Abstract
Natural killer (NK) cells are lymphocytes of innate immunity that respond to virus infected and tumor cells. After allogeneic transplantation, NK cells are the first reconstituting lymphocytes, but are dysfunctional. Manipulating this first wave of lymphocytes could be instrumental in reducing the 40% relapse rate following transplantation with reduced-intensity conditioning. NK cells express numerous activating and inhibitory receptors. Some recognize classical or nonclassical HLA class I ligands, others recognize class I–like ligands or unrelated ligands. Dominant in the NK-cell transplant literature are killer cell immunoglobulin-like receptors (KIRs), encoded on chromosome 19q. Inhibitory KIR recognition of the cognate HLA class I ligand is responsible for NK-cell education, which makes them tolerant of healthy cells, but responsive to unhealthy cells having reduced expression of HLA class I. KIR A and KIR B are functionally distinctive KIR haplotype groups that differ in KIR gene content. Allogeneic transplant donors having a KIR B haplotype and lacking a recipient HLA-C epitope provide protection against relapse from acute myeloid leukemia. Cytomegalovirus infection stimulates and expands a distinctive NK-cell population that expresses the NKG2C receptor and exhibits enhanced effector functions. These adaptive NK cells display immune memory and methylation signatures like CD8 T cells. As potential therapy, NK cells, including adaptive NK cells, can be adoptively transferred with, or without, agents such as interleukin-15 that promote NK-cell survival. Strategies combining NK-cell infusions with CD16-binding antibodies or immune engagers could make NK cells antigen specific. Together with checkpoint inhibitors, these approaches have considerable potential as anticancer therapies.
NK-cell biology and genetics
Natural killer (NK) cells, effector lymphocytes of innate immunity, represent 10% to 20% of peripheral blood mononuclear cells. NK cells respond to virus-infected and malignant cells, without requiring prior sensitization,1 and play key roles in autoimmunity and pregnancy.2 To recognize targets in a specific manner, NK cells integrate signals triggered by interaction of target cell ligands with an array of activating and inhibitory NK-cell receptors (Table 1). These signals activate NK cells to kill target cells, both directly using perforin and granzyme B, and indirectly by antibody-mediated cellular cytotoxicity (ADCC), in which antibody crosslinks the target cell to the Fc receptor of the NK cell (CD16). Secretion of chemokines and cytokines, including tumor necrosis factor-α and interferon-γ (IFN-γ), is also induced by NK-cell activation. By upregulating HLA class I in surrounding tissue, IFN-γ bridges between innate and adaptive immunity.3 It enhances target cell recognition by CD8 T cells and skews CD4 T cells toward a T-cell helper type 1 (TH1) phenotype.4 Further promoting NK-cell cytolysis and IFN-γ secretion are the cytokines: type I IFNs, interleukin-2 (IL-2), IL-18, and IL-15, which are secreted by dendritic cells, macrophages, and infected tissue cells. In all of these ways, NK cells contribute to the immune response against cancer and infection.
Human NK-cell receptors and their ligands
| Receptor . | Ligand . |
|---|---|
| Inhibitory receptors | |
| KIR2DL1 | C2 HLA-C |
| KIR2DL2 | C1 HLA-C |
| KIR2DL3 | C1 HLA-C |
| KIR2DL4 | HLA-G |
| KIR2DL5 | Not known |
| KIR3DL1 | Bw4 HLA-A and -B |
| KIR3DL2 | HLA-A3 and -A11 |
| KIR3DL3 | Not known |
| NKG2A | HLA-E |
| LIR-1 | HLA class I |
| Activating receptors | |
| KIR2DS1 | C2 HLA-C |
| KIR2DS2 | HLA-A11 |
| KIR2DS3 | Not known |
| KIR2DS4 | HLA-A11 |
| KIR2DS5 | C2 HLA-C |
| KIR3DS1 | HLA-F |
| NKG2C | HLA-E |
| NKG2D | MICA, MICB, ULBP1-4 |
| CD16 | IgG 1, 3, 4 |
| NKp30 | B7-H6, BAT3, CMV pp65 |
| NKp44 | Viral hemagglutinins |
| NKp46 | Viral hemagglutinins |
| NKp80 | AICL |
| CD160 | HLA-C |
| DNAM-1 | CD112, CD155 |
| CD96 | CD155, CD111 |
| 2B4 | CD48 |
| CRACC | CRACC |
| NTB-A | NTB-A |
| Receptor . | Ligand . |
|---|---|
| Inhibitory receptors | |
| KIR2DL1 | C2 HLA-C |
| KIR2DL2 | C1 HLA-C |
| KIR2DL3 | C1 HLA-C |
| KIR2DL4 | HLA-G |
| KIR2DL5 | Not known |
| KIR3DL1 | Bw4 HLA-A and -B |
| KIR3DL2 | HLA-A3 and -A11 |
| KIR3DL3 | Not known |
| NKG2A | HLA-E |
| LIR-1 | HLA class I |
| Activating receptors | |
| KIR2DS1 | C2 HLA-C |
| KIR2DS2 | HLA-A11 |
| KIR2DS3 | Not known |
| KIR2DS4 | HLA-A11 |
| KIR2DS5 | C2 HLA-C |
| KIR3DS1 | HLA-F |
| NKG2C | HLA-E |
| NKG2D | MICA, MICB, ULBP1-4 |
| CD16 | IgG 1, 3, 4 |
| NKp30 | B7-H6, BAT3, CMV pp65 |
| NKp44 | Viral hemagglutinins |
| NKp46 | Viral hemagglutinins |
| NKp80 | AICL |
| CD160 | HLA-C |
| DNAM-1 | CD112, CD155 |
| CD96 | CD155, CD111 |
| 2B4 | CD48 |
| CRACC | CRACC |
| NTB-A | NTB-A |
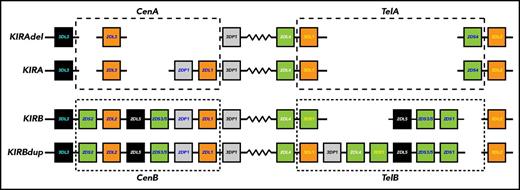
NK cells express combinations of cell surface receptors in stochastic fashion, giving rise to numerous, functionally distinct subsets of cells.5 Such expression is best described for the killer cell immunoglobulin-like receptors (KIRs). The family of 14 polymorphic KIR genes includes 7 encoding inhibitory receptors (3DL1-3, 2DL1-3, 2DL5), 6 encoding activating receptors (3DS1, 2DS1-5), and 1 that has inhibitory and activating potential (2DL4). Diverse KIR haplotypes differ in their gene and allele content and form 2 functional groups (Figure 1). The common KIR A haplotype comprises KIR3DL3-2DL3-2DP1-2DL1-3DP1-2DL4-3DL1-2DS4-3DL2 and a less common variant lacks KIR2DP1 and 2DL1. KIR B haplotypes are characterized by their variable gene content and presence of 1 or more of 7 B-specific genes: KIR2DS1, 2DS2, 2DS3, 2DS5, 2DL2, 2DL5, and 3DS1. Thus, 5 activating KIRs are associated with KIR B, whereas KIR2DS4 is the only activating receptor associated with KIR A. KIR haplotypes include 4 framework genes that define both the centromeric region, with KIR3DL3 at its 5′ end and KIR3DP1 at its 3′ end, and the telomeric region, with KIR2DL4 at its 5′ end and KIR3DL2 at its 3′ end.
KIR A and KIR B haplotypes of the human KIR locus. Human KIR haplotypes differ in their content of KIR genes and in the relative number of genes coding for activating and inhibitory KIR. Although the human population has numerous different KIR haplotypes they divide into 2 functionally distinctive groups. These group A and B KIR haplotypes exhibit different correlations with a spectrum of diseases, as well as the outcomes of HCT and other forms of immunotherapy. Shown are gene maps for 2 A and 2 B KIR haplotypes, which represent the overall diversity of KIR haplotypes. Each box represents a KIR gene, for which the shading gives the nature of the encoded protein: green, activating KIR; orange, inhibitory KIR; black, KIR of unknown function: gray, pseudogene, no KIR. Human KIR are of 4 evolutionary lineages, which are distinguished by the color of the label in the gene box: white, lineage I; yellow, lineage II; dark blue, lineage III; cyan, lineage V. The zigzag joining the centromeric and telomeric regions is an extended repetitive sequence and a hotspot for reciprocal recombination. Within the telomeric and centromeric regions the KIR genes are separated by short homologous sequences of a few hundred base pairs.
KIR A and KIR B haplotypes of the human KIR locus. Human KIR haplotypes differ in their content of KIR genes and in the relative number of genes coding for activating and inhibitory KIR. Although the human population has numerous different KIR haplotypes they divide into 2 functionally distinctive groups. These group A and B KIR haplotypes exhibit different correlations with a spectrum of diseases, as well as the outcomes of HCT and other forms of immunotherapy. Shown are gene maps for 2 A and 2 B KIR haplotypes, which represent the overall diversity of KIR haplotypes. Each box represents a KIR gene, for which the shading gives the nature of the encoded protein: green, activating KIR; orange, inhibitory KIR; black, KIR of unknown function: gray, pseudogene, no KIR. Human KIR are of 4 evolutionary lineages, which are distinguished by the color of the label in the gene box: white, lineage I; yellow, lineage II; dark blue, lineage III; cyan, lineage V. The zigzag joining the centromeric and telomeric regions is an extended repetitive sequence and a hotspot for reciprocal recombination. Within the telomeric and centromeric regions the KIR genes are separated by short homologous sequences of a few hundred base pairs.
Three well-characterized KIR ligands are the HLA-A, -B, -C epitopes arising from polymorphism at residues 80 to 83 of the α1 domain. The C1 epitope is defined by asparagine 80 of HLA-C and is recognized by KIR2DL2 and KIR2DL3; the C2 epitope is defined by lysine 80 of HLA-C and is recognized by KIR2DL1, 2DS1, and 2DS5; the Bw4 epitope, carried by subsets of HLA-A and -B, is defined by arginine 83 and recognized by KIR3DL1. A fourth epitope (A3/11), recognized by KIR3DL2,6,7 comprises a peptide of Epstein-Barr virus bound to HLA-A*03 or HLA-A*11. KIR2DS2 binds HLA-A*11 and KIR3DS1 binds HLA-F.8,9
Important genetic features of KIR and HLA-A, -B, and C genes are their high polymorphism and their independent segregation on chromosomes 19q13.4 (KIR) and 6p21.3 (HLA).10 Consequently, within families and populations, individuals differ widely in the functional combinations of ligand and receptor they inherit. Thus, the genetic diversity created by the compound genotype of KIR and HLA class I is far greater than that due to either HLA or KIR alone. The advantage of such diversity to the human host is that infectious pathogens encounter, and have to adapt to, a different immune system in almost every person they infect.2,11
Presence of KIR A and KIR B in all human populations attests to their functional importance, but their relative frequencies vary considerably.12 Worldwide, the frequencies of KIR A and C2+HLA-C are inversely correlated, which is likely due to the pregnancy disorders associated with poor placentation and the compound genotype of a KIR A, C1+HLA-C homozygous mother carrying a C2+HLA-C fetus.13 In addition to reproductive disorders, a broad spectrum of infectious and autoimmune diseases is correlated with KIR A and B haplotype differences.14 Thus, the relative frequency of KIR A and B in a population is likely to reflect its history of life-threatening disease.
Since KIR typing was first described in 1997,15 assessment for the presence or absence of each KIR gene has been the method used in almost all studies to correlate KIR with disease and clinical outcomes. This method is insensitive to allelic differences, which can have quantitative and qualitative effects. For example, Africans have 11 KIR2DS5 allotypes, 6 are activating C2-specific receptors and provide resistance to preeclampsia; the other 5 neither recognize C2 nor do they provide resistance to preeclampsia.16,17 In the latter category is KIR2DS5*002, the predominant allele in Europeans. Currently, high-throughput, next-generation sequencing methods are being described but they have yet to see general application in clinical histocompatibility and immunogenetics laboratories.
Target recognition, tolerance, missing self, and NK-cell education
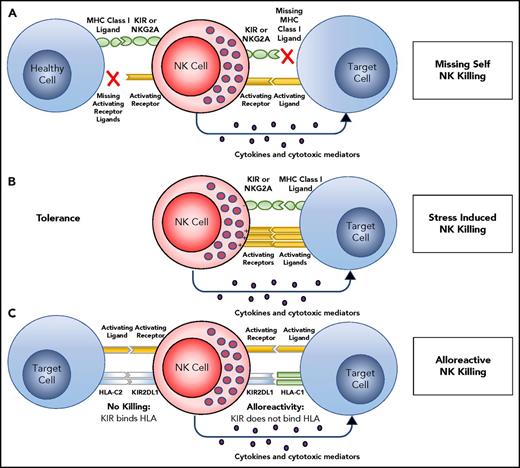
NK cells in peripheral blood are effector cells that can lyse tissue cells subject to infection, malignant transformation, or other forms of stress. To protect healthy cells and tissues from such attack, NK tolerance is mediated by their expression of inhibitory receptors for HLA class I (Figure 2A). One such inhibitory receptor is CD94:NKG2A, which recognizes the complex of HLA-E and a peptide cleaved from the leader sequence of an HLA-A, -B, or -C allotype.18 The amount of HLA-E that reaches the cell surface is thus dependent on the amount of HLA-A, -B, and -C being made by a cell. The other inhibitory receptors are inhibitory KIRs that recognize the A3/11, Bw4, C1, and C2 epitopes of HLA-A, -B, and -C. For an NK cell to be functional, it must acquire an inhibitory receptor that recognizes self-HLA class I. During development, the signaling pathways of activation and inhibition are balanced so that the amount of HLA-A, -B, and -C expressed by healthy cells is sufficient to inhibit all NK cells. In the context of human NK cells, this process of learning to recognize self HLA class I is called “NK-cell education”19-22 or “NK-cell licensing.”23 When infection, cancer, or other disease causes cells to reduce or lose HLA class I expression, NK cells are no longer inhibited and so they attack the unhealthy cells. The NK cells are thus seen to recognize the “missing self” HLA class I (Figure 2A).24 NK-cell education is not like rigid rote learning, but is flexible so that expression of increasing numbers of inhibitory self-receptors is correlated with increased effector function.25 NK cells that fail to express an inhibitory self-receptor or express an activating self-receptor, are usually incapable of responding to HLA class I–deficient cells.5,24,26,27 NK-cell killing of targets may also be mediated by overwhelming activation induced by upregulation of stress ligands on a damaged target, such as MICA, ULBP1, PVR, Nectin-2, or CD48 (Figure 2B).
Target recognition, tolerance, missing self. (A) NK cells recognize and kill their targets by an integrated balance of inhibitory and activating signals to discriminate between healthy cells (tolerance) vs elimination of transformed or virally infected targets (killing). NK-cell tolerance depends on several major histocompatibility complex (MHC) class I inhibitory signals (either classical, HLA-A, -B or -C, or nonclassical, HLA-E) expressed by heathy cells that engage KIR or NKG2A with minimal activation signals resulting in tolerance. Malignant transformation or viral infection promotes target cell killing by downregulation of MHC class I expression and an upregulation of signals from activating NK-cell receptors. (B) Although in some cases, MHC downregulation is variable or incomplete, target cell killing can still occur by changing the balance with activating signals upregulated by stress-induced activating receptor ligands. (C) This balance between inhibition and activation can be uniquely manipulated in the hematopoietic transplant setting by selection of donors who will respond to apparent missing self HLA class I in the HLA-mismatched recipients. For example, reconstitution with a high frequency of donor KIR2DL1+ NK cells would not be inhibited in a HLA-C1 (C1+-HLA-C) recipient (KIR ligand mismatch). Here, NK-cell alloreactivity would kill the recipient’s tumor. In contrast, when the same KIR2DL1+ NK cells reconstitute in an HLA-C2 recipient (C2+-HLA-C) (KIR ligand match), the recipient’s tumor would be seen as having self HLA class I and would not provoke an alloreactive NK-cell response.
Target recognition, tolerance, missing self. (A) NK cells recognize and kill their targets by an integrated balance of inhibitory and activating signals to discriminate between healthy cells (tolerance) vs elimination of transformed or virally infected targets (killing). NK-cell tolerance depends on several major histocompatibility complex (MHC) class I inhibitory signals (either classical, HLA-A, -B or -C, or nonclassical, HLA-E) expressed by heathy cells that engage KIR or NKG2A with minimal activation signals resulting in tolerance. Malignant transformation or viral infection promotes target cell killing by downregulation of MHC class I expression and an upregulation of signals from activating NK-cell receptors. (B) Although in some cases, MHC downregulation is variable or incomplete, target cell killing can still occur by changing the balance with activating signals upregulated by stress-induced activating receptor ligands. (C) This balance between inhibition and activation can be uniquely manipulated in the hematopoietic transplant setting by selection of donors who will respond to apparent missing self HLA class I in the HLA-mismatched recipients. For example, reconstitution with a high frequency of donor KIR2DL1+ NK cells would not be inhibited in a HLA-C1 (C1+-HLA-C) recipient (KIR ligand mismatch). Here, NK-cell alloreactivity would kill the recipient’s tumor. In contrast, when the same KIR2DL1+ NK cells reconstitute in an HLA-C2 recipient (C2+-HLA-C) (KIR ligand match), the recipient’s tumor would be seen as having self HLA class I and would not provoke an alloreactive NK-cell response.
In a manner analogous to the KIR A and KIR B haplotypes, HLA class I haplotypes divide into 2 groups according to whether they promote education through interactions with the CD94:NKG2 self-receptor or a KIR self-receptor. The key feature that distinguishes the 2 HLA haplotype groups is the methionine/threonine14 dimorphism at position −21 in the leader sequence of HLA-B.28 Cleavage of 21M HLA-B leader sequences produces nonamer peptides that bind HLA-E and promote its expression at the cell surface and recognition by CD94:NG2A.29 In contrast, peptides cleaved from −21T HLA-B leader sequences lack these functions.30 Thus, HLA haplotypes encoding 21M HLA-B, but not those encoding −21T HLA-B, specialize in NK-cell education based on recognition of HLA-E by CD94:NKG2A. Reinforcing this specialization, HLA haplotypes encoding −21M HLA-B do not encode the Bw4 and C2 KIR ligands, but are restricted to the A3/11 and C1 KIR ligands. In a complementary manner, HLA haplotypes encoding −21T can encode all 4 of the KIR ligands. Thus, HLA haplotypes encoding −21T HLA-B specialize in NK-cell education mediated by KIR recognition of HLA-A, -B, -C. The −21 HLA-B dimorphism divides all human populations into 3 groups: M/M, M/T, and T/T. Compared with T/T individuals, M/M and M/T individuals have NK cells that are better educated and exhibit more potent cytokine secretion, ADCC, and response to missing self HLA class I.28
NK cells in hematopoietic cell transplantation and donor selection
Although NK cells are the first reconstituting lymphocytes following hematopoietic cell transplantation (HCT), their killing and cytokine-secreting effector functions are impaired compared with the NK cells of their healthy donors.31 Despite being weak, engrafting NK cells could still contribute some protection against graft-versus-host disease, infection, and relapse, as well as facilitating engraftment.
A beneficial alloreactive NK-cell response can be induced in recipients of allogeneic HCT by selecting donors who have a KIR ligand the recipient lacks (Figure 2C). Consider donor NK cells that express the C2 inhibitory receptor KIR2DL1 that have been educated to respond to reduced expression of C2+HLA-C. If the recipient is homozygous for C1+HLA-C, there is no C2+HLA-C to inhibit the donor cells and tumor targets will be killed. Alternately, recipients with C2+HLA-C will inhibit donor NK cells via KIR2DL1. Thus, when recipients lack 1 or more of the 4 epitopes recognized by KIR (ie, missing KIR ligand), it is possible to select donors whose NK cells will give an alloreactive response against the recipient. This is not possible for recipients who have all 4 HLA epitopes that are KIR ligands or in an HLA-matched transplant. This therapeutic strategy was pioneered in the setting of haploidentical HCT as treatment of acute myeloid leukemia (AML), for which it was shown that alloreactive NK cells correlate with less relapse and improved survival for patients transplanted while in complete remission.32,33
Many investigators have now explored the possibility of producing similar alloreactions in transplant settings where the donor and recipient do not differ by a complete HLA haplotype. These studies show that there is no consistent benefit seen for transplants in which the recipient lacks a donor KIR ligand or where the donor and recipient are KIR ligand mismatched. Complicating factors are differences in stem cell source, conditioning regimen, T-cell depletion, and the post-HCT immunosuppression. Despite the differences, the consensus view is that NK cells have high potential for improving posttransplant outcomes.34
Retrospective studies have assessed the impact of donor and recipient KIR genotype on HCT outcome. Donor KIR3DS1 associated with reduced acute graft-versus-host disease in unrelated donor HLA-matched transplants. Protection increased with 2 copies of KIR3DS1.35 In addition, KIR2DS1 and C1+HLA-C conferred protection from AML relapse. AML patients experienced less relapse and improved disease-free survival after receiving transplants from donors having 1 or 2 KIR B haplotypes.36 That KIR3DS1 and KIR2DS1 are B haplotype genes is consistent with an emerging consensus that donor KIR B haplotypes are generally beneficial for HCT outcome.37
Adoptive transfer of NK cells and cytokines to promote their expansion
Another strategy to exploit antileukemia properties of NK cells is adoptive transfer of mature NK cells educated in the donor in a haploidentical nontransplantation setting. In a first trial of this approach, patients received 1 of 3 different preparative regimens: high cyclophosphamide and fludarabine, low cyclophosphamide, and methylprednisone. Following NK-cell infusion, patients received IL-2 daily for 14 days. Donor NK-cell expansion was observed only for patients receiving the preparatory regimen of high cyclophosphamide and fludarabine, which induced a surge of endogenous IL-15/IL-15Rα.38,39 Successful expansion of NK cells was determined by detection of >100 NK cells per microliter of blood 12 to 14 days after infusion. On this protocol, 30% of poor prognosis AML patients achieved a complete remission, which correlated with in vivo NK-cell expansion.
The use of adoptive transfer of NK cells to treat various malignancies has yielded mixed results. Shi et al40 infused haploidentical KIR-mismatched NK cells into 10 patients with relapsed multiple myeloma, followed 14 days later with an autologous stem cell graft. Five patients achieved near complete remission. Bachanova et al41 treated 6 patients with non-Hodgkin lymphoma with infused haploidentical NK cells. They found the NK cells expanded poorly in vivo and host regulatory T cells (Tregs) were significantly increased in number after NK-cell infusion and IL-2 administration. In a mouse model, pretreatment with Ontak (denileukin diftitox) to deplete host Tregs improved the capacity of adoptively transferred NK cells to eliminate tumors.42,43 Applying a similar strategy to AML patients, in a clinical trial combining Ontak treatment with infused haploidentical NK cells, there was increased in vivo expansion of NK cells and improved AML clearance.44
NK cells express an array of cytokine receptors that modulate NK-cell development, proliferation, homeostasis, and effector functions. Several cytokine receptors are constitutively expressed; others are inducible.45 Although stimulation with individual cytokines can enhance NK-cell function, synergistic effects from multiple cytokines result in stronger responses.46 IL-2 and IL-15,47 which activate many downstream signaling molecules including the mammalian target of rapamycin pathway, have been most effective.48 The first human clinical trial with IL-15 showed potent in vivo activation and expansion of NK cells.49 Recombinant human IL-15 (rhIL-15), in monomeric form, is being investigated for therapeutic use in solid tumors as well as to support NK-cell persistence and activity postadoptive cellular transfer in patients with leukemia.50 IL-15N72D/IL-15Rα-Fc superagonist complexes (ALT-803)51,52 and heterodimeric IL-15/IL-15Rα are currently being tested in clinical trials alone and in combinations.53,54 ALT-803 and rhIL-15 are better tolerated when given subcutaneously rather than intravenously. This conclusion is based on lower peak IL-15 concentrations, with potential to induce secondary inflammatory cytokines, thus causing less fevers and constitutional symptoms. Advantages to IL-15 complexes, such as ALT-803, include physiologic presentation to the immune system, better pharmacokinetics as IL-15Rα stabilizes IL-15 (eg, ALT-803 is given weekly rather than daily, like rhIL-15), and better homing to lymphoid tissue, especially when given subcutaneously.
The role of particular NK-cell subsets for relapse protection or treatment
When mice are infected with cytomegalovirus (CMV), NK cells expressing the activating Ly49H receptor proliferate and persist in high numbers after infection subsides.55 Likewise, humans infected with human CMV (HCMV) expand their pool of CD56dimCD57+NKG2C+ NK cells. In HCT recipients, these cells become activated and proliferate following reactivation of HCMV.56,57 These cells are called adaptive NK cells because they adapt to a pathogen and deliver a more potent recall response. Adaptive NK cells have methylation patterns of genomic DNA that are different from those of other NK cells, but similar to cytotoxic T cells. Further distinguishing adaptive NK cells from conventional NK cells is reduced expression of FcεR1γ, SYK, EAT-2, and PLZF.58,59 Compared with other NK-cell subsets, adaptive NK cells produce more tumor necrosis factor-α and IFN-γ on target cell recognition and ADCC. That adaptive NK cells express CD94:NKG2C, the activating HLA-E receptor, whereas other NK cells express its inhibitory counterpart, CD94:NKG2A, could be a critical factor for NK-cell mediation of graft-versus-leukemia effects.
Association of CMV reactivation with relapse protection post-HCT has been the subject of several reports.60 Studying allogeneic HCT recipients, Cichocki et al61 found that those who reactivated CMV had lower leukemia relapse and superior disease-free survival at 1 year. This protective effect strongly correlates with adaptive NK-cell expansion.61 Properties of adaptive NK cells include longer survival, enhanced metabolism, resistance to myeloid-derived suppressor cells, resistance to Treg-mediated suppression, and reduced surface expression of inhibitory checkpoint receptors. This research led to development of an adaptive NK-cell product generated using IL-15 plus a glycogen synthase kinase 3 inhibitor (GSK3i).62
Cytokines can be used to induce memory-like (CIML) NK cells.63 Activation of CD56bright and CD56dim NK cells with IL-15, IL-12, and IL-18 increases CD25 expression and STAT5 signaling64 leading to increased IFN-γ production, cytotoxicity, and proliferation. CIML NK cells have been tested clinically for patients with advanced AML and are being used as an adjunct to haploidentical transplants for patients with advanced AML.65
Checkpoint blockade to enhance NK activity
Checkpoint blockade is 1 of several important discoveries that led to integration of immunotherapy into routine cancer treatment. The principle of checkpoint blockade is to interrupt a receptor’s inhibitory signal with a cognate ligand. The checkpoints can be NK-cell specific or can be common to several types of immune cell. KIR2D blockade has been a focus for investigation. Phase 1 clinical trials in which anti-KIR antibody IPH2101 was given to AML66 and multiple myeloma patients67 demonstrated binding of IPH201 to cells for 2 weeks, some increases in serum cytokines, and NK-cell activation. A phase 2 study investigating smoldering multiple myeloma showed no therapeutic benefit,68 as assessed by the capacity of IPH2101 to mediate trogocytosis of the KIR2D receptors of NK cells.69 Based on a similar rationale,70 blocking of the highly expressed inhibitory receptor CD94:NKG2A is being tested. The anti-NKG2A monoclonal antibody (IPH2201-monalizumab) is currently being evaluated in early phase clinical trials for a variety of tumor types, either alone or as combination therapy in hematologic malignancies.
Other notable checkpoint receptors are T-cell immunoglobulin and mucin-domain–containing-3 (Tim-3) and lymphocyte activation gene 3 (Lag-3). Expressed on T cells and NK cells, Tim-3 is a negative regulator of T-cell–mediated immune responses. On NK cells, this receptor can exhibit activating or inhibitory function, depending on context. Blockade of galectin-9, the ligand for Tim-3, reduced the IFN-γ production of NK cells from healthy donors presented with AML targets. This observation suggests Tim-3 is an activating receptor.71 However, when Tim-3 was crosslinked with antibodies, it suppressed NK-cell–mediated cytotoxicity.72 A phase 1 clinical trial using the therapeutic Tim-3–blocking antibody TSR-022 in patients with advanced tumors is currently in progress.73
Conventional NK cells express an inhibitory receptor called T-cell immunoreceptor with immunoglobulin and immunoreceptor tyrosine-based inhibition motif domains (or TIGIT) that binds PVR and Nectin-2, the same ligands recognized by DNAM-1. Many tumors overexpress the TIGIT ligand, CD155, and have been linked with enhanced tumor proliferation and migration.74,75 TIGIT itself is upregulated on CD8+ T cells and Tregs in many clinical settings.76-78 Blockade of TIGIT in vitro enhances T-cell function.76,79 Similarly, TIGIT blockade on NK cells enhances cytokine secretion and cytotoxicity.80-82
A focus of current research in cancer immunotherapy is the checkpoint receptor programmed death-1 (PD-1). PD-1 is a dominant and relevant checkpoint for tumor-mediated immunosuppression. Several products that block PD-1 or its ligand, PD-L1, are now part of routine cancer care and a variety of combinatorial approaches are being tested in clinical trials. NK-cell expression of PD-1 has yet to be defined, but a recent study proposes a link between NK expression of PD-1 and CMV serostatus.83 Increased PD-1 expression on NK cells from multiple myeloma patients has been described,84 suggesting PD-1 expression could be both activation dependent and important for tumor killing.
Tumor targeting with immune engagers and CD16 stimulation
NK cells express the low-affinity Fc receptor CD16a, which mediates ADCC. Two isoforms of CD16, CD16a and CD16b,85 are differentially expressed.86-89 CD16a has a transmembrane domain and is surface expressed on CD56dim NK cells, macrophages, and placental trophoblasts. CD16b is expressed on neutrophils and anchored to the membrane by a glycophosphatidylinositol domain. Although the extracellular domains of CD16a and CD16b have >90% sequence similarity, CD16b cannot signal. Consequently, only engagement of CD16a causes tumor target cell killing.85,86,90,91 CD16a allotypes vary in their affinity for the Fc of immunoglobulin G (IgG). NK cells expressing CD16a with 158VV or 158VF allotypes have lower affinity for the Fc of rituximab, the therapeutic anti-CD20 monoclonal antibody, than the CD16a 158FF allotype.92 This difference correlates with clinical outcome. Strategies that combine immunotherapies are expected to improve cancer treatment. For example, blockade of inhibitory KIR in combination with tumor-targeted monoclonal antibodies could increase CD16-mediated ADCC. Anti-CD137 antibodies in combination with rituximab are known to increase degranulation and IFN-γ production.93 Cytokines could also enhance NK-cell–mediated ADCC.94 As therapeutic NK-cell activation becomes better optimized, a counterregulatory mechanism that should be considered is the cleavage of CD16 by ADAM17, which removes CD16 from the NK-cell surface.95 Selective inhibition of ADAM17 can prevent such shedding of CD16 and thus enhance the response to ADCC. Fc-induced production of cytokines by NK cells exposed to rituximab-coated B-cell targets can further enhance ADAM17 inhibition, a strategy being tested in a clinical trial for relapse prevention after autologous HCT.
Reagents called bispecific killer engagers (BiKEs) are also being used to direct NK-cell killing at tumor targets.96-98 BiKEs are small molecules containing 2 antibody Fv regions (variable region heavy and variable region light) of different specificity. One is specific for CD16 and the other to a surface component of the tumor cell target. By strongly crosslinking NK cells to tumor targets, BiKEs overcome the naturally low affinity of CD16 for IgG. This approach effectively makes NK cells tumor antigen specific.
The anti-CD16 moiety of the BiKE can be tailored to bind broadly to CD16a/CD16b or be CD16a specific. With this platform, the signals delivered at the immune synapse can be defined and varied. This application has been tested clinically using a bispecific tetravalent antibody targeted to CD16a on NK cells and CD30 on Hodgkin disease targets.99 Using broader CD16a/b and CD33 targeting to AML, anti-CD16x33 BiKE activation can overcome inhibitory signaling mediated by class I HLA to kill primary leukemia cells98 as well as target CD33+ myeloid-derived suppressor cells in patients with myelodysplastic syndrome.96 However, although BiKEs mediate ADCC, they do not deliver a signal that leads to NK-cell proliferation and survival. Such a proliferative signal is likely to be essential for clinical efficacy, as was the case for T-cell chimeric antigen receptor (CAR) strategies targeting CD19 to lymphoid malignancies. To fulfill this need, a trispecific killer engager (TriKE) was made by adding an IL-15 linker to a BiKE.100,101 The first TriKE manufactured for clinical testing in a trial to begin in 2018 (anti-CD16–IL-15–anti-CD33) targets AML, myelodysplastic syndrome, and other CD33+ hematopoietic malignancies. The 161533 TriKE enhances tumor-specific killing of AML with proliferation that is enriched for antigen-specific NK cells.
Summary and future directions
Advances on several fronts in basic NK-cell biology have immediate translational potential. In the clinical setting of allogeneic transplantation and adoptive transfer, NK cells have been shown to exhibit remarkable therapeutic properties, particularly in their capacity to eliminate myeloid leukemia cells. Because KIR interactions with HLA class I clearly affect the outcomes of hematopoietic transplantation, the application of high-resolution KIR typing would refine strategies for choosing an optimal donor. More recently, the tremendous potential for cytokines, checkpoint modulators, and immune engagers to activate NK cells is being tested with new therapeutic agents. The potency of these strategies could diminish the clinical relevance of more subtle KIR/HLA interactions for NK-cell–based immunotherapy. Further characterization of the tumor microenvironment and of NK-cell receptors, their ligands, and checkpoints is needed to improve understanding of how NK cells recognize tumors and how this can be optimized in the clinical setting. Clinical trials with adoptive transfer of expanded NK cells, NK cells from hematopoietic progenitors, adaptive NK cells from CMV+ donors, and off-the-shelf NK cells from induced pluripotent stem cells have either started or will be in the clinic within the next year. Various NK-cell products, delivered alone or in combination with cytokines, checkpoint inhibitors, and CD16-mediated targeting agents, have promise for excellent clinical efficacy. In addition to academic centers, more than a dozen companies are interested in commercializing NK-cell products or proteins designed specifically to affect NK cells. On the clinicaltrials.gov Web site, one can find 149 interventional trials under the search term of “NK cells.” Selected agents in active development are listed in Table 2 (taken from clinicaltrials.gov, the Innate Killer Summit meeting 2017, and Society for Natural Immunity [SNI] 2015/2016 meetings). Allogeneic HCT affords a good setting to test these reagents given the high rates of relapse, especially after reduced-intensity conditioning. Moreover, allogeneic transplantation offers a unique clinical setting for registration trials with a short follow-up time that can be limited to the first year. NK-cell–directed therapies have already changed the standard of care, most notably for lymphoma, with the approval of the anti-CD20 agent rituximab that mediates ADCC via CD16. The approval of the first chimeric antigen receptor therapy further highlights the excitement about cellular immune therapy. Rapid scientific progress and an explosion of clinical trials testing NK cells or agents that work through NK cells should lead to approval of an NK-cell–mediated therapy within the next 5 years.
Selected list of NK-cell–based therapies under commercial development
| Company (in alphabetical order) . | Agent/therapy . | Trial details . |
|---|---|---|
| NK-cell therapies | ||
| Celgene | PNK-007 | Trials in AML and multiple myeloma |
| Cyto-Sen | PM21 particles: Off-the-shelf NK-cell expansion particles derived from the K562 cell line | Making good manufacturing practice material; clinical trials planned |
| Fate | FATE-NK100 (adaptive NK cells) | Trials for AML, ovarian cancer, and solid tumors with commercially available antibodies |
| NK cells derived from native and genetically modified induced pluripotent stem cells | Clinical trials planned, allowing multiple doses, off-the-shelf cryopreserved NK cells | |
| Fortress/Coronado | CNDO-109 NK-cell product | Phase 1 trial in advanced AML |
| Gamida-Cell | Nicotinamide-based NK-cell expansion (NAM-NK) | Phase 1 trial for lymphoma and multiple myeloma in combination with rituximab and elotuzumab |
| GlycoStem | Umbilical cord blood progenitor expanded NK cells | Phase 1 trial for AML, complete |
| Green Cross | MG4104 expanded random donor NK cells (no HLA matching) | Phase 1 trial including lymphoma and solid tumors |
| Miltenyi | NK-cell selection devices (CD3 and CD19 depletion and CD56 selection) | At least 12 studies listed for various cancers and combinations |
| NantKWest | NK-92 cell line–derived modified products | 5 trials for various cancers alone, gene modified, and in combinations |
| NKarta | NK-cell expansion with membrane-bound IL-15/41BB-L K562 feeders | Clinical trials in development |
| PersonGen Bio | CD33-CAR, CD19-CAR, and CD7-CAR in NK-92 cell lines | Phase 1 in AML, B-cell malignancies, and other CD7+ cancers |
| Targeted NK-cell proteins | ||
| Affimed | Tetravalent CD30-targeted CD16A engagers; other targets in development | 5 trials for lymphoma/leukemia; 1 with checkpoint combination |
| Altor Bioscience | IL-15/IL-15Ra-Fc (ALT-803) | 29 studies for various cancers, alone and in combination with NK cells, antibodies, and checkpoints |
| Bristol-Myers Squibb | Lirilumab (anti-KIR) | 9 trials for various cancers in various combinations |
| GT Pharma | CD33-targeted CD16A/CD16B engager with IL-15 linker (TriKE); other targets in development | Phase 1 trial for advanced CD33+ myeloid malignancies |
| Innate Pharma | Monalizumab (anti-NKG2A) | 3 trials for various cancers, 1 with antibody combination |
| Nektar | NKTR-255 polymer IL-15 | Clinical trials in development |
| Novartis | HetIL-15 | Phase 1 trial, alone and combination with checkpoint blockade |
| OncoMed | OMP-313M32 (anti-TIGIT) | Phase 1 trial in cancer patients |
| Tesaro | TSR-022 (anti-TIM-3) | Phase 1 trial in cancer patients |
| Company (in alphabetical order) . | Agent/therapy . | Trial details . |
|---|---|---|
| NK-cell therapies | ||
| Celgene | PNK-007 | Trials in AML and multiple myeloma |
| Cyto-Sen | PM21 particles: Off-the-shelf NK-cell expansion particles derived from the K562 cell line | Making good manufacturing practice material; clinical trials planned |
| Fate | FATE-NK100 (adaptive NK cells) | Trials for AML, ovarian cancer, and solid tumors with commercially available antibodies |
| NK cells derived from native and genetically modified induced pluripotent stem cells | Clinical trials planned, allowing multiple doses, off-the-shelf cryopreserved NK cells | |
| Fortress/Coronado | CNDO-109 NK-cell product | Phase 1 trial in advanced AML |
| Gamida-Cell | Nicotinamide-based NK-cell expansion (NAM-NK) | Phase 1 trial for lymphoma and multiple myeloma in combination with rituximab and elotuzumab |
| GlycoStem | Umbilical cord blood progenitor expanded NK cells | Phase 1 trial for AML, complete |
| Green Cross | MG4104 expanded random donor NK cells (no HLA matching) | Phase 1 trial including lymphoma and solid tumors |
| Miltenyi | NK-cell selection devices (CD3 and CD19 depletion and CD56 selection) | At least 12 studies listed for various cancers and combinations |
| NantKWest | NK-92 cell line–derived modified products | 5 trials for various cancers alone, gene modified, and in combinations |
| NKarta | NK-cell expansion with membrane-bound IL-15/41BB-L K562 feeders | Clinical trials in development |
| PersonGen Bio | CD33-CAR, CD19-CAR, and CD7-CAR in NK-92 cell lines | Phase 1 in AML, B-cell malignancies, and other CD7+ cancers |
| Targeted NK-cell proteins | ||
| Affimed | Tetravalent CD30-targeted CD16A engagers; other targets in development | 5 trials for lymphoma/leukemia; 1 with checkpoint combination |
| Altor Bioscience | IL-15/IL-15Ra-Fc (ALT-803) | 29 studies for various cancers, alone and in combination with NK cells, antibodies, and checkpoints |
| Bristol-Myers Squibb | Lirilumab (anti-KIR) | 9 trials for various cancers in various combinations |
| GT Pharma | CD33-targeted CD16A/CD16B engager with IL-15 linker (TriKE); other targets in development | Phase 1 trial for advanced CD33+ myeloid malignancies |
| Innate Pharma | Monalizumab (anti-NKG2A) | 3 trials for various cancers, 1 with antibody combination |
| Nektar | NKTR-255 polymer IL-15 | Clinical trials in development |
| Novartis | HetIL-15 | Phase 1 trial, alone and combination with checkpoint blockade |
| OncoMed | OMP-313M32 (anti-TIGIT) | Phase 1 trial in cancer patients |
| Tesaro | TSR-022 (anti-TIM-3) | Phase 1 trial in cancer patients |
Technology selected based on known developmental plan (this list does not include all technologies). Sources: clinicaltrials.gov, Innate Killer Summit San Diego 2017, and Society for Natural Immunity Annual Meeting Abstracts 2016.
Acknowledgments
The authors thank Lisbeth Guethlein for help in the design and drawing of Figure 1, and Paul Robertson for Figure 2.
This work was supported by National Institutes of Health, National Cancer Institute grant P01 CA111412.
Authorship
Contribution: S.C., P.P., and J.S.M. designed and wrote paper.
Conflict-of-interest disclosure: J.S.M. serves on the Scientific Advisory Boards of Celgene, Fate Therapeutics, and GT Pharma, and has received research funds and/or clinical trials support from these relationships. These relationships have been reviewed and managed by the University of Minnesota in accordance with its conflict-of-interest polices. These relationships had no role in funding this research for this review, which has been funded by the National Institutes of Health grants. The remaining authors declare no competing financial interests.
Correspondence: Jeffrey S. Miller, Division of Hematology, Oncology and Transplantation, University of Minnesota Cancer Center, MMC 806, Harvard St at East River Rd, Minneapolis, MN 55455; e-mail: mille011@umn.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal