TO THE EDITOR:
Acute myeloid leukemia (AML) is a heterogeneous disease for which genetic profiles dictate clinical outcomes. Cytogenetic and molecular profiling in AML is a mandatory diagnostic and prognostic requirement, yet interpretation of these results is becoming increasingly complex. Next generation sequencing (NGS) technologies have enabled comprehensive characterization of the genomic landscape of AML, revealing complex patterns of clonal evolution during development and treatment.1,2 The datasets generated represent invaluable resources to interrogate the genomic information from large patient cohorts and identify clinically relevant molecular biomarkers of disease initiation, progression, and response to treatment.3,4
The 2017 update of the European LeukemiaNet recommendations (ELN 2017) on genetic risk stratification provides a comprehensive genomic classification and prognostication schema, updating the European LeukemiaNet 2010 recommendations (ELN 2010).5 A key change in ELN 2017 has been to reclassify nucleophosmin 1 mutant (NPM1mut) AML with a low–allelic-ratio FLT3-internal tandem duplication (FLT3ITD-L; allelic ratio [AR] < 0.5) mutation as favorable risk, along with AML with NPM1mut (not containing a FLT3-internal tandem duplication [FLT3ITD] mutation), biallelic CEBPA mutations, and core-binding factor AMLs. The evidence supporting NPM1mutFLT3ITD-L as favorable risk is conflicting, with several groups showing a favorable outcome comparable with NPM1mutFLT3 wild-type (FLT3wt) disease,6-9 and others demonstrating an intermediate prognosis,10-12 although none of these studies have compared survival of NPM1mutFLT3ITD-L within an overall-risk stratification schema. The published survival differences may be explained by varying FLT3ITD AR thresholds, patient characteristics, clinical treatment algorithms, or methods of FLT3ITD detection and quantification (supplemental Methods; supplemental Table 1; available on the Blood Web site), but in aggregate, the literature does not provide a consistent message regarding the outcome or optimal management of patients with NPM1mutFLT3ITD-L AML. The gold standard assay to identify FLT3ITD uses polymerase chain reaction–amplified products processed by capillary electrophoresis13 and conventionally uses genomic DNA, but can use complementary DNA with good concordance.14 FLT3ITD detection and quantification by NGS are challenging due to short DNA sequence fragments, hence the frequency or AR may be underestimated and poorly correlates with fragment based analysis.15
We therefore sought to compare the performance of ELN 2017 with ELN 2010 in 2409 AML patients from 3 publicly available data sets (German-Austrian AML Study Group1 [AMLSG], n = 1316; The Cancer Genome Atlas2 [TCGA], n = 150, and Therapeutically Applicable Research To Generate Effective Treatments15 [TARGET] pediatric AML, n = 943) with comprehensive genomic and clinical data. In particular, we aimed to examine the prognosis of the NPM1mutFLT3ITD-L subgroup in the overall context of ELN 2017.
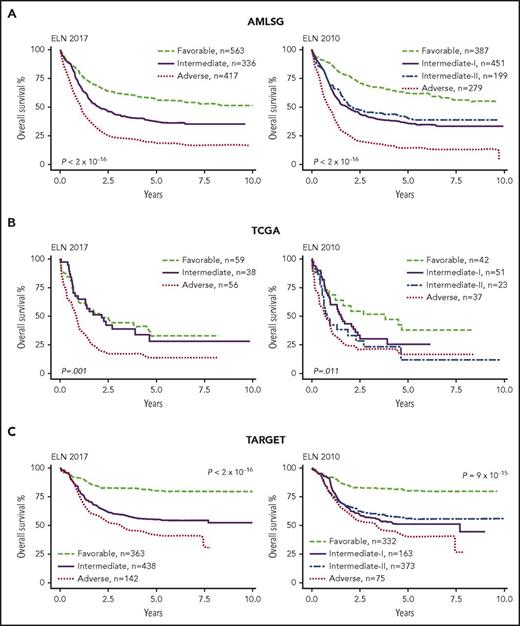
We first compared Kaplan-Meier overall survival (OS) curves between ELN 2010 and ELN 2017. ELN 2017 stratified the AMLSG cohort into significantly different favorable, intermediate, and adverse survival risk groups (Figure 1A). In the TCGA cohort, ELN 2017 failed to identify a favorable-risk group, with overlapping favorable and intermediate risk survival curves (P = .8, Figure 1B). Across both adult cohorts, the favorable-risk group had a 5% to 6% inferior 5-year OS rate in ELN 2017 compared with ELN 2010 (supplemental Table 2). Interestingly, the TCGA cohort had inferior survival compared with AMLSG across all risk groups, especially in the favorable category (5-year OS, 57% [AMLSG] vs 33% [TCGA]) with the TCGA favorable group performing similarly to the AMLSG intermediate group (5-year OS, 33%). Given these stark differences in survival, the 2 adult cohorts were not pooled for further analysis. Although ELN 2017 was designed for adult AML, it was able to significantly separate 3 risk groups in the pediatric TARGET population with only minor differences in OS between ELN 2010 and ELN 2017 (Figure 1C; supplemental Table 2). TARGET risk groups had superior OS compared with adult counterparts. These data validate ELN 2017 and ELN 2010 across a broad spectrum of patient ages and AML subtypes.
ELN 2017 and ELN 2010 Kaplan-Meier OS curves for AMLSG, TCGA, and TARGET data. (A) AMLSG ELN 2017 and ELN 2010 stratification. Cox regression on ELN 2017 revealed a significantly different HR using the Wald test statistic for all 3 risk groups (favorable vs intermediate: P = 3 × 10−12; HR, 1.7; 95% CI, 1.4-1.9; favorable vs adverse: P < 2 × 10−16; HR, 3.2; 95% CI, 2.8-3.7; intermediate vs adverse: P < 2 × 10−16; HR, 2; 95% CI, 1.7-2.2). ELN 2010 was also able to stratify the cohort into risk groups (favorable vs intermediate-I: P = 2 × 10−14; HR, 2.2; 95% CI, 1.8-2.7; favorable vs intermediate-II: P = 3 × 10−7; HR, 1.9; 95% CI, 1.5-2.4; favorable vs adverse: P < 2 × 10−16; HR, 4.2; 95% CI, 3.5-5.2; intermediate-I vs adverse: P = 2 × 10−14; HR, 2; 95% CI, 1.7-2.3; intermediate-II vs adverse: P = 2 × 10−12; HR, 2; 95% CI, 1.8-2.8); without difference between the intermediate-I and -II groups (P = .23). (B) In the TCGA cohort, ELN 2017 could stratify the favorable and intermediate risk groups from the adverse risk group (favorable vs adverse: P = .001; HR, 2.1; 95% CI, 1.3-3.3; intermediate vs adverse: P = .008; HR, 2; 95% CI, 1.2-3.2), but failed to stratify the favorable from the intermediate risk groups (P = .8). ELN 2010 successfully stratified the favorable from the intermediate-II and adverse risk groups (P = .03; HR, 2.2; 95% CI, 1.1-3.8; P = .002; HR, 2.4; 95% CI, 1.4-4.1), but not the favorable from the intermediate-I risk group (P = .2) or the intermediate-I and intermediate-II from the adverse risk group (P = .06; P = .6). (C) In the TARGET pediatric cohort, the ELN 2017 was again able to separate the 3 risk groups (favorable vs intermediate: P = 2 × 10−12; HR, 2.7; 95% CI, 2.1-3.6; favorable vs adverse: P = 3 × 10−16; HR, 3.8; 95% CI, 2.8-5.2; intermediate vs adverse: P = .008; HR, 1.4; 95% CI, 1.1-1.8). ELN 2010 was also able to stratify the favorable from the intermediate and adverse groups (favorable vs intermediate-I: P = 3 × 10−10; HR, 2.9; 95% CI, 2.1-4.1; favorable vs intermediate-II: P = 7 × 10−11; HR, 2.6; 95% CI, 2-3.5; favorable vs adverse: P = 7 × 10−12; HR, 3.8; 95% CI, 2.6-5.7; and intermediate-II vs adverse: P = .02; HR, 1.5; 95% CI, 1-2), but not the intermediate-I from the adverse risk group (P = .14) or the intermediate-I from the intermediate-II risk group (P = .46).
ELN 2017 and ELN 2010 Kaplan-Meier OS curves for AMLSG, TCGA, and TARGET data. (A) AMLSG ELN 2017 and ELN 2010 stratification. Cox regression on ELN 2017 revealed a significantly different HR using the Wald test statistic for all 3 risk groups (favorable vs intermediate: P = 3 × 10−12; HR, 1.7; 95% CI, 1.4-1.9; favorable vs adverse: P < 2 × 10−16; HR, 3.2; 95% CI, 2.8-3.7; intermediate vs adverse: P < 2 × 10−16; HR, 2; 95% CI, 1.7-2.2). ELN 2010 was also able to stratify the cohort into risk groups (favorable vs intermediate-I: P = 2 × 10−14; HR, 2.2; 95% CI, 1.8-2.7; favorable vs intermediate-II: P = 3 × 10−7; HR, 1.9; 95% CI, 1.5-2.4; favorable vs adverse: P < 2 × 10−16; HR, 4.2; 95% CI, 3.5-5.2; intermediate-I vs adverse: P = 2 × 10−14; HR, 2; 95% CI, 1.7-2.3; intermediate-II vs adverse: P = 2 × 10−12; HR, 2; 95% CI, 1.8-2.8); without difference between the intermediate-I and -II groups (P = .23). (B) In the TCGA cohort, ELN 2017 could stratify the favorable and intermediate risk groups from the adverse risk group (favorable vs adverse: P = .001; HR, 2.1; 95% CI, 1.3-3.3; intermediate vs adverse: P = .008; HR, 2; 95% CI, 1.2-3.2), but failed to stratify the favorable from the intermediate risk groups (P = .8). ELN 2010 successfully stratified the favorable from the intermediate-II and adverse risk groups (P = .03; HR, 2.2; 95% CI, 1.1-3.8; P = .002; HR, 2.4; 95% CI, 1.4-4.1), but not the favorable from the intermediate-I risk group (P = .2) or the intermediate-I and intermediate-II from the adverse risk group (P = .06; P = .6). (C) In the TARGET pediatric cohort, the ELN 2017 was again able to separate the 3 risk groups (favorable vs intermediate: P = 2 × 10−12; HR, 2.7; 95% CI, 2.1-3.6; favorable vs adverse: P = 3 × 10−16; HR, 3.8; 95% CI, 2.8-5.2; intermediate vs adverse: P = .008; HR, 1.4; 95% CI, 1.1-1.8). ELN 2010 was also able to stratify the favorable from the intermediate and adverse groups (favorable vs intermediate-I: P = 3 × 10−10; HR, 2.9; 95% CI, 2.1-4.1; favorable vs intermediate-II: P = 7 × 10−11; HR, 2.6; 95% CI, 2-3.5; favorable vs adverse: P = 7 × 10−12; HR, 3.8; 95% CI, 2.6-5.7; and intermediate-II vs adverse: P = .02; HR, 1.5; 95% CI, 1-2), but not the intermediate-I from the adverse risk group (P = .14) or the intermediate-I from the intermediate-II risk group (P = .46).
The recategorization of NPM1mutFLT3ITD-L as favorable in ELN 2017, compared with intermediate-I risk in ELN 2010 likely explains the inferior survival for favorable-risk adults in ELN 2017. In the AMLSG ELN 2017 favorable-risk cohort (n = 564), 159 patients (28%) had NPM1mutFLT3ITD-L and 207 (38%) had NPM1mut alone. NPM1mutFLT3ITD-L patients overall exhibited high-risk features, including raised lactate dehydrogenase, white blood cell (WBC) counts, and peripheral blood and bone marrow (BM) blast percentage. In general, WBC counts were FLT3ITD AR dependent, increasing from FLT3wt to high–allelic ratio FLT3ITD (supplemental Figure 1). In the TCGA ELN 2017 favorable-risk cohort, 26 patients (17%) had NPM1mut and 20 (13%) had NPM1mutFLT3ITD-L. Patients were generally older in the TCGA cohort compared with the AMLSG cohort. NPM1mutFLT3ITD-L patients had higher WBC counts and BM blasts, although this difference was not statistically significant. Interestingly, the median WBC count in the TCGA cohort was unexpectedly high in the NPM1mutFLT3ITD-L subgroup at 64 × 109/L (supplemental Figure 1). Concomitant DNMT3A mutations were found in approximately half of both NPM1mutFLT3ITD-L and NPM1mut patients from each cohort. Mutations resulting in activation of signaling pathways (NRAS, PTPN11, and FLT3 tyrosine kinase domain) were enriched in NPM1mut compared with NPM1mutFLT3ITD-L in both cohorts (AMLSG, P = 2.7 × 10−10, TCGA, P = .003). The pediatric cohort included 44 (4.7%) patients with the NPM1mut alone and 17 (1.8%) NPM1mutFLT3ITD-L patients. Differences in leukemia burden were not apparent between the 2 groups. Patients with NPM1mutFLT3ITD-L were significantly older (median age, 15.9 vs 12.8 years, P = .023). Consistently, we observed an age-related increase in the incidence of NPM1 and FLT3ITD mutations, and patients with both mutations were the oldest (supplemental Figure 2), confirming findings in other cohorts.16,17 The characteristics of each cohort are further described in supplemental Tables 3 and 4.
We examined OS and relapse risk (RR) of patients with NPM1mutFLT3ITD-L and NPM1mut alone. The presence of NPM1mutFLT3ITD-L was associated with inferior OS (Figure 2A) and increased RR (supplemental Figure 3) in the AMLSG cohort, but not in the TCGA or TARGET cohorts. This was confirmed in a multivariate analysis (adjusted P = .005; hazard ratio [HR] = 1.7; 95% confidence interval [CI], 1.2-2.3; supplemental Figure 4). In the TCGA cohort, the outcomes were unexpectedly poor in both groups with median survival <2 years and, together with the higher WBC counts, suggests the selection of AML patients with relatively high biological risk. NPM1mut and FLT3ITD are found at a lower frequency in childhood (range, 5%-8%) compared with adults with AML.18,19 In the TARGET cohort, both NPM1mut and NPM1mutFLT3ITD-L patients had equally good prognoses. Other significant adverse risk factors identified in multivariate analysis of AMLSG included increasing age and WBC count in intermediate–cytogenetic risk patients and DNMT3A mutations specifically within the NPM1mutFLT3ITD-L group, with the latter finding confirmed in survival analyses (supplemental Figure 5). Lower numbers limited the power of multivariate analysis in the TCGA and TARGET cohorts.
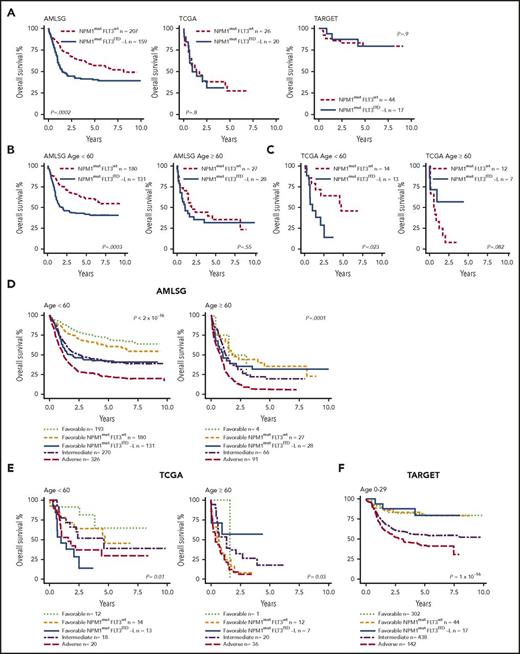
Impact of FLT3ITD-L and age in NPM1-mutated AML on OS and relapse-free survival in ELN 2017 favorable-risk patients. (A) In the AMLSG cohort, there was a significant difference in survival between NPM1mutFLT3wt patients vs NPM1mutFLT3ITD-L patients (AR < 0.5). This difference is not seen in the TCGA and TARGET cohorts. The AMLSG (B) and TCGA (C) cohorts were stratified by age. Younger (<60 years) patients with NPM1mutFLT3ITD-L had outcomes inferior to NPM1mutFLT3wt patients in both cohorts. In older (≥60 years) patients in both cohorts, NPM1mutFLT3wt appeared to lose its favorable influence, with both groups having similarly poor outcomes. (D) Comparison of ELN 2017 favorable NPM1mut FLT3ITD-L patients with other ELN 2017 risk groups in the AMLSG cohorts. NPM1mutFLT3ITD-L patients <60 years showed an increased risk of death compared with other favorably classified patients (P = 8.7 × 10−8; HR, 2.5; 95% CI, 1.8-3.5), whereas there was no difference compared with the intermediate risk group (P = .76). NPM1mutFLT3wt patients showed no difference from other favorably classified patients (P = .07), but a significantly decreased risk compared with the intermediate risk group (P = 8.8 × 10−5; HR, 0.6; 95% CI, .4-.8). In patients ≥60 years, no statistical difference was observed between favorable, intermediate, NPM1mutFLT3wt, and NPM1mutFLT3ITD subgroups. (E) Similar trends were observed for the TCGA cohort in patients <60 years, with NPM1mutFLT3ITD-L patients having a significantly poorer survival compared with the favorable (P = .004; HR = 6.9; 95% CI, 1.9-25.7) and intermediate groups (P = .03; HR, 2.8; 95% CI, 1.1-7). In this cohort, survival of NPM1mutFLT3wt patients was not statistically different to favorable (P = .3) or intermediate risk groups (P = .8). In patients ≥60 years, there were also no significant differences between the favorable, intermediate, and NPM1mutFLT3wt and NPM1mutFLT3ITD-L subgroups. (F) Conversely, in the TARGET pediatric cohort, NPM1mutFLT3wt (P = .98) and NPM1mutFLT3ITD-L (P = .87) patients had similar survival to other favorably classified patients. Although the NPM1mutFLT3wt group had a significantly decreased risk of death compared with intermediate-classified patients (P = .006; HR, 0.37; 95% CI, 0.28-0.5), the difference between the intermediate and NPM1mutFLT3ITD-L groups did not reach significance (P = .06). All P values displayed in the graphs are the result of a log rank test statistic to assess the global statistical significance of the model.
Impact of FLT3ITD-L and age in NPM1-mutated AML on OS and relapse-free survival in ELN 2017 favorable-risk patients. (A) In the AMLSG cohort, there was a significant difference in survival between NPM1mutFLT3wt patients vs NPM1mutFLT3ITD-L patients (AR < 0.5). This difference is not seen in the TCGA and TARGET cohorts. The AMLSG (B) and TCGA (C) cohorts were stratified by age. Younger (<60 years) patients with NPM1mutFLT3ITD-L had outcomes inferior to NPM1mutFLT3wt patients in both cohorts. In older (≥60 years) patients in both cohorts, NPM1mutFLT3wt appeared to lose its favorable influence, with both groups having similarly poor outcomes. (D) Comparison of ELN 2017 favorable NPM1mut FLT3ITD-L patients with other ELN 2017 risk groups in the AMLSG cohorts. NPM1mutFLT3ITD-L patients <60 years showed an increased risk of death compared with other favorably classified patients (P = 8.7 × 10−8; HR, 2.5; 95% CI, 1.8-3.5), whereas there was no difference compared with the intermediate risk group (P = .76). NPM1mutFLT3wt patients showed no difference from other favorably classified patients (P = .07), but a significantly decreased risk compared with the intermediate risk group (P = 8.8 × 10−5; HR, 0.6; 95% CI, .4-.8). In patients ≥60 years, no statistical difference was observed between favorable, intermediate, NPM1mutFLT3wt, and NPM1mutFLT3ITD subgroups. (E) Similar trends were observed for the TCGA cohort in patients <60 years, with NPM1mutFLT3ITD-L patients having a significantly poorer survival compared with the favorable (P = .004; HR = 6.9; 95% CI, 1.9-25.7) and intermediate groups (P = .03; HR, 2.8; 95% CI, 1.1-7). In this cohort, survival of NPM1mutFLT3wt patients was not statistically different to favorable (P = .3) or intermediate risk groups (P = .8). In patients ≥60 years, there were also no significant differences between the favorable, intermediate, and NPM1mutFLT3wt and NPM1mutFLT3ITD-L subgroups. (F) Conversely, in the TARGET pediatric cohort, NPM1mutFLT3wt (P = .98) and NPM1mutFLT3ITD-L (P = .87) patients had similar survival to other favorably classified patients. Although the NPM1mutFLT3wt group had a significantly decreased risk of death compared with intermediate-classified patients (P = .006; HR, 0.37; 95% CI, 0.28-0.5), the difference between the intermediate and NPM1mutFLT3ITD-L groups did not reach significance (P = .06). All P values displayed in the graphs are the result of a log rank test statistic to assess the global statistical significance of the model.
Given the older age and relative poor survival of the favorable group in the TCGA cohort, we performed separate analyses for patients ≥60 years of age and patients <60 years of age in the AMLSG and TCGA cohorts (Figure 2B-C). In patients <60 years, NPM1mutFLT3ITD-L conferred a poorer prognosis in both the AMLSG (P = .0003) and the TCGA cohort (P = .023), similar to or worse than ELN 2017 intermediate-risk patients in the respective cohorts (Figure 2D-E). Conversely, for patients ≥60 years, NPM1mutFLT3ITD-L and NPM1mut alone groups had similar, poor outcomes. The poor outcome in NPM1mutFLT3wt patients ≥60 years highlights the absence of an absolute favorable-risk cohort and is consistent with other cohorts.20 There were limited numbers of adolescent and young adult (AYA; age, 18-30 years) patients with AML available for review, and results did not reach statistical significance, however, there was a trend toward inferior outcomes in AYA patients with NPM1mutFLT3ITD-L compared with NPM1mutFLT3wt AML (data not shown). In pediatric AML, we observed that NPM1mutFLT3ITD-L did not confer adverse prognosis compared with other favorable genotypes (Figure 2F). Although this may reflect the high rate of allogeneic stem cell transplantation (SCT) in the NPM1mutFLT3ITD-L group (52.9% in TARGET vs 11.4% in NPM1mutFLT3wt, P = .0017), the favorable prognosis of the FLT3ITD-L subgroup even without NPM1mut in pediatric cohorts, but not in adults, requires further study to determine whether this reflects disease biology or treatment.
Targeted inhibitors of FLT3 have been developed in an attempt to overcome the negative prognostic impact of FLT3ITD in AML. Despite substantial single-agent activity, early randomized studies of FLT3 inhibitors together with chemotherapy in elderly patients failed to demonstrate a survival advantage of this approach.21 The recent RATIFY study22 combined the multikinase inhibitor midostaurin with intensive induction and consolidation chemotherapy in FLT3mut AML, and followed this with ≤12 months of maintenance midostaurin treatment. In this cohort (age <60 years), midostaurin led to a survival benefit in FLT3ITD AML, leading to the international registration of midostaurin for the treatment of FLT3mut AML in combination with intensive chemotherapy and maintenance.
Altogether, these data raise important considerations regarding the application of ELN 2017 that have significant implications for patient selection for allogeneic SCT or alternative novel therapies. Adults with NPM1mutFLT3ITD-L have inferior survival to other favorable-risk patients, and concurrent DNMT3A mutations, as a potential risk modifier in this group, should be prospectively examined. Furthermore, patients ≥60 years of age with NPM1 mutations, with or without FLT3ITD, have adverse outcomes and should not be considered favorable risk. More broadly, this study demonstrates the power of using publicly available datasets, strengthening the call for increased responsible data sharing.
The online version of this article contains a data supplement.
Acknowledgments
The authors thank Elli Papaemmanuil (Memorial Sloan Kettering Cancer Center), Nic Waddell (QIMR Berghofer Medical Research Institute), and Cameron Curley (Royal Brisbane and Women’s Hospital) for critical review and helpful comments. The results shown here are, in part, based on data generated by the TCGA Research Network (http://cancergenome.nih.gov/).
S.W.L. is a CSL Centenary Fellow and acknowledges the support of the Gordon and Jessie Gilmour Leukaemia Research Fund.
Authorship
Contribution: J.S. performed the analysis and wrote the manuscript; V.Y.L. and G.R.H. wrote the manuscript; and S.W.L. designed the research and wrote the manuscript.
Conflict-of-interest disclosure: S.W.L. is a consultant for Novartis (midostaurin). The remaining authors declare no competing financial interests.
Correspondence: Steven W. Lane, Translational Leukaemia Research Laboratory, QIMR Berghofer Medical Research Institute, Herston Rd, Herston, QLD 4006, Australia; e-mail: steven.lane@qimrberghofer.edu.au.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal