optical sections colocalizing K13 and PfEMP1 (labeled by anti-ATS) in Pf3D7 segmenter and merozoites. Pearson’s correlation coefficients are as indicated. Experimental replicates, n = 3. Scale bars are as indicated. Parasite nucleus (blue) is stained with Hoechst 33242. HuBand3, Human band 3; P, pellet; PC, Pearson’s correlation coefficient; S, supernatant.
Because our prior studies suggested that PI3P targets PfEMP1 to the red cell,29 we examined the distribution of K13 in relation to PfEMP1. A monospecific “pan-PfEMP1” antibody was developed to the conserved acidic C-terminal sequence or ATS present in all members of this antigenic family (supplemental Figure 2D-E). As is shown in Figure 2F-H, in IFAs, PfEMP1 protein appeared closely apposed to K13 (Pearson correlation coefficient of 0.7-0.8; supplemental Figure 2F-G) in merozoites inside segmenters and released extracellularly, suggesting that K13/PI3P vesicles may affect PfEMP1 biology at these stages and new rings (which form minutes after merozoite release).
K13 copurifies with a PfEMP1 immunoproteome enriched in proteostasis systems that include the UPR associated with clinical artemisinin-resistant strains
Pf3D7 lysates prepared from segmenters were optimized for extraction of PfEMP1 (as is summarized in supplemental Figure 3A-D; supplemental Methods). Immunopurification using anti-ATS antibodies (or mock without primary antibodies) was carried out under high (and stringent) concentrations of both nonionic and chaotropic detergents (1% Tx-100 and 1.0% deoxycholate; supplemental Figure 3D, supplemental Methods) to minimize nonspecific protein-protein interactions with beads coated with protein G (Figure 3A; supplemental Figure 3A-D, supplemental Table 1). Two experimental replicates revealed an immunoproteome of 503 shared proteins (Figure 3B), which included K13 and comprised 88% of the total protein mass identified (Figure 3B; supplemental Table 1).
K13 copurified in a PfEMP1-immunoproteome enriched in proteostatic pathways that include the UPR, implicated in clinical artemisinin resistance. (A) Detection of PfEMP1 (arrow, left panel) and total protein content (right panel) captured by anti-ATS antibodies (but not in its absence at −1°) on covalently conjugated protein G beads. Molecular weight standards are in kDa. *Heavy IgG; **Light IgG. (B) Intensity properties of 2 replicate PfEMP1 immunoproteomes. (C) Distribution of proteins in the “503-proteome” comparing their abundance in each proteome expressed as a ratio (abundance ratio [AR] = abundance in Proteome 1/abundance in Proteome 2). X-axis: AR interval range; y-axis: number of proteins. (D) Hypergeometric analyses of 503-proteome and artemisinin-resistant transcriptome associated with clinical strains12 (for proteostasis enrichment and more detail, see supplemental Tables 2-4; Figure 3E-G). (E) Distribution of normalized intensity (ni; intensity divided by molecular weight) of the 207-proteome (AR = 0.5-1.99). Reactive oxidative stress complex components are BiP and PDI-11 (ni 5.17e8 and 1.2e7, respectively; supplemental Table 5). For detailed annotation and proteostasis enrichment, see supplemental Table 4 and supplemental Figure 3H-I. (F) Model integrating K13-PI3P tubules/vesicles (black-gray spheres based on findings of Figures 1 and 2 and supplemental Figures 1 and 2) and K13-PfEMP1 immunoproteome (green; based on findings in Figure 3; supplemental Figure 3) in proteostatic mechanisms of vesicular export and protein quality control and folding in the ER and cytoplasm (dotted circle). Experimental replicates, n = 2. Other organelles and key are as shown. HT, host targeting sequence; PEXEL, plasmodium export element; PNEP, PEXEL-negative proteins; PPM, parasite plasma membrane; PVM, parasitophorous vacuolar membrane; ROSC, reactive oxidative stress complex.
K13 copurified in a PfEMP1-immunoproteome enriched in proteostatic pathways that include the UPR, implicated in clinical artemisinin resistance. (A) Detection of PfEMP1 (arrow, left panel) and total protein content (right panel) captured by anti-ATS antibodies (but not in its absence at −1°) on covalently conjugated protein G beads. Molecular weight standards are in kDa. *Heavy IgG; **Light IgG. (B) Intensity properties of 2 replicate PfEMP1 immunoproteomes. (C) Distribution of proteins in the “503-proteome” comparing their abundance in each proteome expressed as a ratio (abundance ratio [AR] = abundance in Proteome 1/abundance in Proteome 2). X-axis: AR interval range; y-axis: number of proteins. (D) Hypergeometric analyses of 503-proteome and artemisinin-resistant transcriptome associated with clinical strains12 (for proteostasis enrichment and more detail, see supplemental Tables 2-4; Figure 3E-G). (E) Distribution of normalized intensity (ni; intensity divided by molecular weight) of the 207-proteome (AR = 0.5-1.99). Reactive oxidative stress complex components are BiP and PDI-11 (ni 5.17e8 and 1.2e7, respectively; supplemental Table 5). For detailed annotation and proteostasis enrichment, see supplemental Table 4 and supplemental Figure 3H-I. (F) Model integrating K13-PI3P tubules/vesicles (black-gray spheres based on findings of Figures 1 and 2 and supplemental Figures 1 and 2) and K13-PfEMP1 immunoproteome (green; based on findings in Figure 3; supplemental Figure 3) in proteostatic mechanisms of vesicular export and protein quality control and folding in the ER and cytoplasm (dotted circle). Experimental replicates, n = 2. Other organelles and key are as shown. HT, host targeting sequence; PEXEL, plasmodium export element; PNEP, PEXEL-negative proteins; PPM, parasite plasma membrane; PVM, parasitophorous vacuolar membrane; ROSC, reactive oxidative stress complex.
Because the PfEMP1 immunopurification was carried out under stringent conditions, if a protein was represented in each proteome, it was included for analyses in the shared set of 503. To facilitate comparative analyses, we determined the abundance of a protein in each immunoproteome and calculated an abundance ratio (AR; supplemental Methods) as a measure of the fidelity of protein association across replicate proteomes. As is shown in Figure 3C (and supplemental Table 1), over 40% of proteins were accommodated within a twofold range (AR, 0.5 to 1.99), and ∼70% were within threefold range. Importantly, K13, BiP (a major ER chaperone and UPR sensor), and protein disulfide isomerase (PDI) (a second major ER-redox protein) showed AR’s of 0.89, 1.29, and 1.98, respectively.
Hypergeometric analyses suggested that the “503-proteome” was highly enriched in multiple pathways of protein homeostasis or “proteostasis” that control the biogenesis, folding degradation, and trafficking of proteins within the parasite and exported to the host cell, regulated by protein quality control and folding in the ER and cytoplasm (supplemental Figure 3E-F). There was no significant enrichment of apicoplast and mitochondria: significant overlap was seen with the nucleus, but the P values suggested preferential enrichment of ER and cytoplasmic proteostatic pathways. The 503-proteome showed significant intersection with transcriptomic changes associated with artemisinin-resistant clinical isolates12 (P ≤ 4.7e-3; Figure 3D; supplemental Figure 3G). This overlap was enriched in 487 (top 9.6%) upregulated transcripts12 (P ≤ 3.2e-5; Figure 3D; supplemental Figure 3G) but not with the 511 genes (10.1%) that were downregulated. Finally, in the AR 0.5 to 1.99 core of a 207-proteome, there was no statistically significant intersection with the nucleus or the UPR, but several systems of ER/cytoplasmic proteostasis, including protein translocation export (PTEX) host-export components, continued to be enriched (supplemental Figure 3H-I) and comprised ∼50% of the proteome intensity (Figure 3E; supplemental Table 5). BiP, the key UPR sensor, was a major component of the 207-proteome (Figure 3E; supplemental Table 5; note that hypergeometric analyses do not account for intensity or AR). The data shown in Figures 2 and 3 support the molecular interaction of PfEMP1 with K13: associated PI3P tubules/vesicles of the ER that encompass protein proteostasis functions and augment export to the host.
Synthetic elevation of PI3P that induces resistance in absence of K13 mutation also yields signatures of proteostasis and clinical resistance
We previously reported that increasing PI3P levels by transgenic expression of human VPS34 in Pf3D7 parasites to yield Pf3D7VPS34myc confers artemisinin resistance in the absence of the K13 mutation, whereas Pf3D7VPS34AAAmyc expressing a catalytic dead VPS34 (in which active-site DRH was replaced by AAA) showed no increase in PI3P and remained artemisinin sensitive.10 Therefore, PfEMP1 immunoproteomes were stringently isolated and characterized from Pf3D7VPS34myc and Pf3D7VPS34AAA (Figure 4A-D; supplemental Figure 4A-C; supplemental Tables 7 and 8). The proteomes were similar in composition, with an overlap in 85% of their composite proteins (Figure 4E). Both contained 95% of the (503) PfEMP1 immunoproteome isolated from Pf3D7 but were also larger (705 proteins for Pf3D7VPSmyc and 770 proteins for Pf3D7VPSAAA, primarily due to increased proteosomal components; supplemental Figure 4D-E, supplemental (Tables 7 and 8). Hypergeometric analyses revealed that the Pf3D7VPS34myc immunoproteome was significantly enriched in 487 (top 9.6%) upregulated genes of the clinical transcriptome12 (P < 1.6e-7; Figure 4G; supplemental Figure 4F). Notably, this enrichment included 94% of 3D7 immunoproteome, which showed a significant intersection with the clinical proteome. Moreover, as with its 3D7 counterpart, the immunoproteome of Pf3D7VPS34myc was not enriched in the 511 genes (10.1%) downregulated in the clinical transcriptome (Figure 4G; supplemental Figure 4F). Similar results were obtained for the PfEMP1 immunoproteome of Pf3D7VPSAA (as was expected on the basis of its similarity in composition to that of Pf3D7VPS34myc; supplemental Figure 4G). These findings suggested that the major proteostatic characteristics of PfEMP1 immunoproteome of Pf3D7 are preserved in those from the Pf3D7VPS transgenic strains. Moreover, the composition of the vesicle immunoproteome is similar in resistant and sensitive (Pf3D7VPS) strains. However, elevation of PI3P may expand vesicles to confer resistance in Pf3D7VPSmyc, implicating vesicular expansion (of proteostatic mechanisms, including UPR, via chaperone complexes; Figure 4H) in artemisinin resistance.
Synthetic elevation of PI3P that induces resistance in absence of K13 mutation yields vesicle immunoproteomes enriched in signatures of proteostasis and clinical resistance. (A-B) Detection of PfEMP1 (arrow) captured by anti-ATS antibodies (but not in its absence; −1°) from PfVPS34myc and mass intensity properties of 2 replicate immunoproteomes. (C-D) Detection of PfEMP1 (arrow) captured by anti-ATS antibodies (but not in its absence; −1°) from immunoproteome from PfVPS34AAAmyc (catalytic dead enzyme in which AAA replaces the VPS34-catalytic active-site residues glutamic acid, arginine, and histidine DRH) and mass intensity properties of 2 replicate proteomes. Molecular weight standards are in kDa. (E) Comparative distribution of proteins in proteomes of VPS34myc and VPS34AAAmyc. (F) Hypergeometric analyses showing enrichment of VPS34myc immunoproteome and upregulated (but not downregulated) transcripts of the clinical artemisinin-resistant transcriptome.12 (G) Protein distribution analyses indicating that 95% of Pf3D7 of proteins enriched in the clinical transcriptome are present in the PfVPS34myc PfEMP1 immunoproteome. (H) Number and percentage of hits for indicated chaperone networks in the vesicle immunoproteome from PfVPSmyc reveals substantial association with reactive oxidative stress complex and T-complex chaperones also known as TCP1 ring complex of the UPR. Mu, mass units.
Synthetic elevation of PI3P that induces resistance in absence of K13 mutation yields vesicle immunoproteomes enriched in signatures of proteostasis and clinical resistance. (A-B) Detection of PfEMP1 (arrow) captured by anti-ATS antibodies (but not in its absence; −1°) from PfVPS34myc and mass intensity properties of 2 replicate immunoproteomes. (C-D) Detection of PfEMP1 (arrow) captured by anti-ATS antibodies (but not in its absence; −1°) from immunoproteome from PfVPS34AAAmyc (catalytic dead enzyme in which AAA replaces the VPS34-catalytic active-site residues glutamic acid, arginine, and histidine DRH) and mass intensity properties of 2 replicate proteomes. Molecular weight standards are in kDa. (E) Comparative distribution of proteins in proteomes of VPS34myc and VPS34AAAmyc. (F) Hypergeometric analyses showing enrichment of VPS34myc immunoproteome and upregulated (but not downregulated) transcripts of the clinical artemisinin-resistant transcriptome.12 (G) Protein distribution analyses indicating that 95% of Pf3D7 of proteins enriched in the clinical transcriptome are present in the PfVPS34myc PfEMP1 immunoproteome. (H) Number and percentage of hits for indicated chaperone networks in the vesicle immunoproteome from PfVPSmyc reveals substantial association with reactive oxidative stress complex and T-complex chaperones also known as TCP1 ring complex of the UPR. Mu, mass units.
K13C580Y, the major mutation of artemisinin resistance, expands ER-PI3P tubules and vesicles to propagate them throughout the parasite and in export to the host red cell
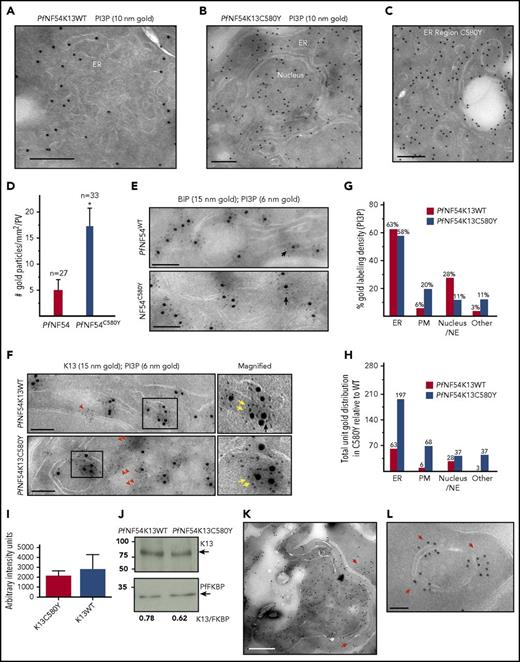
To directly examine vesicle expansion upon K13 mutation, we returned to cryo-IEM. Because our analyses of ER-PI3P (Figures 1 and 2) was initiated in the PfNF54 (wild-type PfNF54WT) strain, we compared findings in the matched-resistant counterpart PfNF54K13C580Y (carrying the major mutation reported in 80% of isolates in southeast Asia and created by CRISPR/Cas9 insertion of a single, chromosomal point mutation in PfNF54K13WT8 ). In comparison with their wild-type counterparts, K13C580Y mutants show an intense PI3P label in the ER and all over the parasite (Figure 5A-C; supplemental Figure 5A-C, including inside nuclei; Figure 5B). ER tubules and vesicles of mutants become amplified and heavily labeled with PI3P (Figure 5C). Quantitation revealed a threefold increase in total gold label at schizont stages (Figure 5D: 0-3 hours). The 0- to 3-hour ring stages of PfNF54K13C580Y contain 2.7 times more PI3P than do wild-type parasites,10 suggesting that mutants show comparable PI3P elevation in both schizont and new rings. There was no change in the labeling of ER marker BiP (Figure 5E) or K13 (Figure 5F; K13 stereological analyses in mutants are not shown). Rather, mutants contained large regions of dense PI3P label, which are devoid of PfK13 (Figure 5F, double red arrows) not seen in wild-type parasites (in which K13-free PI3P are small clusters, shown by the single red arrow in Figure 5F). Stereological analyses suggested that most of the PI3P (∼60%) remains associated with the ER (Figure 5G; although its relative amount was threefold higher in the mutant; Figure 5H). PI3P is seen at the nuclear envelope (an extension of the ER)/nucleus in wild-type of parasites. Nuclei of emerging daughter merozoites concentrated PI3P in mutants (Figure 5B). But the biggest fold change in PI3P in mutants was in the ER (threefold; Figure 5H) plasma membrane and other regions (which include the apicoplast and fv; 10-fold; Figure 5H). RNA sequencing analyses and Western blots confirmed that transcript and total cellular K13 protein levels are not altered between wild-type and mutants (even in 0- to 3-hour parasites; Figure 5I-J; consistent with findings that K13 transcripts were unchanged in clinical isolates12 ). Together, these data suggest that an increase in PI3P tubules and vesicles in the major K13C580Y mutant closely corresponds to fold changes in cellular levels of this lipid, suggesting that they are parasite major effectors of artemisinin resistance. Furthermore, in addition to its distribution through the entire parasite, PI3P was exported to vesicular structures and “Maurer’s clefts” in the cytoplasm of the infected red cell (Figure 5K-L, red arrows), known to be intermediates of parasite-protein export to the red cell membrane.32,33
K13C580Y, the major mutation of artemisinin resistance, amplifies PI3P tubovesicles, propagating them throughout the parasite and into the red cell. (A-B) Cryo-IEM of artemisinin-sensitive PfNF54K13WT (A) and artemisinin-resistant PfNF54K13C580Y (B) late trophozoite/schizont parasites probed for PI3P (10 nm gold). (C) Region of ER tubules/vesicles. Scale bars, 100 nm (bar sizes differ on basis of magnification). (D) Quantitation of gold particles associated per parasite vacuole for indicated numbers of parasites for each strain. (E-F) PfNF54K13WT and PfNF54K13C580Y ER dually probed for PI3P (6 nm gold) and the secretory ER marker BiP (15 nm gold) (E) or PI3P (6 nm gold) and K13 (15 nm gold) (F). Boxes in left-hand panels are shown magnified in the right-hand panels. Black arrows indicate luminal (space) PI3P; double yellow arrows indicate PI3P vesicular clusters adjacent to K13; red arrowheads indicate small PI3P clusters, and double red arrowheads indicate large PI3P clusters devoid of K13. Scale bars, 150 nm. (G-H) Stereological analyses of percentage of PI3P gold particle density in PfNF54K13WT or PfNF54K13C580Y with each strain normalized to itself (G) and comparative fraction of PI3P-gold particles in each strain (H), accounting for threefold increase in PfNF54K13C580Y in relation to PfNF54K13WT (total number of gold particles in PfNF54K13WT set to 100). (I) RNA sequence analyses for K13C580Y in PfNF54K13WT and PfNF54K13C580Y. Y-axis indicates arbitrary intensity units. (J) Western blots showing levels of K13, PfFKBP protein (a parasite cytosolic protein that serves as a loading control), and associated ratios, indicating no significant difference in levels of K13 protein expression in PfNF54K13C580Y in comparison with PfNF54K13WT. (K-L) PfNF54K13C580Y-infected red cells probed for PI3P (10 nm gold). Red arrows show PI3P at the parasite plasma and vacuolar membranes, in vesicles in the host red cell (scale bar, 500 nm) (K) and associated with “Maurer’s clefts,” intermediates in export to the red cell membrane (scale bar, 100 nm) (L). PI3P is not detected in the host in PfNF54K13WT-infected red cells. Experimental replicates, n = 2. IEM imaged in a Philips CM120 electron microscope (Eindhoven, The Netherlands) under 80 kV. PM, plasma membrane; WT, wild-type.
K13C580Y, the major mutation of artemisinin resistance, amplifies PI3P tubovesicles, propagating them throughout the parasite and into the red cell. (A-B) Cryo-IEM of artemisinin-sensitive PfNF54K13WT (A) and artemisinin-resistant PfNF54K13C580Y (B) late trophozoite/schizont parasites probed for PI3P (10 nm gold). (C) Region of ER tubules/vesicles. Scale bars, 100 nm (bar sizes differ on basis of magnification). (D) Quantitation of gold particles associated per parasite vacuole for indicated numbers of parasites for each strain. (E-F) PfNF54K13WT and PfNF54K13C580Y ER dually probed for PI3P (6 nm gold) and the secretory ER marker BiP (15 nm gold) (E) or PI3P (6 nm gold) and K13 (15 nm gold) (F). Boxes in left-hand panels are shown magnified in the right-hand panels. Black arrows indicate luminal (space) PI3P; double yellow arrows indicate PI3P vesicular clusters adjacent to K13; red arrowheads indicate small PI3P clusters, and double red arrowheads indicate large PI3P clusters devoid of K13. Scale bars, 150 nm. (G-H) Stereological analyses of percentage of PI3P gold particle density in PfNF54K13WT or PfNF54K13C580Y with each strain normalized to itself (G) and comparative fraction of PI3P-gold particles in each strain (H), accounting for threefold increase in PfNF54K13C580Y in relation to PfNF54K13WT (total number of gold particles in PfNF54K13WT set to 100). (I) RNA sequence analyses for K13C580Y in PfNF54K13WT and PfNF54K13C580Y. Y-axis indicates arbitrary intensity units. (J) Western blots showing levels of K13, PfFKBP protein (a parasite cytosolic protein that serves as a loading control), and associated ratios, indicating no significant difference in levels of K13 protein expression in PfNF54K13C580Y in comparison with PfNF54K13WT. (K-L) PfNF54K13C580Y-infected red cells probed for PI3P (10 nm gold). Red arrows show PI3P at the parasite plasma and vacuolar membranes, in vesicles in the host red cell (scale bar, 500 nm) (K) and associated with “Maurer’s clefts,” intermediates in export to the red cell membrane (scale bar, 100 nm) (L). PI3P is not detected in the host in PfNF54K13WT-infected red cells. Experimental replicates, n = 2. IEM imaged in a Philips CM120 electron microscope (Eindhoven, The Netherlands) under 80 kV. PM, plasma membrane; WT, wild-type.
Dynamics of PfEMP1 suggest PI3P vesicular amplification in 0- to 3-hour ring stage of laboratory and clinical K13 mutants
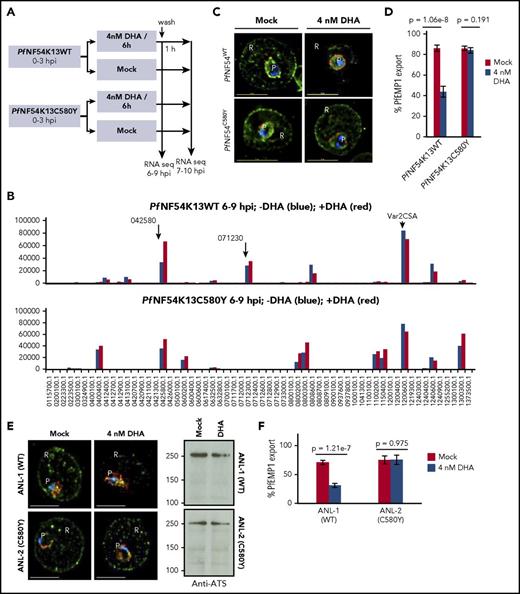
Dissemination of PI3P to intraerythrocytic vesicles and Maurer’s clefts in Figure 5K-L suggested that PfNF54K13C580Y mutants may affect export of parasite proteins to the host red cell. This led to the idea of tracking parasite protein export as a surrogate for PI3P vesicular amplification. This was particularly important for analyses of 0- to 3-hour rings, the stages of peak artemisinin resistance, but not amenable to quantitative IEM needed to assess quantitative distribution of the lipid PI3P. On the basis of our findings, PfEMP1 is a marker for K13/PI3P vesicles, but little is known about its expression in newly formed rings or modulation by short-term exposure to DHA, a potent PfPI3Kinase inhibitor that suppresses ring PI3P.10 Therefore (and as summarized in Figure 6A), studies were initiated in which 0- to 3-hour rings were exposed to DHA treatment (6 hours at 4 nM). We detected no significant effect on expression of either the main PfEMP1 (VAR2CSA) or additional minor PfEMP1 genes owing to mutation or drug (Figure 6B; supplemental Figure 6A-B). PfEMP1 export to the red cell was detected in the young rings (Figure 6C). Export was substantially reduced by 4 nM DHA in PfNF54K13WT parasites (Figure 6C-D). Quantitative analyses of 400 optical sections suggested a greater than 50% reduction in export (note that PI3P levels cannot be entirely depleted without compromising cell viability). In contrast, in PfNF54K13C580Y, 4 nM DHA failed to block PfEMP1 export (Figure 6C-D), suggesting sustained (and resistant) vesicular export in a strain in which PI3P is elevated. DHA inhibited PfEMP1 export in a sensitive Cambodian clinical strain (ANL-1) but not the clinically resistant counterparts ANL-2 K13C580Y and ANL-9 R539T (Figure 6E-F; supplemental Figure 6C). Removal of DHA restored export in sensitive ANL1 strains (supplemental Figure 6D). Additional PI3K inhibitors mimicked the effect of DHA on PfEMP1 export in sensitive and resistant clinical strains (supplemental Figure 6E).
Effect of K13 mutation and drug exposure on PfEMP1 expression and export. (A) Schematic of parasite drug exposure. (B) RNA sequence analyses of PfEMP1 expression in 0- to 3-hour PfNF54K13WT and PfNF54K13C580Y rings exposed to 4 nM DHA (positive, red) or mock treated (negative, blue) for 6 hours. Intensity units are as follows: y-axis, PfEMP1 gene id; x-axis, black arrows indicate a major transcript seen in WT parasites. The main PfEMP1 transcript 1200600 (var2csa) was expressed in both wild-type and mutant parasites +/−DHA. Second and third K13WT transcripts (0425800, 0712300) were also expressed in DHA. In C580Y, 0425800, 0400400, 0600200, 0800200, and 0300300 were expressed +/−DHA. PfEMP1 transcript levels were sustained an hour after DHA was washed out (supplemental Figure 5A-B), and parasites successfully matured through subsequent stages of the asexual life cycle (not shown). (C-D) DHA 4 nM potently inhibits PfEMP1 export (green; detected by ATS antibodies in IFA) to the red cell in artemisinin-sensitive PfNF54K13WT but not resistant PfNF54K13C580Y (C). Quantitative analyses of 400 optical sections through 30 infected red cells (D). (E-F) Quantitative analyses of sensitivity of PfEMP1 export (green) to DHA in the artemisinin-sensitive clinical strain ANL-1 but not its resistant clinical counterpart ANL-2 (C580Y). Western blots reveal equivalent PfEMP1 protein levels with or without DHA. For all samples, pixel intensity at 100% exceeded 6.5 × 106 (AU) (panels C-F). Experimental replicates: n = 2 (panels A-B); n = 2 (panels C-F). PfExp2 (Pf-exported protein 2, red), a parasitophorous vacuolar membrane maker, was used to stain the parasite periphery (panels C,E). Scale bars, 5 µm. Imaged with a 100×, NA-1.4 objective on an Olympus IX inverted fluorescence microscope using DeltaVision Deconvolution microscopy software.25 P, parasite; R red cell.
Effect of K13 mutation and drug exposure on PfEMP1 expression and export. (A) Schematic of parasite drug exposure. (B) RNA sequence analyses of PfEMP1 expression in 0- to 3-hour PfNF54K13WT and PfNF54K13C580Y rings exposed to 4 nM DHA (positive, red) or mock treated (negative, blue) for 6 hours. Intensity units are as follows: y-axis, PfEMP1 gene id; x-axis, black arrows indicate a major transcript seen in WT parasites. The main PfEMP1 transcript 1200600 (var2csa) was expressed in both wild-type and mutant parasites +/−DHA. Second and third K13WT transcripts (0425800, 0712300) were also expressed in DHA. In C580Y, 0425800, 0400400, 0600200, 0800200, and 0300300 were expressed +/−DHA. PfEMP1 transcript levels were sustained an hour after DHA was washed out (supplemental Figure 5A-B), and parasites successfully matured through subsequent stages of the asexual life cycle (not shown). (C-D) DHA 4 nM potently inhibits PfEMP1 export (green; detected by ATS antibodies in IFA) to the red cell in artemisinin-sensitive PfNF54K13WT but not resistant PfNF54K13C580Y (C). Quantitative analyses of 400 optical sections through 30 infected red cells (D). (E-F) Quantitative analyses of sensitivity of PfEMP1 export (green) to DHA in the artemisinin-sensitive clinical strain ANL-1 but not its resistant clinical counterpart ANL-2 (C580Y). Western blots reveal equivalent PfEMP1 protein levels with or without DHA. For all samples, pixel intensity at 100% exceeded 6.5 × 106 (AU) (panels C-F). Experimental replicates: n = 2 (panels A-B); n = 2 (panels C-F). PfExp2 (Pf-exported protein 2, red), a parasitophorous vacuolar membrane maker, was used to stain the parasite periphery (panels C,E). Scale bars, 5 µm. Imaged with a 100×, NA-1.4 objective on an Olympus IX inverted fluorescence microscope using DeltaVision Deconvolution microscopy software.25 P, parasite; R red cell.
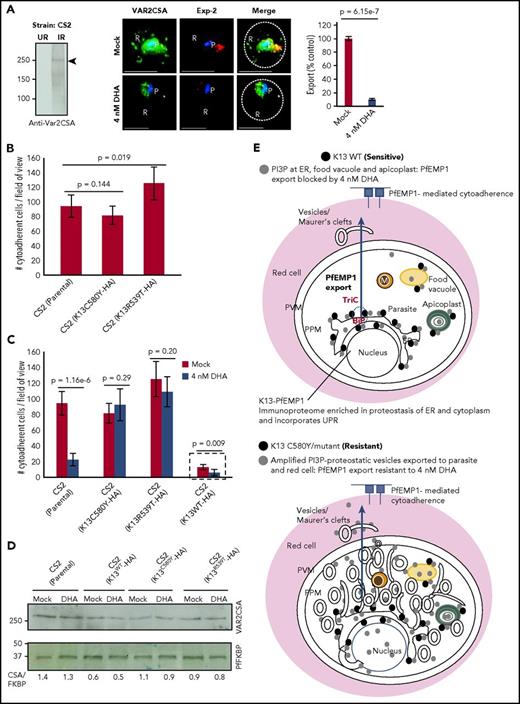
In the P falciparum CS2 strain,34,35 the export of VAR2CSA (a ligand for host chondroitin sulfate; CSA) was blocked by DHA and other PfPI3Kinase inhibitors (Figure 7A; supplemental Figure 7A-B). The conservation of the VAR2CSA promoter and gene suggest that its lack of responsiveness to DHA at a transcriptional level in PfNF54K13WT and PfNF54K13C580Y (Figure 6B; supplemental Figure 6A-B) should be conserved across strains.36 Because expression of VAR2CSA at the infected red cell surface confers adherence to chondroitin sulfate, we were interested in testing the effects of K13 mutation on cytoadherence of infected red cells. To do so, we created transgenic CS2 parasites expressing dominant-negative forms of HA-tagged K13C80Y and K13R539T (a strategy shown to be active in Pf3D710 ; construct design in supplemental Figure 7). Neither mutation had a significant effect on adhesin levels. Cytoadherence was unaffected by C580Y and may have been slightly increased by R539T (Figure 7B-C). DHA blocked cytoadherence of the parent CS2 strain but not of either K13 mutant and was without effect on VAR2CSA protein levels across all 3 strains. Transgenic expression of a second K13WT gene in the CS2 strain reduced levels of both cytoadherence and VAR2CSA. One explanation may be that K13 increases proteasomal targeting of PfPI3K to affect PI3P turnover, which in turn decreases VAR2CSA levels. DHA further depressed binding of transgenic K13WT strains (Figure 7C, hatched box; supplemental Figure 7) but without additional suppression of the VAR2CSA protein (Figure 7D). VAR2CSA levels in K13C580Y and K13R539T were not elevated in comparison with parental CS2 parasites, suggesting that PfEMP1 is not likely to be a substrate for K13. Mok et al12 reported that resistant clinical isolates progress from rings through trophozoite and schizont stages more slowly than do their sensitive counterparts. Because PfEMP1 export and adhesion increase at later stages of growth, the slower progression of mutants does not account for their higher levels of DHA-insensitive export of PfEMP1. Rather, it appears that PfEMP1-export dynamics in mutants with elevated PI3P are no longer suppressed by K13 regulation or PfPI3K inhibition. Together, our findings (shown in Figures 6 and 7) suggest that PfEMP1-export dynamics provide a surrogate for detecting PI3P-amplified vesiculation in early ring stages of K13 mutants across strains: both export of PfEMP1 to the red cell and cytoadherence of the infected red cells (later in life cycle) gain artemisinin resistance.
Effect of K13 mutation and drug exposure on export of VAR2CSA, cytoadherence, and models. (A) Antibodies to VAR2CSA detect ∼250 kDa in infected red cells but not in uninfected red cells by Western blots (left; see also supplemental Figure 6A). IFA and fluorescence quantitation (right) show inhibition of VAR2CSA (green) export to the red cell by 4nM DHA in artemisinin-sensitive CS2 strain, in relation to mock treatment. Red, Pf exported protein 2 marker of the parasitophorous vacuolar membrane; Hoechst (blue), parasite nucleus; dotted line, red cell periphery. (B) Parental CS2 parasites or transgenic CS2 expressing K13C580Y show similar levels of adherence to CSA; transgenic expression of K13R539T increases adherence slightly (see supplemental Figure 6C for construction of transgenic lines). (C) Potent inhibition of cytoadherence in parental CS2 by DHA was blocked by in trans expression of K13C580Y and K13R539T. Trans expression of K13WT in CS2 reduced cytoadherence, but DHA further decreased adherence (hatched box; see also supplemental Figure 6D; that HA-tagged K13WT and dominant-negative genes are functional was established by Mbengue et al in 201510 ). Means (±SDs) from 2 experimental replicates are shown (each with triplicate data points; P values as shown) (panels B-C). Imaged with a 100×, NA-1.4 objective on an Olympus IX inverted fluorescence microscope and quantified using DeltaVision Deconvolution microscopy software.25 (D) Western blots show 4 nM DHA does not block VAR2CSA expression in the CS2 strain (parental) or in transgenic CS2-expressing HA-tagged K13C580Y or K13R539T. Although transgenic K13WT reduces VAR2CSA levels by 50%, DHA does not cause further reduction. Experimental replicates: n = 2 (panels A-D). (E) Models integrate study findings of K13-dependent PI3P tubovesicular action in parasites sensitive and resistant to artemisinins. Key as shown. Green solid circles are apicoplasts; yellow solid circles are food vacuoles; orange solid circles are mitochondria. Scale bars, 5 µm. Exp-2, Pf exported protein 2 marker of the parasitophorous vacuolar membrane; IR, infected red cell; P, parasite; R, red cell; TriC, TCP1 ring complex; UR, uninfected red cell.
Effect of K13 mutation and drug exposure on export of VAR2CSA, cytoadherence, and models. (A) Antibodies to VAR2CSA detect ∼250 kDa in infected red cells but not in uninfected red cells by Western blots (left; see also supplemental Figure 6A). IFA and fluorescence quantitation (right) show inhibition of VAR2CSA (green) export to the red cell by 4nM DHA in artemisinin-sensitive CS2 strain, in relation to mock treatment. Red, Pf exported protein 2 marker of the parasitophorous vacuolar membrane; Hoechst (blue), parasite nucleus; dotted line, red cell periphery. (B) Parental CS2 parasites or transgenic CS2 expressing K13C580Y show similar levels of adherence to CSA; transgenic expression of K13R539T increases adherence slightly (see supplemental Figure 6C for construction of transgenic lines). (C) Potent inhibition of cytoadherence in parental CS2 by DHA was blocked by in trans expression of K13C580Y and K13R539T. Trans expression of K13WT in CS2 reduced cytoadherence, but DHA further decreased adherence (hatched box; see also supplemental Figure 6D; that HA-tagged K13WT and dominant-negative genes are functional was established by Mbengue et al in 201510 ). Means (±SDs) from 2 experimental replicates are shown (each with triplicate data points; P values as shown) (panels B-C). Imaged with a 100×, NA-1.4 objective on an Olympus IX inverted fluorescence microscope and quantified using DeltaVision Deconvolution microscopy software.25 (D) Western blots show 4 nM DHA does not block VAR2CSA expression in the CS2 strain (parental) or in transgenic CS2-expressing HA-tagged K13C580Y or K13R539T. Although transgenic K13WT reduces VAR2CSA levels by 50%, DHA does not cause further reduction. Experimental replicates: n = 2 (panels A-D). (E) Models integrate study findings of K13-dependent PI3P tubovesicular action in parasites sensitive and resistant to artemisinins. Key as shown. Green solid circles are apicoplasts; yellow solid circles are food vacuoles; orange solid circles are mitochondria. Scale bars, 5 µm. Exp-2, Pf exported protein 2 marker of the parasitophorous vacuolar membrane; IR, infected red cell; P, parasite; R, red cell; TriC, TCP1 ring complex; UR, uninfected red cell.
Discussion
Our studies reveal several new and unexpected findings on the mechanisms and consequences of the causal marker for artemisinin-resistant K13’s action in P falciparum and its host red blood cell. We show that K13 is a marker for parasite PI3P and associates with the virulence-determinant PfEMP1 exported to the red cell. K13 mutations of artemisinin resistance that increase PI3P also regulate the dynamics of PfEMP1 export to the host cell. The PfEMP1 K13-containing proteome was enriched in host-targeted PTEX machinery,37,38 and several convey cargo with relevant export signals.39-41 However, it inexplicably lacked low molecular weight and large antigenic families of rifins and stevors (instead of enriching them), suggesting that there may be additional steps of export selectivity that remain poorly understood.32,33,42 Importantly, PTEX may not regulate PI3P elaboration within the parasite, a primary resistance survival effect of K13 mutation. Dissemination of PI3P vesicles of proteostasis may also provide mechanisms for mitigating artemisinin damage to the host red cell. Further PfEMP1 export and cytoadherence become DHA resistant in K13 mutants (as detected in static assays, suggesting that even greater differences are expected between wild-type and mutant parasites under circulatory conditions). Therefore, in addition to protecting the infected red cell against proteopathy, resistant mutations may provide better adherence to host receptors and immune evasion, even in the presence of a drug. This may contribute to the persistence of resistant parasites at low and asymptomatic parasitemias despite mass drug administration.43,44
We also find that K13 and the parasite PI3P enrich at the ER and to a lesser degree fv and apicoplast. K13 is cytoplasmic, whereas PI3P also locates in the lumen. Because the parasite encodes for a single cytosolic PfPI3K,10 luminal import of PI3P may result from endovesiculation or yet-to-be-defined ATP-dependent lipid translocases (in yeast, PI3P is detected in the lumen of the autophagosome31 ). K13’s physiological function may be to control PI3P levels and turnover in the ER (and secondarily at the fv and apicoplast). PfEMP1, K13, and K13-mutant PI3P-tubovesicular enrichment in schizonts may be transferred via daughter merozoites into newly formed 0- to 3-hour rings, the stage of clinical artemisinin resistance at which PI3P is correspondingly increased and PfEMP1 export provides a protein surrogate to detect amplified vesicular export. K13 mutants at late trophozoite/schizont stages do not manifest artemisinin resistance in the RSA, but their proteostasis pathways are altered, and killing by artemisinins is maximal, suggesting the need to understand properties of K13 mutants at these stages (or correlates thereof) in in vivo human infections.
Finally, our data support the idea that amplified PI3P tubovesicles encompass protein proteostasis pathways and UPR and confer resistance, presumably by widely disseminating resistance intermediates to mitigate promiscuous artemisinin-mediated “proteopathy” (via protein alkylation and aggregation) throughout the infected red cell.15,22 In higher eukaryotes, PI3P expansion in the ER stimulates membrane curvature needed for macroautophagy to efficiently remove misfolded/toxic protein aggregates45-47 as well as unconventional autophagic secretion.48 However, ER autophagy remains poorly understood in malaria parasites,49,50 and its mechanistic analyses need further study, as do the functions of additional phosphoinositide genes recently identified as artemisinin resistance-associated malarial-gene loci.51 Elaborated PI3P tubules/vesicles reach all destinations in mutant infected red cells (Figure 7E), which may explain the complexity of artemisinin resistance as well as how a single K13-proteostatic determinant may reflect selective pressure in multiple organellar systems and hundreds of parasite genes implicated in resistance.15-23 Because PI3P elevation is sufficient for inducing resistance, expansion of PI3P vesicles may also account for K13-independent resistance.52 The vesicular resistance mechanism is likely to be effective against new drugs that induce toxicity through protein aggregates/proteopathy53,54 and may present many targets to facilitate resistance to partner drugs in multidrug-resistant malaria.55,56 Because PI3P also targets the red cell in K13 mutants, host factors may modulate artemisinin resistance, unlike known resistance mechanisms of major antimalarials presently in clinical use.57
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
All parasite gene/protein sequences were obtained from PlasmoDB (www.plasmodb.org). The authors would like to thank the technical competence of Kimberley Zichichi from the Microscopy Facility at Yale University.
This work was supported by grants from the National Institutes of Health, National Heart, Lung, and Blood Institute (HL069630, HL130330 [K.H.]) and National Institute of Allergy and Infectious Diseases (AI103548) (I.C.), the government of India (DST ECR/2015/000387), Department of Biotechnology Ramalingaswami Re-entry Fellowship (BT/HRD/35/02/2006), and University for Potential Excellence–II (Project ID 245) (S.B.). Support of the Wistar Proteomics and Metabolomics Core Facility was provided by a grant from the Cancer Center Support to the Wistar Institute (CA010815).
Authorship
Contribution: S.B., I.C., A.M., N.S., M.G., Z.S., K.H., H.-Y.T., and D.W.S. designed, performed, and interpreted the experimental work; R.V.S. helped design and interpret the experimental work; K.H., S.B., I.C., A.M., N.S., H.-Y.T., D.W.S., R.V.S., and N.M. wrote the manuscript; I.S. and N.M. provided key intellectual insight into aspects of this study; I.S. conducted and reviewed all of the statistical analyses; and all authors commented on the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for A.M. is Institut Pasteur of Dakar, Dakar, Senegal.
The current affiliation for M.G. is Wellcome Trust Sanger Institute, Cambridge, United Kingdom.
The current affiliation for Z.S. is Department of Chemical Engineering, University of Chemistry and Technology Prague, Prague, Czech Republic.
The current affiliation for R.V.S. is Department of Medicinal Chemistry and Molecular Pharmacology, Purdue University, West Lafayette, IN.
Correspondence: Kasturi Haldar, Boler-Parseghian Center for Rare and Neglected Diseases, University of Notre Dame, 103 Galvin Life Sciences, Notre Dame, IN 46556; e-mail: khaldar@nd.edu; and Souvik Bhattacharjee, Special Centre for Molecular Medicine, Jawaharlal Nehru University, New Delhi 110067, India; e-mail: souvik@jnu.ac.in.
References
Author notes
I.C., A.M., and N.S. contributed equally to this study.


![Figure 3. K13 copurified in a PfEMP1-immunoproteome enriched in proteostatic pathways that include the UPR, implicated in clinical artemisinin resistance. (A) Detection of PfEMP1 (arrow, left panel) and total protein content (right panel) captured by anti-ATS antibodies (but not in its absence at −1°) on covalently conjugated protein G beads. Molecular weight standards are in kDa. *Heavy IgG; **Light IgG. (B) Intensity properties of 2 replicate PfEMP1 immunoproteomes. (C) Distribution of proteins in the “503-proteome” comparing their abundance in each proteome expressed as a ratio (abundance ratio [AR] = abundance in Proteome 1/abundance in Proteome 2). X-axis: AR interval range; y-axis: number of proteins. (D) Hypergeometric analyses of 503-proteome and artemisinin-resistant transcriptome associated with clinical strains12 (for proteostasis enrichment and more detail, see supplemental Tables 2-4; Figure 3E-G). (E) Distribution of normalized intensity (ni; intensity divided by molecular weight) of the 207-proteome (AR = 0.5-1.99). Reactive oxidative stress complex components are BiP and PDI-11 (ni 5.17e8 and 1.2e7, respectively; supplemental Table 5). For detailed annotation and proteostasis enrichment, see supplemental Table 4 and supplemental Figure 3H-I. (F) Model integrating K13-PI3P tubules/vesicles (black-gray spheres based on findings of Figures 1 and 2 and supplemental Figures 1 and 2) and K13-PfEMP1 immunoproteome (green; based on findings in Figure 3; supplemental Figure 3) in proteostatic mechanisms of vesicular export and protein quality control and folding in the ER and cytoplasm (dotted circle). Experimental replicates, n = 2. Other organelles and key are as shown. HT, host targeting sequence; PEXEL, plasmodium export element; PNEP, PEXEL-negative proteins; PPM, parasite plasma membrane; PVM, parasitophorous vacuolar membrane; ROSC, reactive oxidative stress complex.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/131/11/10.1182_blood-2017-11-814665/4/m_blood814665f3.jpeg?Expires=1767712774&Signature=gDlXplDrqjM-j-Vj8iFprenZslNkdznwAkLEF3NhxYnoH5kSDKFBw7yp~HrzXIfQ1rQTwEx1ML5xmzD~zulugc0r0-vT731EuoKRgOgsxKMFHH5QDfGnyBCaklap34wuOuOtqvtGCwB7lWnt8gbiSjhdTjaFfRGFbq-Ai0AVcp9FEzCWnLUAwDi-mRzSXvhUQz5BeQkuliOSEoSvxwXMPx9fKh7VhUqGgjeitLdX8yFsitNnSd1Lqde4rB9~MmhJvsPWBLTxYDwKPK2xwvjzBD3FeL8F9w3I6rL78~6EEhtX2zBYoVc28L6pjmOgHWfUf5rJj1BV2YGU6NLPl2CpQA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal