Key Points
T-cell activation induces TCR transactivation of CXCR4 to stabilize cytokine mRNA transcripts via a PREX1-Rac1–signaling pathway.
Inhibition of the TCR-CXCR4–signaling pathway impairs TCR-dependent and TCR-independent cytokine secretion by CTCL cells.
Abstract
As with many immunopathologically driven diseases, the malignant T cells of cutaneous T-cell lymphomas (CTCLs), such as Sézary syndrome, display aberrant cytokine secretion patterns that contribute to pathology and disease progression. Targeting this disordered release of cytokines is complicated by the changing cytokine milieu that drives the phenotypic changes of CTCLs. Here, we characterize a novel signaling pathway that can be targeted to inhibit the secretion of cytokines by modulating either CXCR4 or CXCR4-mediated signaling. We demonstrate that upon ligation of the T-cell antigen receptor (TCR), the TCR associates with and transactivates CXCR4 via phosphorylation of S339-CXCR4 in order to activate a PREX1-Rac1–signaling pathway that stabilizes interleukin-2(IL-2), IL-4, and IL-10 messenger RNA (mRNA) transcripts. Pharmacologic inhibition of either TCR-CXCR4 complex formation or PREX1-Rac1 signaling in primary human T cells decreased mRNA stability and inhibited secretion of IL-2, IL-4, and IL-10. Applying this knowledge to Sézary syndrome, we demonstrate that targeting various aspects of this signaling pathway blocks both TCR-dependent and TCR-independent cytokine secretion from a Sézary syndrome–derived cell line and patient isolates. Together, these results identify multiple aspects of a novel TCR-CXCR4–signaling pathway that could be targeted to inhibit the aberrant cytokine secretion that drives the immunopathogenesis of Sézary syndrome and other immunopathological diseases.
Introduction
Immunopathogenesis often involves the aberrant release of T-lymphocyte–derived cytokines that promote autoimmunity, immunosuppression, immunodeficiency, or tumor progression. The cutaneous T-cell lymphomas (CTCLs), mycosis fungoides and Sézary syndrome, are characterized by a specific pattern of cytokine release that drives disease progression. High interleukin-2 (IL-2) levels, found early in disease, promote proliferation and survival of CTCL cells, adoption of a regulatory T-cell (Treg) phenotype by effector T cells, and expression of FoxP3 in CTCL cells.1-3 Increased IL-4 levels later in disease promote eosinophilia, immunosuppression, and susceptibility to infections.2-4 CTCL cells at end stages of disease develop a Treg phenotype that leads to immunosuppression, T-cell exhaustion, and suppression of antitumor immunity within lesions by the release of IL-10.2-5 Identifying a signaling pathway that mediates an aspect of cytokine release common to multiple cytokines could provide new targets for treating the immunopathogenesis of CTCLs.
The T-cell antigen receptor (TCR) is essential for the recognition of foreign peptides and for initiating the activation of T cells that leads to the cytokine production critical for an immune response. CXCR4, a 7-transmembrane G-protein coupled receptor, mediates T-cell migration toward antigen-presenting cells producing its sole endogenous ligand, CXCL12 (also known as SDF-1), thereby enhancing TCR’s exposure to foreign antigens. Signaling via either TCR or CXCR4 is often critically affected by the presence or the activation state of the other receptor. TCR expression is essential for CXCL12-induced gene expression in T cells.6-10 Conversely, CXCL12/CXCR4 signaling is necessary for TCR-initiated immune synapse formation, enhanced phosphorylation of early signaling molecules, and thymic β selection.11-15 Because various receptor tyrosine kinases transactivate CXCR4 in order to mediate cell motility, cell growth, and tumorigenesis,16-19 we explored the possibility that TCR might similarly transactivate CXCR4 in order to mediate cytokine production.
Messenger RNA (mRNA) stability of cytokine transcripts is tightly regulated by activated T cells to carefully modulate an immune response. Dysregulation of mRNA turnover may lead to immunopathology including autoimmunity, immunosuppression, or tumor progression. mRNA decay is regulated by cis elements intrinsic to the mRNA and trans-acting factors such as RNA-binding proteins.20-22 The GTPase, Rac1, has previously been linked to mRNA stabilization in various cell types.23-27 In T cells, Rac1 has been mainly studied for its role in migration and cellular signaling.28,29 CXCR4 has been shown to activate Rac1 via the Rac-GEF, PREX1, in various cell types,17,19,30,31 however, a role for PREX1 in T-cell signaling had not been previously described.
Here, we used primary human T cells to characterize a novel signaling pathway that regulates cytokine mRNA stability, and, importantly, we used this new knowledge to disrupt this pathway and inhibit cytokine secretion in malignant T cells derived from Sézary syndrome. We show that upon ligation of the TCR, the TCR associates with and transactivates CXCR4 in order to activate a PREX1-Rac1–signaling pathway that stabilizes IL-2, IL-4, and IL-10 mRNA transcripts. Importantly, we show, in a Sézary syndrome–derived cell line and patient isolates, that inhibition of various aspects of this signaling pathway blocks both inducible TCR-dependent and constitutive TCR-independent cytokine secretion. Together, these results identify multiple steps of a novel signaling pathway that can be targeted as a means to reduce the aberrant cytokine secretion of CTCLs or other forms of T-cell–driven immunopathology.
Methods
Materials
A complete list of materials can be found in supplemental Methods (available on the Blood Web site).
Cells
Normal human peripheral blood T cells (peripheral blood mononuclear cell [PBMC] T cells) from healthy volunteers and T cells from residual diagnostic patient specimens were isolated with ∼98% purity (supplemental Figure 3D) and maintained as described.6 Blood and patient specimens were obtained and used with informed consent and approval by the Mayo Institutional Review Board. Jurkat T cells were maintained as described.6 HUT-78 cells were maintained in Iscove modified Dulbecco medium, 20% fetal calf serum, 1% penicillin-streptomycin, and 2 mM l-glutamine.
Cytokine production
Cells were treated with AMD3100 or NSC23766 or transfected with 750 nM control small interfering RNA (siRNA), CXCR4 siRNA-1, PREX1 siRNA-1 (Dharmacon), or CXCR4 siRNA-2 or PREX1 siRNA-2 (Ambion) utilizing the Human T-cell nucleofector kit (Lonza) with program U-014 prior to analysis of cytokine production. Cytokine production was analyzed via intracellular cytokine staining and enzyme-linked immunosorbent assay (ELISA) as described6,32 or via cytokine bead array analysis (BD Biosciences).
FRET
CXCR4–yellow fluorescent protein (YFP) and CD3ζ–cyan fluorescent protein (CFP) were described previously.6 CCR7-YFP was prepared by subcloning CCR7 from pcDNA-CCR7 (Missouri S&T cDNA Resource Center) into pEYFP-N1 (Clontech). Cells were prepared for fluorescence resonance energy transfer (FRET) analysis6,7 and placed in 96-well clear-bottom black tissue-culture plates (Corning). Fluorescent emission spectra in response to 433 nm of light were assayed using the Varioskan Flash (Thermo Scientific) before and after stimulation. Spectra were read from the top of the plate at 37°C and used 12-nm bandwidth ×150 msec read with agitation between reads. Spectra were analyzed as described.6,7 Percent change in YFP or CFP in response to OKT3 was determined by calculating the difference in emission intensity over the CFP range (460-500) or the YFP range (525-550) and normalizing this change to the total fluorescence of the unstimulated sample.
Proximity ligation assay
Jurkat T cells were transfected with CXCR4-YFP, cultured for 16 hours, pretreated with AMD3100 for 1 hours, centrifuged onto fibronectin- and OKT3-coated coverslips, and incubated for 30 minutes at 37°C. Cells were then fixed with 3% paraformaldehyde, permeabilized with 0.15% Triton Surfact-amps (Thermo Fisher), blocked with 5% bovine serum albumin/0.1% glycine/phosphate-buffered saline, incubated with goat anti–green fluorescent protein and rabbit anti-CD3ζ overnight at 4°C, and processed using the DUOLINK In SITU kit (Sigma).
Rac1 activation, ERK activation, NFAT localization, luciferase activity, and mRNA stability
For Rac1 activation, GST-PAK1-PBD (gift from Vijay Shah, Mayo Clinic, Rochester, MN) were prepared as described.33 PBMC T cells were stimulated, lysed in MLB buffer (Millipore) with protease inhibitors, and centrifuged to remove nuclei. Supernatants were rotated with 10 μg of GST-PAK1-PBD for 15 minutes at 4°C, washed, and boiled in sample buffer. Extracellular signal-regulated kinase (ERK) activation10 and IL-2 luciferase activity6,34 were assayed as described. For NFAT localization, subcellular fractionation was performed as described.35 The 3′ untranslated region (UTR) luciferase reporter vector was generated by cloning the complete IL-2 3′UTR into pmirGLO (Promega) using the following primers: 5′GGCGCCGCTAGCTAATTAAGTGCTTCCCAC3′ and 5′GCCCGCGTCGACTTTTTTTTATATTTATCAAATTTATTAAATAGTTTTACTAACC3′. To assess mRNA stability, PBMC T cells were stimulated and then treated with 8 μg/mL actinomycin D for the indicated time. mRNA was reverse transcribed via the iScript cDNA synthesis kit (Bio-Rad) and measured via RT2 SYBR Green Fluor quantitative reverse transcription–polymerase chain reaction (qRT-PCR) (Qiagen) on a Roche Light Cycler 480 utilizing PrimeTime qPCR primer sets (IDT). Transcript levels were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (which remained relatively constant for all treatments; supplemental Figure 4B-C) and quantified using the 2ΔΔCt method.
Statistical analysis
Two-tailed paired Student t tests (Microsoft Excel) were used for analysis unless otherwise indicated. The means of 2 distributions were considered significantly different if P was ≤.05.
Results
CXCR4 is required for TCR-initiated production of IL-2, IL-4, and IL-10
Because TCR and CXCR4 crossregulate each other’s functions,6-15 we sought to determine the role of CXCR4 in TCR-initiated cytokine production. We used crosslinked CD3 monoclonal antibody (mAb) with soluble CD28 mAb to activate primary human T cells purified from peripheral blood of healthy donors (PBMC T cells). Exogenous CXCL12 was not added to these cultures, and these T cells expressed 10 000-fold less CXCL12 mRNA than IL-2 or IL-10 mRNA (Figure 1A). Interestingly, depletion of CXCR4 with distinct CXCR4 siRNAs resulted in a significant decrease in the amount of IL-2, IL-4, and IL-10 produced by T cells upon stimulation, but the effects on interferon-γ (IFN-γ) varied from donor to donor (Figure 1B-E). Results from T cells from multiple donors are summarized in Figure 1D-E with the average response represented as a black line. Depletion of CXCR4 did not induce apoptosis (supplemental Figure 1A-B). These results indicate that CXCR4 expression is required for IL-2, IL-4, and IL-10 production in response to T-cell activation.
CXCR4 is required for TCR-initiated production of IL-2, IL-4, and IL-10. (A) Purified, human PBMC T cells were stimulated with 1 μg/mL plate-bound OKT3 and soluble CD28, cultured for 24 hours, and harvested for analysis via qRT-PCR for CXCL12 mRNA transcript levels. A human bone marrow mesenchymal stromal stem cell line (BMSC) was used as a positive control for CXCL12 expression. The results shown are normalized to the reference gene GAPDH, where GAPDH is set to 100. Each point denotes the mean mRNA transcript level ± standard deviation (SD) for 5 independent donors (n = 2 for BMSC samples). (B-E) Human PBMC T cells were purified, transfected with either control siRNA, a pool of CXCR4 siRNAs (CXCR4 siRNA-1) or CXCR4 siRNA-2 (single siRNA), and cultured for 24 hours. (B-C) CXCR4 cell surface levels were assayed via flow cytometry. Mean fluorescent intensities (MFI) are shown for a representative experiment. (C) Summarizes the results with each bar denoting the mean ± standard error of the mean (SEM). *Significantly different from control siRNA-transfected cells (P < .05; n = 7-9). (D-E) Twenty-four hours after transfection, cells were stimulated as in panel A, then cultured for an additional 24 hours prior to harvest of supernatants for cytokine analysis. Results from 7 to 9 donors, with the black line denoting the average of all donors tested ± SEM. *Significant difference compared with control siRNA-transfected cells (P < .05).
CXCR4 is required for TCR-initiated production of IL-2, IL-4, and IL-10. (A) Purified, human PBMC T cells were stimulated with 1 μg/mL plate-bound OKT3 and soluble CD28, cultured for 24 hours, and harvested for analysis via qRT-PCR for CXCL12 mRNA transcript levels. A human bone marrow mesenchymal stromal stem cell line (BMSC) was used as a positive control for CXCL12 expression. The results shown are normalized to the reference gene GAPDH, where GAPDH is set to 100. Each point denotes the mean mRNA transcript level ± standard deviation (SD) for 5 independent donors (n = 2 for BMSC samples). (B-E) Human PBMC T cells were purified, transfected with either control siRNA, a pool of CXCR4 siRNAs (CXCR4 siRNA-1) or CXCR4 siRNA-2 (single siRNA), and cultured for 24 hours. (B-C) CXCR4 cell surface levels were assayed via flow cytometry. Mean fluorescent intensities (MFI) are shown for a representative experiment. (C) Summarizes the results with each bar denoting the mean ± standard error of the mean (SEM). *Significantly different from control siRNA-transfected cells (P < .05; n = 7-9). (D-E) Twenty-four hours after transfection, cells were stimulated as in panel A, then cultured for an additional 24 hours prior to harvest of supernatants for cytokine analysis. Results from 7 to 9 donors, with the black line denoting the average of all donors tested ± SEM. *Significant difference compared with control siRNA-transfected cells (P < .05).
TCR associates with and transactivates CXCR4; this interaction is disrupted by AMD3100
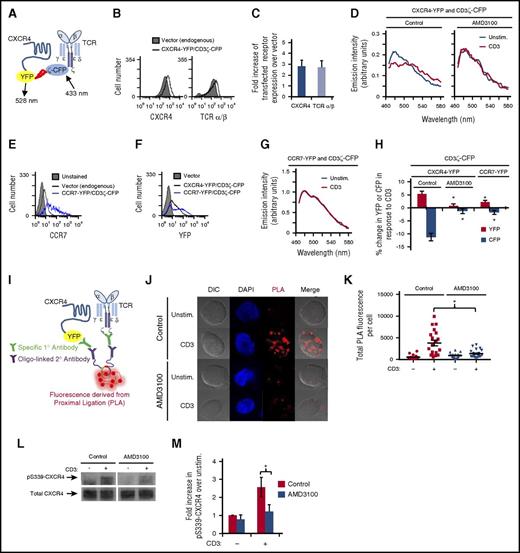
Several tyrosine kinase receptors transactivate CXCR4 in order to mediate signal transduction in various cell types.16-19 Here, we asked whether ligation of TCR induces an association between TCR and CXCR4 and/or transactivation of CXCR4 in order to mediate cytokine production. To determine whether this protein-protein interaction occurs, we used FRET6,7 to detect TCR-CXCR4 complex formation (Figure 2A). YFP-tagged CXCR4 and CFP-tagged CD3ζ were cotransfected into Jurkat T cells resulting in a twofold increase in CXCR4 and TCR expression levels (Figure 2B-C). If these proteins come within 10 nm of each other, FRET occurs with CFP donating energy to YFP when exposed to 433 nm of light. Indeed, upon T-cell activation, the YFP emission (525-550 nm) increased and the CFP emission (460-500 nm) decreased, indicating that CD3ζ-CFP associates with CXCR4-YFP upon T-cell activation resulting in the formation of TCR-CXCR4 complexes (Figure 2D,H). Addition of CXCL12 and/or CD28 did not enhance TCR-CXCR4 complex formation beyond CD3 stimulation (supplemental Figure 2A). We next sought to disrupt the formation of this complex. Previous studies indicate that the CXCR4 antagonist AMD3100 binds to CXCR4 and alters the conformation of CXCR4 in a manner that modifies its functions independent of CXCL12.36-38 We hypothesized that AMD3100 might alter the conformation of CXCR4 and thereby prevent TCR from interacting with CXCR4 upon T-cell activation. Indeed, pretreatment with AMD3100 prevented the donation of energy from CD3ζ-CFP to CXCR4-YFP upon TCR ligation (Figure 2D,H; supplemental Figure 2B), indicating inhibition of complex formation. Moreover, AMD3100 inhibited TCR-CXCR4 complex formation without altering CXCR4 cell surface levels (supplemental Figure 2C-F). As a control, coexpression of CCR7-YFP with CD3ζ-CFP did not result in a change in YFP and CFP fluorescence upon TCR ligation (Figure 2G-H), despite similar levels of CXCR4-YFP and CCR7-YFP expression (Figure 2E-F). To confirm the formation of TCR-CXCR4 complexes upon T-cell activation, we used a proximity ligation assay (PLA).39 PLA detects intimate protein-protein interactions spanning <40 nm. After first binding specific primary antibodies to proteins of interest, oligo-linked secondary antibodies are applied that, if in close proximity, act as a template for the formation and amplification of DNA circles detected via fluorescent probe hybridization (Figure 2I). Figure 2J-K show that few TCR-CXCR4 complexes, detectable by PLA fluorescence, are seen in the absence of T-cell activation. In contrast, ligation of TCR significantly increased PLA fluorescence, indicating that T-cell activation increases the formation of TCR-CXCR4 complexes. AMD3100 prevented the increase in PLA fluorescence upon T-cell activation (Figure 2J-K), consistent with AMD3100 also preventing TCR-CXCR4 complex formation detectable by FRET. Thus, Figure 2A-K show that TCR ligation enhances formation of TCR-CXCR4 complexes and that this complex formation can be inhibited by AMD3100.
TCR associates with and transactivates CXCR4; this interaction is disrupted by AMD3100. (A) Schematic diagram of FRET between fluorescent fusion proteins of CXCR4 and CD3ζ. (B-H) Jurkat T cells were transfected with the indicated fluorescent fusion proteins and cultured for 16 to 18 hours. (B,E-F) Transfected cells were analyzed via flow cytometry to assess cell surface levels of the indicated receptors or YFP. (C) Graph summarizing the mean fold increase of cell surface levels of the indicated receptor in cells transfected with CXCR4-YFP and CD3ζ-CFP compared with vector control transfected cells, ± SEM (n = 3). (D,G-H) Sixteen to 18 hours posttransfection, the cells were pretreated with 120 μM AMD3100 where indicated for 1 hour and then stimulated with 1 μg/mL OKT3 crosslinked with 0.1 mg/mL goat anti-mouse immunoglobulin G (IgG) for 20 minutes. Spectra of the same cells were obtained before and after stimulation. (D,G) Representative spectra are shown. (H) Summary of 3 to 4 independent experiments. Each bar denotes the percentage change in CFP or YFP in response to OKT3 stimulation. *Significant difference compared with CXCR4-YFP/CD3ζ-CFP, vehicle sample (P < .05). (I) Schematic diagram of a PLA analyzing interactions between CXCR4-YFP and CD3ζ. (J-K) Jurkat T cells were transfected with CXCR4-YFP, cultured for 16 to 18 hours, pretreated with AMD3100 for 1 hour, centrifuged onto fibronectin- and OKT3-coated coverslips, and incubated for 30 minutes at 37°C. Cells were then fixed and stained as described in “Methods.” PLA was visualized using an LSM780 laser-scanning confocal microscope (Carl Zeiss) with a 100×/1.46 oil objective and laser/emission filter: 488/500-554 for CXCR4-YFP to identify transfected cells, 405/411-481 for 4′,6-diamidino-2-phenylindole (DAPI) (blue) and 594/624 for PLA (red). ZEN software was used for acquisition of images. FIJI was used to assess total PLA fluorescence. (J) Representative results are shown (original magnification ×100). (K) Summary of images acquired in 3 independent experiments for a total of 17 to 30 cells per condition, ± SEM (P < .05). (L-M) Jurkat T cells were transfected with CXCR4-YFP, incubated for 16 to 18 hours, treated with 60 μM AMD3100 for 1 hour, stimulated with crosslinked OKT3 as in panel D for 5 minutes, lysed, harvested for immunoprecipitation for CXCR4, and immunoblotted for pS339-CXCR4 and total CXCR4. (L) Representative results are shown. (M) Summary of the mean fold increase in pS339-CXCR4 upon CD3 stimulation compared with unstimulated cells, ± SEM (n = 3; P < .05). DIC, differential interference contrast; Unstim., unstimulated.
TCR associates with and transactivates CXCR4; this interaction is disrupted by AMD3100. (A) Schematic diagram of FRET between fluorescent fusion proteins of CXCR4 and CD3ζ. (B-H) Jurkat T cells were transfected with the indicated fluorescent fusion proteins and cultured for 16 to 18 hours. (B,E-F) Transfected cells were analyzed via flow cytometry to assess cell surface levels of the indicated receptors or YFP. (C) Graph summarizing the mean fold increase of cell surface levels of the indicated receptor in cells transfected with CXCR4-YFP and CD3ζ-CFP compared with vector control transfected cells, ± SEM (n = 3). (D,G-H) Sixteen to 18 hours posttransfection, the cells were pretreated with 120 μM AMD3100 where indicated for 1 hour and then stimulated with 1 μg/mL OKT3 crosslinked with 0.1 mg/mL goat anti-mouse immunoglobulin G (IgG) for 20 minutes. Spectra of the same cells were obtained before and after stimulation. (D,G) Representative spectra are shown. (H) Summary of 3 to 4 independent experiments. Each bar denotes the percentage change in CFP or YFP in response to OKT3 stimulation. *Significant difference compared with CXCR4-YFP/CD3ζ-CFP, vehicle sample (P < .05). (I) Schematic diagram of a PLA analyzing interactions between CXCR4-YFP and CD3ζ. (J-K) Jurkat T cells were transfected with CXCR4-YFP, cultured for 16 to 18 hours, pretreated with AMD3100 for 1 hour, centrifuged onto fibronectin- and OKT3-coated coverslips, and incubated for 30 minutes at 37°C. Cells were then fixed and stained as described in “Methods.” PLA was visualized using an LSM780 laser-scanning confocal microscope (Carl Zeiss) with a 100×/1.46 oil objective and laser/emission filter: 488/500-554 for CXCR4-YFP to identify transfected cells, 405/411-481 for 4′,6-diamidino-2-phenylindole (DAPI) (blue) and 594/624 for PLA (red). ZEN software was used for acquisition of images. FIJI was used to assess total PLA fluorescence. (J) Representative results are shown (original magnification ×100). (K) Summary of images acquired in 3 independent experiments for a total of 17 to 30 cells per condition, ± SEM (P < .05). (L-M) Jurkat T cells were transfected with CXCR4-YFP, incubated for 16 to 18 hours, treated with 60 μM AMD3100 for 1 hour, stimulated with crosslinked OKT3 as in panel D for 5 minutes, lysed, harvested for immunoprecipitation for CXCR4, and immunoblotted for pS339-CXCR4 and total CXCR4. (L) Representative results are shown. (M) Summary of the mean fold increase in pS339-CXCR4 upon CD3 stimulation compared with unstimulated cells, ± SEM (n = 3; P < .05). DIC, differential interference contrast; Unstim., unstimulated.
To determine whether ligation of TCR alters the activation state of CXCR4, we used antibodies specific for phosphorylated serine 339 of CXCR4 (pS339-CXCR4). Phosphorylation of S339-CXCR4 correlates with CXCR4 internalization and signaling.18,40,41 Interestingly, ligation of TCR significantly increased phosphorylation of S339-CXCR4 (Figure 2L-M). Furthermore, AMD3100 inhibited this phosphorylation, consistent with the inhibition of TCR-CXCR4 complex formation preventing TCR transactivation of CXCR4 (Figure 2L-M). Together, results in Figure 2 show that ligation of TCR induces the formation of TCR-CXCR4 complexes and activation of CXCR4 via phosphorylation of S339, and that AMD3100 inhibits both TCR-CXCR4 complex formation and activation of CXCR4.
AMD3100 inhibits IL-2, IL-4, and IL-10 production by multiple T-cell subsets
Because AMD3100 blocks TCR-CXCR4 complex formation, we sought to determine whether AMD3100, similar to CXCR4 depletion (Figure 1), would also block cytokine production induced by ligation of TCR. Indeed, IL-2 production was significantly inhibited over a range of AMD3100 doses (from 1 to 30 μM) (Figure 3A; supplemental Figure 3A). AMD3100 also significantly decreased the amount of IL-4 and IL-10 produced by activated T cells. In contrast, AMD3100 failed to consistently inhibit IFN-γ production, but showed a minute increase in IFN-γ (Figure 3A). Importantly, AMD3100 reduced IL-2, IL-4, and IL-10 production without inducing apoptosis or altering CXCR4 cell surface levels (supplemental Figures 1C and 3B-C).
AMD3100 inhibits TCR-initiated production of IL-2, IL-4, and IL-10. Human PBMC T cells were treated with 60 μM AMD3100 for 1 hour and stimulated with plate-bound OKT3 and soluble CD28 as in Figure 1 for 24 hours. (A) Graphs summarize cytokine production assayed via ELISA from 9 donors, with the black line denoting the average of all donors tested ± SEM. *Significant difference compared with control cells (P < .05). (B-F) Following stimulation, cells were stained for CD3, CD4, CD45RO, and the indicated intracellular cytokine and analyzed by flow cytometry. Graphs summarize results from 5 donors, with the black line denoting the average of all donors tested ± SEM. *Significant difference compared with control cells (P < .05). FSC, forward scatter.
AMD3100 inhibits TCR-initiated production of IL-2, IL-4, and IL-10. Human PBMC T cells were treated with 60 μM AMD3100 for 1 hour and stimulated with plate-bound OKT3 and soluble CD28 as in Figure 1 for 24 hours. (A) Graphs summarize cytokine production assayed via ELISA from 9 donors, with the black line denoting the average of all donors tested ± SEM. *Significant difference compared with control cells (P < .05). (B-F) Following stimulation, cells were stained for CD3, CD4, CD45RO, and the indicated intracellular cytokine and analyzed by flow cytometry. Graphs summarize results from 5 donors, with the black line denoting the average of all donors tested ± SEM. *Significant difference compared with control cells (P < .05). FSC, forward scatter.
To determine whether both naive and memory CD4 and CD8 T-cell subsets require TCR-CXCR4 complex formation for cytokine production, we used intracellular cytokine staining of various T-cell subsets ± AMD3100. Unfortunately, IL-4–producing cells were not detectable due to low production of this cytokine. Nonetheless, AMD3100 significantly decreased the percentage of both CD4 naive (CD4+CD45RO−) and memory (CD4+CD45RO+) T cells producing IL-2 in response to CD3 + CD28 activation (Figure 3B-D). AMD3100 significantly decreased the percentage of CD4 memory cells producing IL-10 in response to CD3 + CD28 stimulation and had a modest effect on CD8 memory cells (Figure 3F). The percentage of CD4 memory cells producing IFN-γ was significantly inhibited by AMD3100, however, CD4 naive and CD8 T cells were not (Figure 3E). The results in Figure 3 indicate that inhibition of TCR-CXCR4 complex formation by AMD3100 consistently impairs IL-2, IL-4, and IL-10 production, and that various T-cell subsets including both naive and memory CD4 T cells use TCR-CXCR4 complex formation to mediate cytokine production.
AMD3100 inhibits cytokine mRNA stability without altering TCR-initiated ERK, NF-κB, or NFAT-signaling pathways
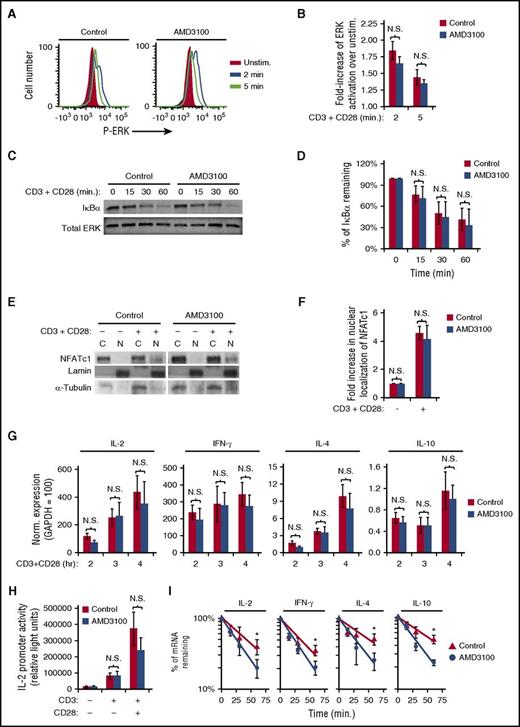
To identify the role of CXCR4 in TCR-mediated cytokine production, we first assayed TCR-activated signaling pathways focusing on AP-1–, NF-κB–, and NFAT-mediated pathways involved in cytokine transcriptional regulation. Surprisingly, AMD3100 treatment of primary human T cells did not alter TCR-initiated ERK activation, IκBα degradation, or NFAT nuclear localization (Figure 4A-F). These results suggested that CXCR4 is not required for cytokine gene transcription. Indeed, AMD3100 did not significantly alter transcript levels of IL-2, IFN-γ, IL-4, or IL-10 at 2, 3, or 4 hours following T-cell activation (Figure 4G). In addition, AMD3100 did not significantly reduce either CD3- or CD3 + CD28–stimulated IL-2 promoter luciferase activity in the Jurkat T-cell line (Figure 4H), also suggesting that inhibition of TCR-CXCR4 complex formation with AMD3100 does not prevent transcription of cytokine mRNAs. We next addressed whether TCR-CXCR4 complex formation is required for cytokine mRNA stability. Inhibition of TCR-CXCR4 complex formation via AMD3100 significantly increased the rate of degradation of IL-2, IFN-γ, IL-4, and IL-10 mRNA transcripts (Figure 4I). AMD3100 decreased the half-life from 41 to 26 minutes for IL-2 transcripts, from 53 to 29 minutes for IL-4 transcripts, and from 53 to 28 minutes for IL-10 transcripts, consistent with less IL-2, IL-4, and IL-10 secretion. Despite the minimal effect of AMD3100 on IFN-γ production, the half-life for IFN-γ mRNA decreased from 35 minutes to 24 minutes with AMD3100 (Figure 4I), suggesting that other mechanisms may partially compensate for decreased IFN-γ transcript stability in AMD3100-treated cells.42-44 Although CXCL12 has the ability to enhance CD3 + CD28–induced cytokine production via increasing AP-1 activity,6,32,45 CXCL12 did not increase stability of IL-2, IFN-γ, IL-4, or IL-10 transcripts (supplemental Figure 4A). The results in Figure 4 demonstrate that inhibition of TCR-CXCR4 complex formation by AMD3100 inhibits cytokine mRNA stability without altering all TCR-initiated signaling pathways including pathways required for cytokine mRNA transcription.
AMD3100 inhibits cytokine mRNA stability without altering TCR-initiated ERK-, NF-κB–, or NFAT-signaling pathways. (A-G) PBMC T cells were pretreated with AMD3100 for 1 hour and stimulated as indicated. (A-D) T cells were stimulated with 1 μg/mL biotinylated OKT3 crosslinked with avidin and soluble CD28 for the indicated time. (A) Representative results of ERK activation assayed by flow cytometry. (B) Summary of results in panel A; each bar denotes the mean fold increase of ERK activation over unstimulated cells, ± SEM (n = 4). (C) Representative results of cellular lysates that were isolated and blotted for IκBα degradation. (D) Summary of results in panel C; each bar denotes the mean percentage of IκBα remaining after stimulation compared with unstimulated cells (n = 3). (E-I) Cells were stimulated with CD3 + CD28 as in Figure 1. (E) Representative results of subcellular fractions isolated after 6 hours of CD3 + CD28 stimulation and immunoblotted for NFATc1. (F) Summary of panel E; each bar denotes mean fold increase in NFAT nuclear localization after stimulation compared with unstimulated cells (n = 3). (G) After stimulation with CD3 + CD28 for the indicated time, the cells were harvested for analysis via qRT-PCR for the indicated mRNA transcript levels. The results shown are normalized to the reference gene GAPDH, where GAPDH is set to 100. Each point denotes the mean mRNA transcript level ± SEM for 4 independent donors. (H) Jurkat T cells were transfected with an IL-2 promoter luciferase reporter construct, incubated for 16 to 18 hours, treated with 60 μM AMD3100 for 1 hour, stimulated as in Figure 1 for 16 hours, and assayed for luciferase activity. Representative experiment is shown. Each bar denotes the mean relative light units, ± SD (n = 4; P > .05) (unpaired Student t test). (I) PBMC T cells were pretreated with AMD3100 for 1 hour and stimulated as in Figure 1 for 5.5 hours prior to addition of actinomycin D. mRNA levels were assayed via qRT-PCR following actinomycin D treatment for the indicated times. Each point denotes the mean percentage of mRNA remaining ± SEM. *Significantly different from control cells (n = 3-4). C, cytoplasmic; N, nuclear; N.S., no significant difference (P > .05).
AMD3100 inhibits cytokine mRNA stability without altering TCR-initiated ERK-, NF-κB–, or NFAT-signaling pathways. (A-G) PBMC T cells were pretreated with AMD3100 for 1 hour and stimulated as indicated. (A-D) T cells were stimulated with 1 μg/mL biotinylated OKT3 crosslinked with avidin and soluble CD28 for the indicated time. (A) Representative results of ERK activation assayed by flow cytometry. (B) Summary of results in panel A; each bar denotes the mean fold increase of ERK activation over unstimulated cells, ± SEM (n = 4). (C) Representative results of cellular lysates that were isolated and blotted for IκBα degradation. (D) Summary of results in panel C; each bar denotes the mean percentage of IκBα remaining after stimulation compared with unstimulated cells (n = 3). (E-I) Cells were stimulated with CD3 + CD28 as in Figure 1. (E) Representative results of subcellular fractions isolated after 6 hours of CD3 + CD28 stimulation and immunoblotted for NFATc1. (F) Summary of panel E; each bar denotes mean fold increase in NFAT nuclear localization after stimulation compared with unstimulated cells (n = 3). (G) After stimulation with CD3 + CD28 for the indicated time, the cells were harvested for analysis via qRT-PCR for the indicated mRNA transcript levels. The results shown are normalized to the reference gene GAPDH, where GAPDH is set to 100. Each point denotes the mean mRNA transcript level ± SEM for 4 independent donors. (H) Jurkat T cells were transfected with an IL-2 promoter luciferase reporter construct, incubated for 16 to 18 hours, treated with 60 μM AMD3100 for 1 hour, stimulated as in Figure 1 for 16 hours, and assayed for luciferase activity. Representative experiment is shown. Each bar denotes the mean relative light units, ± SD (n = 4; P > .05) (unpaired Student t test). (I) PBMC T cells were pretreated with AMD3100 for 1 hour and stimulated as in Figure 1 for 5.5 hours prior to addition of actinomycin D. mRNA levels were assayed via qRT-PCR following actinomycin D treatment for the indicated times. Each point denotes the mean percentage of mRNA remaining ± SEM. *Significantly different from control cells (n = 3-4). C, cytoplasmic; N, nuclear; N.S., no significant difference (P > .05).
Activation of TCR leads to CXCR4-mediated stabilization of cytokine mRNA via activation of a PREX1-Rac1–signaling pathway
Rac1 has previously been described to mediate mRNA stability in various cell types23-27 and is activated in response to both TCR- and CXCR4-induced signaling.8,28,29 Therefore, we hypothesized that TCR-CXCR4 signaling activates Rac1 in order to stabilize cytokine mRNA. Significantly, inhibiting TCR-CXCR4 complex formation with AMD3100 impaired Rac1 activation arising from CD3 + CD28 stimulation (Figure 5A-B), suggesting that TCR transactivates CXCR4 in order to activate Rac1. Pretreatment with NSC23766, a Rac-specific inhibitor,19,46,47 inhibited Rac1 activation as expected (Figure 5C-D) and also increased the degradation rate of IL-2, IFN-γ, IL-4, and IL-10 mRNA transcripts (Figure 5E), suggesting a role for Rac1 in stabilizing cytokine transcripts. To confirm these results, we inserted the 3′UTR of IL-2 into pmirGLO, a luciferase reporter plasmid driven by a constitutive promotor, and then assessed stabilization of this heterologous reporter upon TCR activation. Indeed, CD3 + CD28 stimulation increased luciferase expression, and, importantly, both AMD3100 and NSC23766 significantly inhibited CD3 + CD28–induced luciferase expression (Figure 5F), indicating that CXCR4 and Rac1 signaling mediates stabilization of cytokine transcripts. NSC23766 also significantly inhibited the production of IL-2, IL-4, and IL-10, but not IFN-γ (Figure 5G) similar to either AMD3100 pretreatment or CXCR4 depletion. Thus, the results in Figure 5A-G indicate that T-cell activation induces TCR to transactivate CXCR4 in order to activate a Rac1-signaling pathway that stabilizes cytokine mRNA.
Activation of TCR leads to CXCR4-mediated stabilization of cytokine mRNA by activation of a PREX1-Rac1–signaling pathway. (A-D) PBMC T cells were treated with 60 μM AMD3100 or 50 μM NSC23766 for 1 hour, stimulated with 5 μg/mL biotinylated OKT3 crosslinked with avidin and soluble CD28 for 5 minutes, and assayed for active, GTP-bound Rac1, or total Rac1. Vertical white lines between bands indicate removal of an irrelevant lane from the gel image. (B,D) Summary of results as in panels A and C, respectively; each bar denotes the fold increase in activated Rac1 in stimulated cells compared with unstimulated cells, (n = 3-5; P < .05). (E) PBMC T cells were treated with 50 μM NSC23766 for 1 hour, stimulated with 1 μg/mL plate-bound OKT3 and soluble CD28 for 5.5 hours prior to addition of actinomycin D and assessment of mRNA levels as in Figure 4I (n = 7-10; P < .05). Donor samples were included in this analysis only if there was less than a twofold difference in starting transcript levels between treated and untreated samples at 1 minute of actinomycin D treatment. (F) PBMC T cells were transfected with pmir-GLO empty vector or pmir-GLO-3′UTR at 350 V on a BTX square wave electroporator, treated with 60 μM AMD3100 or 50 μM NSC23766 for 1 hour, stimulated with 1 μg/mL plate-bound OKT3 and soluble CD28 Ab for 5.5 hours, harvested, lysed, and assayed for luciferase activity. Luciferase activity of pmir-GLO-3′UTR was normalized to pmir-GLO empty vector. Representative experiment is shown with each bar denoting mean relative light units ± SD (n = 3; P < .05) (unpaired t test). (G) PBMC T cells were treated with 50 μM NSC23766 for 1 hour, stimulated as in panel F for 24 hours, and supernatants were analyzed for the indicated cytokines as in Figure 1D-E (n = 6). (H-M) PBMC T cells were transfected with PREX1 siRNA-1 (pool of siRNAs), PREX1 siRNA-2 (single siRNA) or control siRNA, cultured for 48 hours, and stimulated as indicated below. (H-I,K-L) Cells were stimulated as in panel A and harvested to either immunoblot PREX1 and actin expression or assay GTP-bound Rac activation as in panels A and B (n = 3). (J,M) Forty-eight hours after transfection, cells were stimulated as in panel G for 24 hours and supernatants were analyzed for the indicated cytokines as in Figure 1D-E (n = 6).
Activation of TCR leads to CXCR4-mediated stabilization of cytokine mRNA by activation of a PREX1-Rac1–signaling pathway. (A-D) PBMC T cells were treated with 60 μM AMD3100 or 50 μM NSC23766 for 1 hour, stimulated with 5 μg/mL biotinylated OKT3 crosslinked with avidin and soluble CD28 for 5 minutes, and assayed for active, GTP-bound Rac1, or total Rac1. Vertical white lines between bands indicate removal of an irrelevant lane from the gel image. (B,D) Summary of results as in panels A and C, respectively; each bar denotes the fold increase in activated Rac1 in stimulated cells compared with unstimulated cells, (n = 3-5; P < .05). (E) PBMC T cells were treated with 50 μM NSC23766 for 1 hour, stimulated with 1 μg/mL plate-bound OKT3 and soluble CD28 for 5.5 hours prior to addition of actinomycin D and assessment of mRNA levels as in Figure 4I (n = 7-10; P < .05). Donor samples were included in this analysis only if there was less than a twofold difference in starting transcript levels between treated and untreated samples at 1 minute of actinomycin D treatment. (F) PBMC T cells were transfected with pmir-GLO empty vector or pmir-GLO-3′UTR at 350 V on a BTX square wave electroporator, treated with 60 μM AMD3100 or 50 μM NSC23766 for 1 hour, stimulated with 1 μg/mL plate-bound OKT3 and soluble CD28 Ab for 5.5 hours, harvested, lysed, and assayed for luciferase activity. Luciferase activity of pmir-GLO-3′UTR was normalized to pmir-GLO empty vector. Representative experiment is shown with each bar denoting mean relative light units ± SD (n = 3; P < .05) (unpaired t test). (G) PBMC T cells were treated with 50 μM NSC23766 for 1 hour, stimulated as in panel F for 24 hours, and supernatants were analyzed for the indicated cytokines as in Figure 1D-E (n = 6). (H-M) PBMC T cells were transfected with PREX1 siRNA-1 (pool of siRNAs), PREX1 siRNA-2 (single siRNA) or control siRNA, cultured for 48 hours, and stimulated as indicated below. (H-I,K-L) Cells were stimulated as in panel A and harvested to either immunoblot PREX1 and actin expression or assay GTP-bound Rac activation as in panels A and B (n = 3). (J,M) Forty-eight hours after transfection, cells were stimulated as in panel G for 24 hours and supernatants were analyzed for the indicated cytokines as in Figure 1D-E (n = 6).
Transactivation of CXCR4 by EGFR can lead to activation of the Rac-GEF PREX1.17,19 Therefore, we sought to determine whether transactivation of CXCR4 by TCR ligation utilizes PREX1 to activate Rac1. Human T cells transfected with distinct PREX1 siRNAs failed to activate Rac1 upon T-cell activation compared with cells transfected with control siRNA (Figure 5H-I,K-L). Moreover, depletion of PREX1 resulted in a significant decrease in IL-2, IL-4, and IL-10 production (Figure 5J,M). Together, the results in Figure 5 suggest that upon ligation of TCR, TCR transactivates CXCR4 in order to activate a PREX1-Rac1–signaling pathway that stabilizes cytokine mRNA transcripts to increase cytokine secretion.
Inhibition of TCR-CXCR4–mediated Rac1 signaling blocks cytokine production by the Sézary syndrome cell line, HUT-78, and patient isolates
Malignant T cells of CTCLs, including Sézary syndrome, secrete cytokines that contribute to disease progression by providing a microenvironment that promotes malignancy.1-5 To determine whether the cytokine production of these malignant T cells can be targeted by inhibiting TCR-CXCR4 complex formation or its downstream Rac1 signaling, we used the HUT-78 cell line derived from a patient with Sézary syndrome, a variant of CTCL with widespread systemic involvement. HUT-78 cells do not produce IL-2 or IFN-γ constitutively,48 however, IL-2 and IFN-γ production were both significantly increased upon stimulation with CD3 (Figure 6A). HUT-78 cells lack CD28,48 therefore, we observed no increased IL-2 production with CD28 costimulation (data not shown). Pretreatment with either AMD3100 or NSC23766 significantly decreased both IL-2 and IFN-γ production in response to TCR ligation (Figure 6A). Similarly, AMD3100 and NSC23766 inhibited CD3-induced luciferase expression from the pmirGLO-3′UTR heterologous reporter, indicating that CXCR4 and Rac1 activity are required to stabilize CD3-induced cytokine mRNA via the 3′UTR (Figure 6B). Interestingly, HUT-78 cells constitutively secrete IL-10,49 and CD3 stimulation does not increase the amount of IL-10 secreted (Figure 6C), suggesting that IL-10 production is TCR-independent. Accordingly, AMD3100 failed to inhibit the constitutive IL-10 production by HUT78 cells (Figure 6C). In contrast, targeting this pathway further downstream with NSC23766 significantly decreased constitutive IL-10 production (Figure 6C). IL-4 production was not detectable in HUT78 cells. In Figure 6D-G, we show data obtained from rare residual diagnostic patient specimens that were limited by the small amount of sample received. Nonetheless, we assessed the effect of NSC23766 on the cells from a patient with peripheral T-cell lymphoma (TCL-1) showing clinical features and phenotype consistent with transformed mycosis fungoides or Sézary syndrome. In addition, we assessed the effect of NSC23766 on cells from bone marrow (TCL-3) and peripheral blood (TCL-4) specimens, both of which contained abnormal CD4+ T-cell populations phenotypically consistent with mycosis fungoides or Sézary syndrome. IL-2 production was increased in the presence of CD3 and CD28 mAbs in specimens TCL-1 and TCL-4, with a more limited effect on TCL-4 which had lower expression of CD3 but higher levels of constitutive IL-2 production (Figure 6D,G). Interestingly, NSC23766 significantly impaired CD3 + CD28–induced IL-2 production in all 3 patient samples tested (Figure 6E-F). Thus, targeting either inducible cytokine production or constitutive cytokine production by malignant T cells appears feasible by inhibiting different points of this TCR-CXCR4 to PREX1-Rac1–signaling pathway. Together, the results in this article support the model shown in Figure 7; activation of TCR induces the TCR to bind and activate CXCR4 on S339, which leads to CXCR4-mediated stabilization of cytokine mRNA via activation of a PREX1-Rac1–signaling pathway. Moreover, we show that targeting this pathway in malignant T cells inhibits cytokine production.
Inhibition of the TCR-CXCR4–mediated Rac1 signaling blocks cytokine production by the Sézary syndrome cell line, HUT-78, and patient isolates. (A) TCR-dependent IL-2 and IFN-γ secretion in HUT78 cells was assayed following 1 hour pretreatment with vehicle, 60 μM AMD3100 or 50 μM NSC22766 and stimulation with 2 μg/mL plate-bound OKT3 for 24 hours as in Figure 1D-E. Each bar denotes the mean cytokine production, ± SEM (n = 3; P < .05). (B) HUT78 cells were transfected with pmir-GLO empty vector or pmir-GLO-3′UTR via electroporation at 315 V, treated with 60 μM AMD3100 or 50 μM NSC23766 for 1 hour, stimulated with 2 μg/mL plate-bound OKT3 for 3.5 hours, harvested, lysed, and assayed for luciferase activity as in Figure 5F. Representative experiment is shown with each bar denoting mean relative light units ± SD (n = 3; P < .05) (unpaired Student t test). (C) HUT78 cells were treated with 60 μM AMD3100 or 100 μΜ NSC23766 for 1 hour, cultured ± 2 μg/mL plate-bound OKT3 for 24 hours, and assayed for IL-10 secretion as in Figure 1 (n = 3). (D-G) T cells isolated from residual diagnostic patient specimens (TCL-1, 3, 4) were pretreated where indicated with 50 μM NSC23766, stimulated with 1 μg/mL plate-bound OKT3 and soluble CD28 for 24 hours, and assayed for IL-2 production as in Figure 1D. (F) Graph summarizes results from panel E. Each bar denotes the percentage change in cytokine production normalized to the corresponding cytokine production by control-treated cells for each stimulation condition (n = 3; ± SEM; P < .05). (G) Patient information.
Inhibition of the TCR-CXCR4–mediated Rac1 signaling blocks cytokine production by the Sézary syndrome cell line, HUT-78, and patient isolates. (A) TCR-dependent IL-2 and IFN-γ secretion in HUT78 cells was assayed following 1 hour pretreatment with vehicle, 60 μM AMD3100 or 50 μM NSC22766 and stimulation with 2 μg/mL plate-bound OKT3 for 24 hours as in Figure 1D-E. Each bar denotes the mean cytokine production, ± SEM (n = 3; P < .05). (B) HUT78 cells were transfected with pmir-GLO empty vector or pmir-GLO-3′UTR via electroporation at 315 V, treated with 60 μM AMD3100 or 50 μM NSC23766 for 1 hour, stimulated with 2 μg/mL plate-bound OKT3 for 3.5 hours, harvested, lysed, and assayed for luciferase activity as in Figure 5F. Representative experiment is shown with each bar denoting mean relative light units ± SD (n = 3; P < .05) (unpaired Student t test). (C) HUT78 cells were treated with 60 μM AMD3100 or 100 μΜ NSC23766 for 1 hour, cultured ± 2 μg/mL plate-bound OKT3 for 24 hours, and assayed for IL-10 secretion as in Figure 1 (n = 3). (D-G) T cells isolated from residual diagnostic patient specimens (TCL-1, 3, 4) were pretreated where indicated with 50 μM NSC23766, stimulated with 1 μg/mL plate-bound OKT3 and soluble CD28 for 24 hours, and assayed for IL-2 production as in Figure 1D. (F) Graph summarizes results from panel E. Each bar denotes the percentage change in cytokine production normalized to the corresponding cytokine production by control-treated cells for each stimulation condition (n = 3; ± SEM; P < .05). (G) Patient information.
Proposed model of TCR-CXCR4–mediated stabilization of cytokine mRNA transcripts. Ligation of TCR induces the TCR to associate with and transactivate CXCR4 on S339 which leads to stabilization of IL-2, IL-4, and IL-10 mRNA transcripts via activation of a PREX1-Rac1–signaling pathway. CDS, coding sequence.
Proposed model of TCR-CXCR4–mediated stabilization of cytokine mRNA transcripts. Ligation of TCR induces the TCR to associate with and transactivate CXCR4 on S339 which leads to stabilization of IL-2, IL-4, and IL-10 mRNA transcripts via activation of a PREX1-Rac1–signaling pathway. CDS, coding sequence.
Discussion
As with most immunopathological diseases, targeting the aberrant immune responses of CTCLs has proven to be more effective than cytotoxic chemotherapy. Immunotherapies that target CTCLs via T-cell–specific mAbs have shown some efficacy, however, relapse usually occurs within months to a few years.3,4,50 Targeting the disordered immunological activity of CTCLs is complicated by the changing cytokine milieu within the microenvironment that drives phenotypic changes of CTCL cells.1-5 Therefore, inhibiting the aberrant release of the cytokines that drive the T-cell phenotypic changes of CTCL cells would likely slow or inhibit the immunopathological progression of the disease.
Here, we describe a novel signaling pathway that mediates the mRNA stability of IL-2, IL-4, and IL-10 transcripts and, therefore, promotes the production and secretion of these cytokines. Utilizing primary human T cells, we show that ligation of the TCR induces the TCR to associate with and transactivate CXCR4 on S339, which leads to stabilization of IL-2, IL-4, and IL-10 mRNA transcripts via activation of a PREX1-Rac1–signaling pathway. Importantly, we showed that the CXCR4 antagonist, AMD3100, inhibited TCR-CXCR4 complex formation and thus Rac1 activation, mRNA stabilization, and secretion of these cytokines. Additionally, the Rac1 inhibitor, NSC23766, also inhibited the mRNA stabilization and secretion of these cytokines. Applying this new knowledge to CTCLs, we show that inhibition of the TCR-CXCR4 to the PREX1-Rac1–signaling pathway inhibited the release of TCR-dependent and TCR-independent cytokine production in the Sézary syndrome–derived cell line, HUT78, and patient isolates. Thus, we describe here a novel signaling pathway that can be targeted to disrupt the secretion of cytokines to treat various immunopathological diseases including CTCLs.
We have identified the transactivation of CXCR4 upon ligation of TCR as a targetable phenomenon to regulate the secretion of cytokines in multiple T-cell subsets. Transactivation of CXCR4 by various receptor tyrosine kinases has been linked to both normal and disease-related signaling in multiple cell types.16-19 This transactivation often involves a physical association between receptors and site-specific phosphorylation of CXCR4 that activates various signaling outcomes.16-18,40,51 Similarly, we found that TCR stimulation resulted in both TCR-CXCR4 complex formation and phosphorylation of CXCR4 on serine 339. Previous studies showed that AMD3100 binds to and alters the conformation of CXCR4 in a manner that modifies its function.36-38 Applying that knowledge, we show here that AMD3100 disrupts TCR transactivation of CXCR4, most likely by altering the conformation of CXCR4 such that TCR cannot associate. Remarkably, this inhibition occurred in both naive and memory CD4 T cells, suggesting that a similar pathway is intact in these T-cell subsets. In contrast, ERK-, NFAT-, and NF-κB–signaling pathways were unaffected by inhibition of TCR-CXCR4 complex formation, suggesting that this complex does not alter all TCR-induced signaling pathways. Additionally, IFN-γ production was not consistently altered by targeting TCR-CXCR4–specific signaling, despite a decrease in IFN-γ mRNA stability, suggesting that transcriptional or translational mechanisms unique to IFN-γ may compensate for decreased mRNA stability. IFN-γ expression levels are affected transcriptionally by polymorphisms in the IFN-γ gene as well as translationally by metabolic mechanisms.42-44,52 Indeed, multiple factors contribute to a T-cell cytokine expression profile, however, a common link for IL-2, IL-4, and IL-10 production is the transactivation of CXCR4 by the TCR to maintain cytokine mRNA stability.
We describe here a novel role for a PREX1-Rac1–signaling pathway that mediates the mRNA stability of cytokines. Previous studies demonstrate that Rac1 mediates the stability of various mRNA transcripts in multiple normal and cancerous cell types.23-27 Ramgolam et al showed that Rac1 activation of p38MAPK stabilized IFN-γ mRNA stability,26 however, p38MAPK activity was unaffected upon inhibition of TCR-CXCR4 complex formation (data not shown). Therefore, future studies are necessary to identify the Rac1-mediated mRNA-stabilizing mechanism. Nonetheless, we identified the RacGEF PREX1 as an activator of Rac1 upon TCR ligation. Interestingly, EGFR transactivation of CXCR4 leads to PREX1-mediated activation of Rac1 in a tumor cell line.17,19 PREX1, Rac1, and CXCR4 expression are often upregulated in cancerous cells,19,53-57 suggesting that transactivation of CXCR4 by other receptor tyrosine kinases could drive this mRNA-stabilizing pathway in malignancy as well.
Targeting different aspects of this TCR-CXCR4 mRNA-stabilizing pathway has demonstrated that we can block both TCR-dependent and TCR-independent cytokine secretion from a Sézary syndrome–derived cell line and patient isolates. AMD3100 only blocked TCR-dependent IL-2 and IFN-γ secretion whereas NSC23766 blocked both TCR-dependent and constitutive TCR-independent IL-10 secretion. Inhibition of constitutive IL-10 secretion is critical as the CTCLs begin to develop a Treg phenotype as the disease progresses.2-5 By characterizing the signaling pathway downstream of TCR-CXCR4 complex formation, we have identified a targetable signaling component, Rac1, which remains intact in CTCL cells. Together, these results indicate that targeting the TCR-CXCR4–signaling pathway may be a viable approach to blocking aberrant cytokine secretion that leads to the immunopathology of CTCL.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors are grateful for the assistance of the Mayo Clinic Flow Cytometry Core Facility. The authors thank the Mayo Clinic Department of Laboratory Medicine and Pathology for providing residual diagnostic samples. The authors also thank the healthy donors and patients who provided samples for this research.
This work was supported in part by the Joanne G. and Gary N. Owen Fund in Immunology Research, the Alma B. Stevenson Endowment Fund for Medical Research, and by National Institutes of Health (NIH) National Institute of General Medical Sciences grant RO1GM59763 (K.E.H.) and National Institute of Allergy and Infectious Diseases grant T32-AI07047 (D.G.O.). R.M.S. was supported by the Mayo Clinic Medical Scientist Training Program Robert L. Howell Physician-Scientist Scholarship. B.A.D. was supported by the NIH National Institute of Allergy and Infectious Diseases Ph.D. training grant in Basic Immunology (T32 AI07425) and the Mayo Clinic Graduate School of Biomedical Sciences (MCGSBS).
Authorship
Contribution: K.N.K. and B.A.D. designed, performed, and analyzed experiments; D.J. was critical for the acquisition of residual patient specimens and diagnoses; and K.N.K. wrote the manuscript with critical input from B.A.D., R.M.S., D.G.O., D.J., and K.E.H.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Karen E. Hedin, Department of Immunology, Mayo Clinic, Guggenheim Building, 3rd Floor, 200 First St Southwest, Rochester, MN 55905; e-mail: hedin.karen@mayo.edu.
References
Author notes
K.N.K. and B.A.D. contributed equally to this study.