Key Points
Genomic analysis of 220 CTCLs identifies 55 genes in lymphomagenesis, including 17 genes not previously implicated in CTCL.
RLTPR (p.Q575E) is a novel gain-of-function mutation that potentiates T-cell receptor signaling via selective upregulation of the NF-κB pathway.
Abstract
Cutaneous T-cell lymphoma (CTCL) is an incurable non-Hodgkin lymphoma of the skin-homing T cell. In early-stage disease, lesions are limited to the skin, but in later-stage disease, the tumor cells can escape into the blood, the lymph nodes, and at times the visceral organs. To clarify the genomic basis of CTCL, we performed genomic analysis of 220 CTCLs. Our analyses identify 55 putative driver genes, including 17 genes not previously implicated in CTCL. These novel mutations are predicted to affect chromatin (BCOR, KDM6A, SMARCB1, TRRAP), immune surveillance (CD58, RFXAP), MAPK signaling (MAP2K1, NF1), NF-κB signaling (PRKCB, CSNK1A1), PI-3-kinase signaling (PIK3R1, VAV1), RHOA/cytoskeleton remodeling (ARHGEF3), RNA splicing (U2AF1), T-cell receptor signaling (PTPRN2, RLTPR), and T-cell differentiation (RARA). Our analyses identify recurrent mutations in 4 genes not previously identified in cancer. These include CK1α (encoded by CSNK1A1) (p.S27F; p.S27C), PTPRN2 (p.G526E), RARA (p.G303S), and RLTPR (p.Q575E). Last, we functionally validate CSNK1A1 and RLTPR as putative oncogenes. RLTPR encodes a recently described scaffolding protein in the T-cell receptor signaling pathway. We show that RLTPR (p.Q575E) increases binding of RLTPR to downstream components of the NF-κB signaling pathway, selectively upregulates the NF-κB pathway in activated T cells, and ultimately augments T-cell-receptor-dependent production of interleukin 2 by 34-fold. Collectively, our analysis provides novel insights into CTCL pathogenesis and elucidates the landscape of potentially targetable gene mutations.
Introduction
Cutaneous T-cell lymphoma (CTCL) is a cancer of the mature skin-homing CD4+ T cell. Patients with advanced disease develop intractable skin lesions, with potential spread of the tumor cells to the blood, the lymph nodes, and the visceral organs. At this time, there are no cures for CTCL, and the median survival for patients with stage IV disease remains less than 5 years.1
Recently, we and other groups have sequenced patient-derived samples to improve our understanding of the genetic basis of CTCL.2-10 These efforts have resulted in significant advances; however, important questions persist. In particular, there is limited consensus across the various genomics studies on the identity and the prevalence of putative driver genes.2-10
There are many potential explanations for these discordant findings. Importantly, the CTCL genomics studies employed different analytical methods, which used nonuniform metrics to prioritize putative driver genes.2-10 Perhaps more importantly, the cohort size of each of these genomic analyses was relatively small (between 5 and 66 patients per study). Pan-cancer analyses performed by the Cancer Genome Atlas suggest these studies are underpowered to identify all putative driver genes that are mutated in less than 10% of samples.11 Nearly all the point mutations implicated in CTCL occur in this range.2-5,7-9 Therefore, on the basis of the Cancer Genome Atlas data, confident identification of driver genes requires a cohort size of at least 200 samples,11 which is a cohort ∼3 times larger than the largest CTCL genomics cohort published to date.
To overcome these limitations, we have analyzed the mutation data from 220 CTCLs. These samples represent the aggregate cohort of all CTCLs with publicly available sequencing data.2-10 The size of the patient cohort has enabled us to identify genes with statistically significant mutation burdens. We use a multitiered analytical pipeline and have identified 55 putative driver genes in CTCL, including 17 genes that have not been implicated in CTCL to date. Last, we provide the first genetic and functional data establishing RLTPR, which encodes a scaffolding protein in the T-cell-receptor signaling pathway, as a putative oncogene.
Methods
Data analysis
We performed a search on PubMed on August 1, 2015, updated on October 1, 2016. We identified 9 studies with published sequencing data (supplemental Tables 1 and 2, available on the Blood Web site). We both performed quality control measures to standardize mutation calls and gene nomenclature and performed a multitiered analytical pipeline described in more detail in supplemental Figures 1-7.
Protein 3D structure analysis
Locations of mutated residues were identified on crystal structures of the relevant proteins or their homologs. Structures were analyzed and figures generated using Pymol (https://www.pymol.org).
Cloning and mutagenesis
RLTPR (HsCD00403288), ARHGEF3 (HsCD00456829), and CSNK1A1 (HG10668-M) cDNA were purchased from DNASU, PlasmID Repository, and Sino Biological, respectively. All cDNAs were mutagenized by Gibson cloning and subcloned into the pCDH-CMV-MCS-EF1-copGFP vector (Systems Biosciences; provided by DNA/RNA Delivery Core, Skin Disease Research Center, Northwestern University, Chicago, IL). Relevant primers are listed in supplemental Table 3.
Cell culture
Jurkat cells (clone E6-1; ATCC) and HEK293T cells were maintained in RPMI-1640 and Dulbecco’s modified Eagle medium, respectively. All media were supplemented with 10% fetal bovine serum and antibiotics. Cells were lentivirally transduced, as previously described.9 Lentivirally transduced cells were selected on the basis of green fluorescent protein expression. In all cases, 3 independent cell lines were generated by transduction of HEK293T or Jurkat cells on 3 separate occasions. Each of these cell lines was tested in 2 biological replicates. Therefore, unless otherwise specified, all experiments were performed in 6 biological replicates. For stimulation, cells were cultured with phorbol 12-myristate 13-acetate (PMA; 50 ng/mL, Sigma) and ionomycin (300 ng/mL, Sigma), with or without CD86 (R&D systems), for the times indicated.
Immunoblot analysis
Whole-cell lysates were analyzed by immunoblotting, using the following antibodies: anti-RLTPR (ab122717, Abcam), anti-ARHGEF3 (ab136064, Abcam), anti-β-actin (sc-47778, Santa Cruz Biotechnology), anti-CARMA1 (4440S, Cell Signaling Technology), anti-CK1α (ab206652, Abcam), anti-Flag (F3165, Sigma-Aldrich), and horseradish peroxidase-conjugated secondary antibodies (sc-2004; sc-2005, Santa Cruz Biotechnology).
Immunoprecipitation assay
Jurkat cells expressing either Flag-tagged WT RLTPR or RLTPR (p.Q575E) were cultured with and without PMA and ionomycin stimulation for 30 minutes. Lysates were made from 20 million cells and incubated with anti-Flag beads (Sigma-Aldrich) for 4 h at 4°C. Bead bound proteins were eluted by sample buffer and probed by immunoblotting.
Quantification of RHOA-GTP
Guanosine triphosphate (GTP)–bound RHOA was detected via G-LISA assays, according to the manufacturer’s specifications (BK124, Cytoskeleton). Absorbance levels of vector controls were subtracted from all values. All values were normalized to HEK293T cells lentivirally transduced with wild-type (WT) ARHGEF3.
siRNA transfection
Three hundred thousand Jurkat cells were nucleofected in R buffer with the Neon Transfection system (Invitrogen). To minimize off-target effects,12 cells were nucleofected with SMARTpool siRNA, a mixture of 4 siRNAs against the same target (siCTRL [D-001206-14-05; Dharmacon] or siRLTPR [M-022592-01-0005; Dharmacon]). After transfection, cells were cultured without antibiotics for 3 days before their use in subsequent assays.
CD4+ T cells were isolated from peripheral blood mononuclear cells, using Dynabeads CD4 Positive Isolation kit (Invitrogen) according to the manufacturer’s manual. Two million CD4+ T cells were nucleofected with siCTRL or siRLTPR, using Amaxa Human T cell Nucleofector kit (Lonza).
Interleukin 2 assay
Three hundred thousand Jurkat cells were cultured with vehicle control or PMA and ionomycin ± CD86 for 6 hours. For isolated CD4+ T cells, 1 million cells were incubated with anti-CD3/anti-CD28 activation beads (Gibco) for 6 hours. Relative transcript levels were detected by SYBR Green, as previously described9 (Bio-Rad). Relevant primers are listed in supplemental Table 3.
RNA sequencing
Jurkat cells expressing WT RLTPR or RLTPR (p.Q575E) were cultured with or without PMA and ionomycin treatment of 6 hours. At the Yale Center for Genome Analysis, cDNA libraries were generated from 500 ng mRNA extracted from these cells (Qiagen), using oligo-DT beads (KAPA Biosystems), and sequenced using 75-bp paired-end sequencing on an Illumina HiSequation 2500, according to Illumina protocols. RNA integrity number scores of samples ranged between 9.7 and 10 (mean = 9.97).
RNA-Seq analysis
For RNA-Seq analysis, we aligned the data using STAR,13 quantified gene-specific transcripts using HT-Seq,14 and identified differentially expressed transcripts using DE-Seq2.15 For gene set variation analysis,16 we used published gene sets (http://software.broadinstitute.org/gsea/msigdb/index.jsp). For transcription factor binding site analysis, we used OPOSSUM (http://opossum.cisreg.ca/oPOSSUM3/).17 Volcano plots and dot plots were generated using R (https://cran.r-project.org). All software employed default settings.
Results
Genome sequencing analysis of 220 CTCLs
We hypothesized that a large cohort size would provide the statistical power to enable confident identification of putative driver genes. To assemble this cohort, we collated the published genomic data from all CTCLs sequenced to date. A PubMed search identified 9 studies with publicly available next-generation sequencing data (supplemental Tables 1 and 2).2-10 We downloaded the point mutation calls, filtered the data to eliminate common germline variants, and standardized gene nomenclature. In total, we analyzed next-generation sequencing data from 220 patients with 23 028 point mutations distributed across 9996 genes. The cohort of CTCLs included 186 reported cases of leukemic CTCL (Sezary syndrome [SS]), 25 reported cases of mycosis fungoides (MF), and 9 reported cases of CTCL not otherwise specified (CTCL-NOS; supplemental Table 4). The distribution of mutations was consistent with what we had previously observed (supplemental Figure 8).9 The most common nucleotide change was C>T, which made up 67.5% of all mutations. The median number of nonsynonymous mutations was 42.
Identification of putative driver genes in CTCL
To identify putative driver genes, we used multiple orthogonal algorithms. Specifically, we employed multiple, independent analytical pipelines to look for mutational signatures characteristic for putative oncogenes and tumor suppressors (supplemental Figures 1-7; supplemental Tables 5-16).
Oncogenes harbor recurrent hotspot mutations at identical amino acid positions.18 Therefore, to identify putative oncogenes, we looked for recurrent amino acid substitutions that occurred more often than expected by chance alone. In contrast, tumor suppressors are characterized by recurrent loss-of-function mutations. Therefore, to identify putative tumor suppressors, we looked for genes that had a higher burden of damaging mutations than expected by chance alone.18 We define damaging mutations as mutations that are likely to abolish protein function; that is, truncating nonsense mutations, frameshift mutations, and splice-site mutations.
We noted that multiple T-cell cancers have overlapping genomic features. To increase the power of our analysis, we looked for recurrent hotspot mutations and recurrent damaging mutations that are shared by CTCL and other T-cell cancers; namely, adult T-cell leukemia/lymphoma (ATLL),19 angioimmunoblastic T-cell lymphoma (AITL),20-22 peripheral T-cell lymphoma not otherwise specified ,21,22 anaplastic large cell lymphoma without anaplastic lymphoma kinase,23 enteropathy-associated T-cell lymphoma,22,24 and nasal-type natural killer/T-cell lymphoma.22 The results of our pan-T-cell lymphoma analysis are listed in supplemental Tables 8-11.
Collectively, our analysis identified 55 putative driver genes in CTCL. These include 17 novel putative driver genes in CTCL (ARHGEF3 [see next section], BCOR, CSNK1A1, CD58, KDM6A, MAP2K1, NF1, PIK3R1, PRKCB, PTPRN2, RARA, RFXAP, RLTPR, SMARCB1, TRRAP, U2AF1, and VAV1) (Figure 1). BCOR, SMARCB1, and VAV1 were previously reported in CTCL5,6,10 ; however, without mutational context, they were listed along with other variants of unknown significance in 1 or 2 CTCL samples.
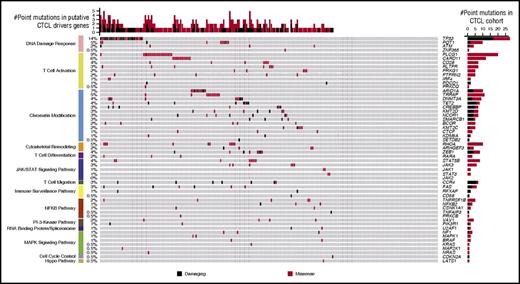
The landscape of cancer-promoting amino acid alterations in putative driver genes in cutaneous T-cell lymphoma. Genomic analysis of 220 CTCLs implicates mutations in 55 genes in CTCL pathogenesis. Genes with protein-altering amino acid alterations are shown here. Damaging mutations include frameshift mutations, splice-site mutations, and truncating nonsense mutations. For clarity, JAK2 is depicted without any mark of point mutation, as it is subject to recurrent amplifications, but no point mutations.
The landscape of cancer-promoting amino acid alterations in putative driver genes in cutaneous T-cell lymphoma. Genomic analysis of 220 CTCLs implicates mutations in 55 genes in CTCL pathogenesis. Genes with protein-altering amino acid alterations are shown here. Damaging mutations include frameshift mutations, splice-site mutations, and truncating nonsense mutations. For clarity, JAK2 is depicted without any mark of point mutation, as it is subject to recurrent amplifications, but no point mutations.
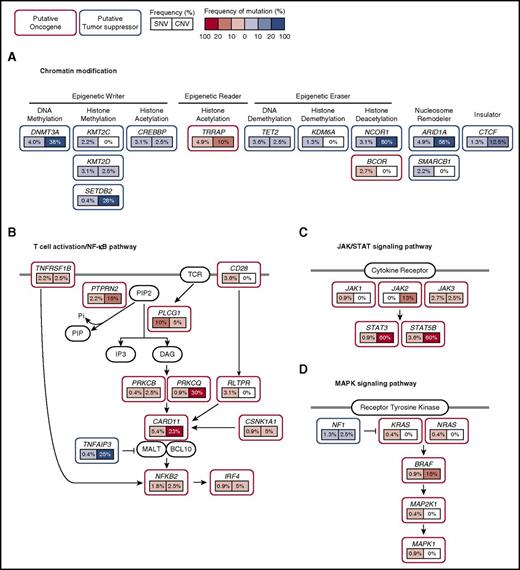
These genes are distributed across 14 biologically relevant pathways (Figure 1). Importantly, 43% of CTCLs harbor potentially targetable point mutations in the chromatin modification, the MAPK signaling pathway, the NF-κB pathway, the PI-3-kinase pathway, the spliceosome, and the JAK/STAT signaling pathway (Figure 2).
Schematic of mutations in recurrently mutated signaling pathways in CTCL. CTCL harbors recurrent mutations that are predicted to affect (A) chromatin, (B) T-cell activation/NF-κB signaling, (C) JAK/STAT signaling, and (D) MAPK signaling. Putative oncogenes and tumor suppressor genes are indicated in red and blue boxes, respectively. Frequencies of single nucleotide variant and copy number variants are indicated as percentages in left and right boxes under the genes, respectively. Prevalence of copy number mutations were obtained from a previous study.9 Point mutations and copy number amplifications in putative oncogenes are red. Point mutations and copy number deletions in putative tumor suppressors are blue. Darker hues indicate higher frequency of mutation. SNV, single nucleotide variant; CNV, copy number variant.
Schematic of mutations in recurrently mutated signaling pathways in CTCL. CTCL harbors recurrent mutations that are predicted to affect (A) chromatin, (B) T-cell activation/NF-κB signaling, (C) JAK/STAT signaling, and (D) MAPK signaling. Putative oncogenes and tumor suppressor genes are indicated in red and blue boxes, respectively. Frequencies of single nucleotide variant and copy number variants are indicated as percentages in left and right boxes under the genes, respectively. Prevalence of copy number mutations were obtained from a previous study.9 Point mutations and copy number amplifications in putative oncogenes are red. Point mutations and copy number deletions in putative tumor suppressors are blue. Darker hues indicate higher frequency of mutation. SNV, single nucleotide variant; CNV, copy number variant.
Subgroup analysis reveals CTCL-specific RHOA mutations
We examined whether the mutations were subtype-specific (supplemental Tables 4 and 17). With the exception of 1 mutation, the mutations were not significantly enriched in MF or SS. MAPK1 (p.E322A; p.E322K) was found exclusively in MF (3 mutations in 25 samples) and not in SS (0 mutation in 186 samples; P = .00149; Fisher’s exact test; supplemental Table 17).
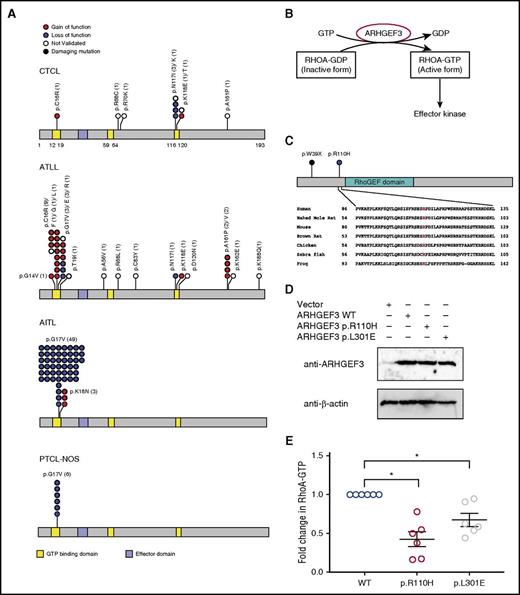
Notably, our pan-T-cell lymphoma analysis revealed a striking disease-specific distribution of RHOA mutations (Figure 3A; supplemental Table 8; supplemental Figure 9). RHOA encodes a GTPase that regulates actin cytoskeleton remodeling, chemotaxis, and signal transduction.25 The most common RHOA alteration in CTCL (p.N117I; p.N117K) makes up 30% of all RHOA alterations in CTCL and only 1.1% of RHOA mutations in AITL, ATLL, and peripheral T-cell lymphoma, collectively. In contrast, there are no p.G17 mutations found in CTCL, although it is commonly mutated in both AITL (94.2% of RHOA mutations) and ATLL (21.2% of RHOA mutations). These differences are unlikely to occur by chance alone and establish CTCL as an outlier among T-cell malignancies (P = .0027 and 6.3 × 105, respectively; Fisher’s exact test).
Characterization of RHOA pathway mutations in CTCL. (A) Distribution of RHOA mutations in malignancies of mature CD4+ T cells. There are no RHOA mutations in anaplastic large cell lymphoma. (B) Schematic of RHOA activation pathways. ARHGEF3 is a RHOA guanine nucleotide exchange factor that activates RHOA by catalyzing the replacement of GDP to GTP. (C) Identification of somatic mutations in ARHGEF3. The arginine at the 110th amino acid position is highly conserved across species. (D) Expression of WT ARHGEF3 (WT) or mutants (p.R110H; p.L301E) in HEK293T cells. We show immunoblot analysis of ARHGEF3 and loading control (β-actin) in lentivirally transduced HEK293T cells. (E) Effects of ARHGEF3 mutations on RHOA activity. HEK293T cells were transduced with WT ARHGEF3 or its mutants. RHOA-GTP levels were assessed by the G-LISA assay (Cytoskeleton). These data represent 6 biological replicates using repeat assays performed in 3 independently made lentivirally transduced cell lines. Each cell line was subject to 2 assays. The horizontal lines indicate the mean ± standard error from 6 independent biological replicates performed. P value was determined by 2-sided paired ratio t test. *P < .05.
Characterization of RHOA pathway mutations in CTCL. (A) Distribution of RHOA mutations in malignancies of mature CD4+ T cells. There are no RHOA mutations in anaplastic large cell lymphoma. (B) Schematic of RHOA activation pathways. ARHGEF3 is a RHOA guanine nucleotide exchange factor that activates RHOA by catalyzing the replacement of GDP to GTP. (C) Identification of somatic mutations in ARHGEF3. The arginine at the 110th amino acid position is highly conserved across species. (D) Expression of WT ARHGEF3 (WT) or mutants (p.R110H; p.L301E) in HEK293T cells. We show immunoblot analysis of ARHGEF3 and loading control (β-actin) in lentivirally transduced HEK293T cells. (E) Effects of ARHGEF3 mutations on RHOA activity. HEK293T cells were transduced with WT ARHGEF3 or its mutants. RHOA-GTP levels were assessed by the G-LISA assay (Cytoskeleton). These data represent 6 biological replicates using repeat assays performed in 3 independently made lentivirally transduced cell lines. Each cell line was subject to 2 assays. The horizontal lines indicate the mean ± standard error from 6 independent biological replicates performed. P value was determined by 2-sided paired ratio t test. *P < .05.
RHOA (p.N117I) is a dominant negative isoform presumably because it, as with similar dominant-negative Rho family isoforms,26 sequesters upstream enzymes (RhoGEFs) that catalyze the formation of the active RHOA isoform (RHOA-GTP) (Figure 3B). Therefore, we hypothesized that CTCLs would harbor inactivating mutations in RhoGEFs, which would similarly lead to decreased levels of RHOA-GTP. We therefore queried our dataset for mutations in all known RhoGEFs and found an isolated case of a damaging mutation (p.W39X) in ARHGEF3. ARHGEF3 is a RHOA-selective guanine nucleotide exchange factor27 that is expressed in CD4+ T cells and upregulated during T-cell activation.9 CTCLs also harbor an additional missense mutation at a highly conserved arginine in the N-terminal domain (p.R110H) (Figure 3C).
On the basis of the biology and mutational signature, we hypothesized that this amino acid substitution would be an inactivating mutation. To test this, we transduced HEK293 T cells with the WT ARHGEF3 or the mutant ARHGEF3 (p.R110H). ARHGEF3 (p.L301E) served as an additional control because the p.L301E has been previously shown to abolish ARHGEF3’s GEF activity.28 To assess the effect of each mutation on RhoGEF function, we used an enzyme-linked immunosorbent assay to quantify the amount of the active RHOA (RHOA-GTP) in HEK293T cells expressing each ARHGEF3 isoform. We found that the CTCL mutation (p.R110H) significantly inhibited GEF activity, in fact more so than the previously validated loss-of-function p.L301E alteration (mean log2 fold change = −1.2 and −0.56, respectively; Figure 3D-E).
Identification of novel recurrent amino acid alterations in putative oncogenes
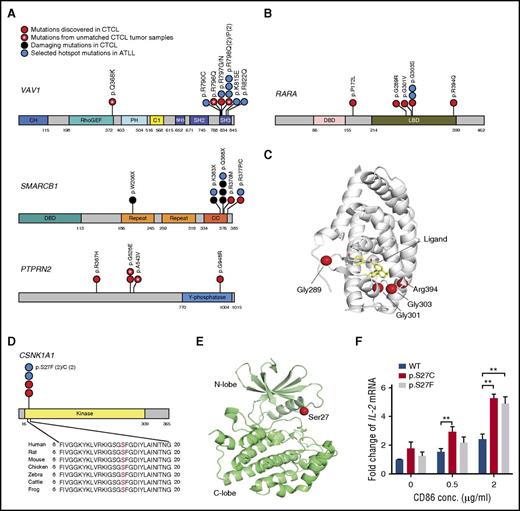
Our analyses identified novel, recurrent amino acid alterations in 4 genes: CSNK1A1 (adjusted P value = 1.86E-07; supplemental Table 9), PTPRN2 (P value = 4.04E-04; supplemental Table 15), RARA (adjusted P value = 1.39E-03; supplemental Table 9), and RLTPR (adjusted P value = 5.91E-20; supplemental Table 6). PTPRN2 encodes a protein tyrosine phosphatase. Similar to PLCG1, PTPRN2 affects the quantities of phosphatidylinositol phospholipids; specifically, PI(3)P and PI(4,5)P2.29 In CTCL, the mutations cluster in a region of unknown significance (Figure 4A). RARA encodes a transcription factor important for T-cell differentiation.30 The mutations in RARA cluster in the ligand binding domain (Figure 4B-C).
Schematics of novel recurrent mutations in CTCL. (A) Distribution of mutations of VAV1, SMARCB1, and PTPRN2 in CTCL and ATLL. If the amino acid alterations occur at the same position, each amino acid substitution is separated by a vertical line. (B) Distribution of mutations in RARA in CTCL and ATLL. (C) Protein structure of RARA (PDB ID: 1DKF),48 showing the locations of some CTCL mutations. The mutations cluster adjacent to the ligand binding pocket. (D) Distribution of mutations in CSNK1A1 in CTCL and ATLL. (E) Protein structure of CK1α (PDB ID: 5FQD), which is encoded by CSNK1A1.49 The location of residue Ser27 is indicated at the tip of the glycine rich P-loop. (F) The CK1α mutations increased CD28-dependent TCR activation. Jurkat cells were transduced with CK1α WT or mutations either of p.S27C or p.S27F. These cells were treated with vehicle control or PMA (50 ng/mL)/ionomycin (300 ng/mL) and CD86 at the indicated concentrations. Data represent as mean ± standard error from 6 independent experiments. P value was determined by 2-sided paired ratio t test. **P < .01 For panels A, B, and D: C1, phorbol esters/diacylglycerol binding domain; CC, coiled-coil domain; CH, indicates calponin homology domain; DBD, DNA binding domain; Kinase, protein kinase domain; LBD, ligand binding domain; PH, pleckstrin homology; Repeat, repeat motif; RhoGEF, RhoGEF domain; SH2, SH2 domain; SH3, SH3 domain; Y-phosphatase, protein-tyrosine phosphatase.
Schematics of novel recurrent mutations in CTCL. (A) Distribution of mutations of VAV1, SMARCB1, and PTPRN2 in CTCL and ATLL. If the amino acid alterations occur at the same position, each amino acid substitution is separated by a vertical line. (B) Distribution of mutations in RARA in CTCL and ATLL. (C) Protein structure of RARA (PDB ID: 1DKF),48 showing the locations of some CTCL mutations. The mutations cluster adjacent to the ligand binding pocket. (D) Distribution of mutations in CSNK1A1 in CTCL and ATLL. (E) Protein structure of CK1α (PDB ID: 5FQD), which is encoded by CSNK1A1.49 The location of residue Ser27 is indicated at the tip of the glycine rich P-loop. (F) The CK1α mutations increased CD28-dependent TCR activation. Jurkat cells were transduced with CK1α WT or mutations either of p.S27C or p.S27F. These cells were treated with vehicle control or PMA (50 ng/mL)/ionomycin (300 ng/mL) and CD86 at the indicated concentrations. Data represent as mean ± standard error from 6 independent experiments. P value was determined by 2-sided paired ratio t test. **P < .01 For panels A, B, and D: C1, phorbol esters/diacylglycerol binding domain; CC, coiled-coil domain; CH, indicates calponin homology domain; DBD, DNA binding domain; Kinase, protein kinase domain; LBD, ligand binding domain; PH, pleckstrin homology; Repeat, repeat motif; RhoGEF, RhoGEF domain; SH2, SH2 domain; SH3, SH3 domain; Y-phosphatase, protein-tyrosine phosphatase.
CSNK1A1 encodes casein kinase 1α (CK1α). Similar to PRKCB,31 CK1α is necessary for NF-κB signaling in response to antigen receptor signaling.32 CSNK1A1 is a putative tumor suppressor in other cancer types.33 However, in CTCL, CSNK1A1 harbors the mutational signature of an oncogene. There are recurrent mutations in CK1α at the p.S27 amino acid position (p.S27F; p.S27C) in both CTCL and ATLLs, which are unlikely to occur by chance alone (adjusted P value = 1.86E-07; supplemental Table 9). Similar to recurrent mutations found in oncogenic kinases, for example, EGFR (p.G719S),34 p.S27 resides at the tip of the P-loop in the kinase domain (Figure 4D-E).
Because of its putative role in antigen receptor signaling, we hypothesized that these mutations may affect T-cell receptor (TCR)-dependent signaling pathways. To test this, we employed a pharmacological model of T-cell receptor signaling in Jurkat cells. We had previously used this system to validate other CTCL mutations in TCR-associated genes.9 We transduced Jurkat cells with lentivirus expressing WT CK1α, CK1α (p.S27C), or CK1α (p.S27F; supplemental Figure 10). We then stimulated these cells with pharmacological TCR mimics, PMA and ionomycin, and costimulatory ligands. For costimulation, we cultured cells with increasing concentrations of a CD28 ligand, CD86-Fc. We found no effect of the mutation in unstimulated Jurkat cells. However, in response to pharmacological TCR mimics and CD86, the mutations increased interleukin 2 (IL-2) expression by 2.16-fold and 2.01-fold, respectively (P = .0033, P = .0062, respectively; 2-sided paired ratio t test; Figure 4F).
Identification of RLTPR as a putative oncogene in T-cell cancers
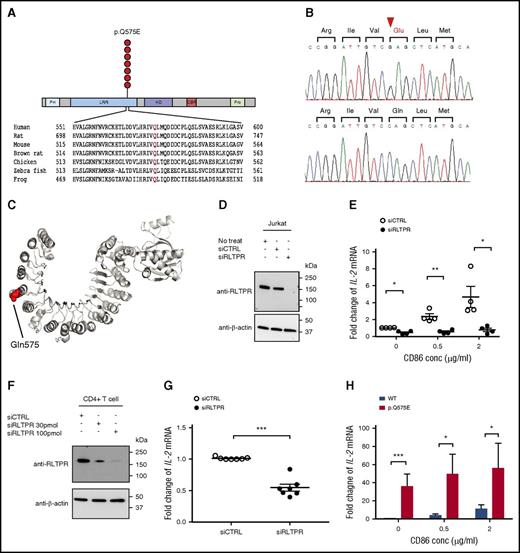
RLTPR encodes a scaffolding protein that has been recently implicated in CD28 signaling.35 The mutations cluster at p.Q575, which resides in the leucine repeat region (LRR) domain. It is the second most common hotspot mutation in CTCL, occurring in 3.1% of CTCLs (Figure 5A-B). It has also been found in 1 case of ATLL.19
Identification of a novel recurrent, gain-of-function RLTPR mutation (p.Q575E) in CTCL. (A) Schematic of the recurrent RLTPR (p.Q575E) amino acid alteration. CBR, capping protein binding region; HD, helical domain; PH, pleckstrin homology; Pro, proline-rich region. (B) Confirmation of the RLTPR mutation by Sanger sequencing. Chromatogram of Sanger sequencing of genomic DNA from CTCL tumor cells (upper) and matched germline controls (ie, monocytes; lower). (C) Protein structure of the RLTPR homolog, CARMIL, showing the location of the p.Q575E amino acid alteration. CARMIL harbors 38% identity more than 670 amino acids when aligned to RLTPR, and p.Q575 is conserved between the 2 proteins. (PDB ID: 4K17).36 (D) Expression of RLTPR in Jurkat cells nucleofected with siCTRL or siRLTPR. Immunoblot analysis with RLTPR antibody, and β-actin as the loading control. (E) Downregulation of RLTPR inhibits TCR signaling in Jurkat cells. IL-2 mRNA expression in Jurkat cells nucleofected with siCTRL or siRLTPR in the presence of pharmacological TCR mimics (PMA/ionomycin) and a CD28 ligand, CD86. Data are shown as mean ± standard error from 4 independent biological replicates. P value was determined by 2-sided paired ratio t test (*P < .05; **P < .01). (F) Expression of RLTPR in isolated CD4+ T cells nucleofected with siCTRL or siRLTPR. (G) Downregulation of RLTPR inhibits TCR signaling in CD4+ T cells. IL-2 mRNA expression in CD4+ T cells nucleofected with siCTRL or siRTLPR in the presence of anti-CD3/anti-CD28 activation beads. Data are shown as mean ± standard error from 7 independent biological replicates from peripheral blood mononuclear cells of 2 different healthy donors. P value was determined by 2-sided paired ratio t test (***P < .001). (H) The RLTPR mutation significantly increases TCR signaling. Jurkat cells were transduced with RLTPR WT or RLTPR p.Q575E. These cells were treated with vehicle control or PMA (50 ng/mL)/ionomycin (300 ng/mL) and CD86 at the indicated concentrations. Data represent as mean ± standard error from 6 independent experiments. P value was determined by 2-sided paired ratio t test. *P < .05; ***P < .001.
Identification of a novel recurrent, gain-of-function RLTPR mutation (p.Q575E) in CTCL. (A) Schematic of the recurrent RLTPR (p.Q575E) amino acid alteration. CBR, capping protein binding region; HD, helical domain; PH, pleckstrin homology; Pro, proline-rich region. (B) Confirmation of the RLTPR mutation by Sanger sequencing. Chromatogram of Sanger sequencing of genomic DNA from CTCL tumor cells (upper) and matched germline controls (ie, monocytes; lower). (C) Protein structure of the RLTPR homolog, CARMIL, showing the location of the p.Q575E amino acid alteration. CARMIL harbors 38% identity more than 670 amino acids when aligned to RLTPR, and p.Q575 is conserved between the 2 proteins. (PDB ID: 4K17).36 (D) Expression of RLTPR in Jurkat cells nucleofected with siCTRL or siRLTPR. Immunoblot analysis with RLTPR antibody, and β-actin as the loading control. (E) Downregulation of RLTPR inhibits TCR signaling in Jurkat cells. IL-2 mRNA expression in Jurkat cells nucleofected with siCTRL or siRLTPR in the presence of pharmacological TCR mimics (PMA/ionomycin) and a CD28 ligand, CD86. Data are shown as mean ± standard error from 4 independent biological replicates. P value was determined by 2-sided paired ratio t test (*P < .05; **P < .01). (F) Expression of RLTPR in isolated CD4+ T cells nucleofected with siCTRL or siRLTPR. (G) Downregulation of RLTPR inhibits TCR signaling in CD4+ T cells. IL-2 mRNA expression in CD4+ T cells nucleofected with siCTRL or siRTLPR in the presence of anti-CD3/anti-CD28 activation beads. Data are shown as mean ± standard error from 7 independent biological replicates from peripheral blood mononuclear cells of 2 different healthy donors. P value was determined by 2-sided paired ratio t test (***P < .001). (H) The RLTPR mutation significantly increases TCR signaling. Jurkat cells were transduced with RLTPR WT or RLTPR p.Q575E. These cells were treated with vehicle control or PMA (50 ng/mL)/ionomycin (300 ng/mL) and CD86 at the indicated concentrations. Data represent as mean ± standard error from 6 independent experiments. P value was determined by 2-sided paired ratio t test. *P < .05; ***P < .001.
As per Ensembl (http://www.ensembl.org/index.html), there are 2 isoforms of RLTPR. To determine the relevant isoform expressed in CTCL, we analyzed RNA-Seq data and found that both CD4+ T cells and CTCL cells express a novel isoform that includes exon 36 and splices out an in-frame segment of exon 14, which we term isoform 3 (supplemental Figure 11). All subsequent experiments assume this is the relevant isoform in CTCL.
p.Q575E replaces a highly conserved uncharged glutamine with a negatively charged glutamic acid in the LRR domain. Using the crystal structure of CARMIL, we conducted simple homology modeling and found that p.Q575 is predicted to be surface exposed and located on the convex surface of the LRR domain (Figure 5C).36
To validate the importance of RLTPR in TCR and/or CD28 signaling, we transfected Jurkat cells with siRLTPR or control siRNA and assessed TCR-dependent cytokine expression. We found that loss of RLTPR led to impairment of IL-2 expression when Jurkat cells are stimulated by PMA and ionomycin (mean downregulation, 2.3-fold; P = .0351; 2-sided paired ratio t test; Figure 5D-E). However, a more dramatic loss of IL-2 production occurred in the presence of the CD28 ligand, CD86 (mean downregulation, 4.2-fold; P = .0284; 2-sided paired ratio t test; Figure 5D-E). We confirmed the role of RLTPR in CD3/CD28 signaling in primary human CD4+ T cells (mean downregulation, 1.9-fold; P = .0005; 2-sided paired ratio t test; Figure 5F-G).
Therefore, we hypothesized that RLTPR (p.Q575E) would selectively potentiate CD28-dependent signaling. To assess this, we lentivirally transduced Jurkat cells with a vector encoding WT RLTPR or mutant RLTPR (p.Q575E). We cultured these cells with vehicle controls or pharmacological stimuli. Strikingly, there appears to be no effect of the RLTPR mutation on unstimulated Jurkat cells. However, in response to PMA and ionomycin alone, the RLTPR mutation increased IL-2 production by 34-fold (P = .0005; 2-sided paired ratio t test; Figure 5H). This effect appears to be largely CD28 independent as the addition of CD28 ligands had a statistically significant but smaller effect on (p.Q575E)-dependent cytokine production (mean of 11.2-fold upregulation, P = .0128; 2-sided paired ratio t test; Figure 5H).
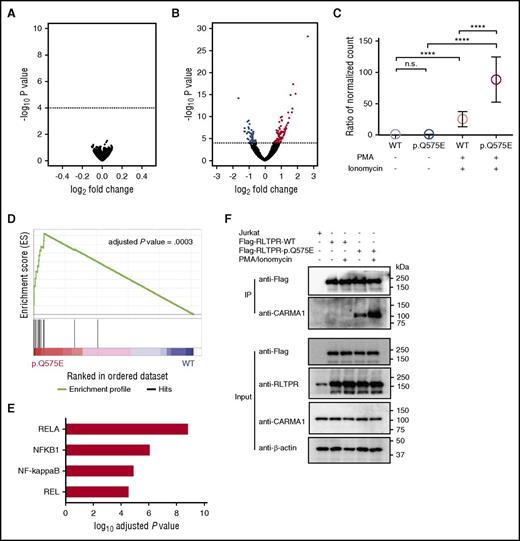
To assess the global effects of the RLTPR mutation, we performed RNA-Seq on unstimulated and stimulated Jurkat cells expressing either WT or mutant RLTPR. Strikingly, we found no statistically significant differences in gene expression between unstimulated RLTPR (WT) and RLTPR (p.Q575E) Jurkats (Figure 6A; adjusted P value threshold of 0.01). In contrast, RLTPR (p.Q575E) had profound effects after stimulation by pharmacologic TCR mimics. RNA-Seq analysis revealed that 96 genes were significantly upregulated and 47 genes significantly downregulated in stimulated RLTPR (p.Q575E) Jurkat cells compared with stimulated RLTPR (WT) (Figure 6B; supplemental Figure 12; supplemental Tables 18-21).
RLTPR selectively activates the NF-κB pathway in a TCR-dependent manner. (A) Volcano plot analysis of differentially expressed RNA transcripts in unstimulated RLTPR (p.Q575E) vs RLTPR WT Jurkat cells. Dotted line indicates adjusted P value = .01. (B) Volcano plots of differentially expressed RNA transcripts in PMA/ionomycin-stimulated RLTPR (p.Q575E) vs RLTPR WT Jurkat cells. Genes that are above the dotted line represent the genes having statistical significance with adjusted P value < .01. Red and blue dots represent upregulated and downregulated genes, respectively. (C) Expression of RLTPR-dependent genes (genes indicated in red in panel B) as a function of RLTPR mutation status and stimulation. Data represent the ratio of normalized mRNA transcript counts in each subgroup compared with unstimulated RLTPR WT Jurkat cells. P value was determined by 2-sided paired ratio t test. n.s., not significant; ****P < .0001. (D) Gene set variation analysis of RNA-seq data comparing signaling pathways differentially activated in PMA-ionomycin activated RLTPR (p.Q575E) Jurkat cells compared with PMA-ionomycin activated WT controls. (E) Identification of transcription factor binding motifs that are selectively upregulated by the RLTPR mutation. OPOSSUM analysis was performed on the 96 RLTPR mutant-dependent gene transcripts; that is, the genes indicated in red in panel B. The graph shows the 4 highest ranked transcription factors, ranked by lowest adjusted P values. (F) Immunoprecipitation assay assessing the interactions between FLAG-tagged RLTPR and CARMA1 according to mutation status and stimulation status. Lysates from Jurkat cells are subjected to co-immunoprecipitation using anti-Flag beads, and blotted with anti-CARMA1 antibody. β-actin was used as the loading control. Immunoblotting for FLAG, RLTPR, and CARMA1 were used to confirm equivalent expression of the FLAG-tagged RLTPR and CARMA1 in each input sample.
RLTPR selectively activates the NF-κB pathway in a TCR-dependent manner. (A) Volcano plot analysis of differentially expressed RNA transcripts in unstimulated RLTPR (p.Q575E) vs RLTPR WT Jurkat cells. Dotted line indicates adjusted P value = .01. (B) Volcano plots of differentially expressed RNA transcripts in PMA/ionomycin-stimulated RLTPR (p.Q575E) vs RLTPR WT Jurkat cells. Genes that are above the dotted line represent the genes having statistical significance with adjusted P value < .01. Red and blue dots represent upregulated and downregulated genes, respectively. (C) Expression of RLTPR-dependent genes (genes indicated in red in panel B) as a function of RLTPR mutation status and stimulation. Data represent the ratio of normalized mRNA transcript counts in each subgroup compared with unstimulated RLTPR WT Jurkat cells. P value was determined by 2-sided paired ratio t test. n.s., not significant; ****P < .0001. (D) Gene set variation analysis of RNA-seq data comparing signaling pathways differentially activated in PMA-ionomycin activated RLTPR (p.Q575E) Jurkat cells compared with PMA-ionomycin activated WT controls. (E) Identification of transcription factor binding motifs that are selectively upregulated by the RLTPR mutation. OPOSSUM analysis was performed on the 96 RLTPR mutant-dependent gene transcripts; that is, the genes indicated in red in panel B. The graph shows the 4 highest ranked transcription factors, ranked by lowest adjusted P values. (F) Immunoprecipitation assay assessing the interactions between FLAG-tagged RLTPR and CARMA1 according to mutation status and stimulation status. Lysates from Jurkat cells are subjected to co-immunoprecipitation using anti-Flag beads, and blotted with anti-CARMA1 antibody. β-actin was used as the loading control. Immunoblotting for FLAG, RLTPR, and CARMA1 were used to confirm equivalent expression of the FLAG-tagged RLTPR and CARMA1 in each input sample.
The relationship between RLTPR (p.Q575E) and concomitant TCR activation led us to hypothesize that RLTPR (p.Q575E) does not activate a TCR-independent pathway or pathways but, instead, potentiates TCR signaling. If true, we would predict that genes induced by RLTPR (p.Q575E) represent genes normally induced by TCR activation in WT cells. To test this, we examined the expression of this gene set as a function of pharmacologic stimulation in the presence and absence of the RLTPR mutation. Indeed, we found that PMA and ionomycin stimulation increased the expression of this gene set in both WT and RLTPR (p.Q575E) Jurkat cells (mean upregulation of 1.61-fold and 3.34-fold, respectively; Figure 6C); however, the magnitude of induction was significantly higher in the presence of the RLTPR mutation (P < .0001; 2-sided paired ratio t test; Figure 6C). There was no difference between unstimulated Jurkat cells expressing RLTPR (p.Q575E) or WT RLTPR (mean fold change = 1.06; Figure 6C; supplemental Table 18).
TCR signaling activates NF-κB, NFAT, and AP1 signaling pathways, all of which are necessary for the production of IL-2.37 To identify which, if any, of these signaling pathways is affected by the RLTPR mutation, we used an unbiased approach (gene set variable analysis).38,39 Our analysis identified only 1 significantly altered pathway, the NF-κB pathway (adjusted P value = .0003; Figure 6D; supplemental Table 22).
To confirm these findings, we used an orthogonal algorithm, OPOSSUM (http://opossum.cisreg.ca/oPOSSUM3/). This software identifies gene set enrichment of consensus transcription factor binding sites in gene promoters. Strikingly, the top 4 transcription factor binding sites implicated by this analysis are all NF-κB binding sites (adjusted P values range from 2.9E-05 to 1.5E-09; Figure 6E; supplemental Figure 13; supplemental Table 23).
RLTPR is thought to be a scaffolding protein and lacks any known enzymatic domains. This led us to hypothesize that the p.Q575E alteration promotes TCR signaling by modifying RLTPR’s ability to form a complex with relevant TCR-dependent enzymes in the NF-κB signaling pathway. Recent mass spectrometry analysis suggested that 1 such binding partner may be CARMA1, a kinase that mediates antigen-receptor-dependent activation of the NF-κB signaling pathway.35 To test this hypothesis, we immunoprecipitated FLAG-tagged WT RLTPR or RLTPR (p.Q575E) and blotted for CARMA1. We found that the p.Q575E alteration dramatically increases interaction of RLTPR with CARMA1 in both unstimulated and stimulated cells (Figure 6F; supplemental Figure 14).
Discussion
We have assembled a carefully curated cohort of CTCLs with publicly available sequencing data. The size of the cohort has provided the statistical power to enable the identification of 55 putative driver genes in CTCL, including novel mutations in 17 genes. Our report includes 5 genes (ARHGEF3, CSNK1A1, PTPRN2, RARA, and RLTPR) with novel mutations that had not been previously reported for any cancer to date. In addition, we provide functional data supporting the role of 3 of these genes: ARHGEF3 as a putative tumor suppressor and CSNK1A1 and RLTPR as putative oncogenes. Importantly, we report the lines of genetic evidence supporting each of these genes in CTCL. In doing so, we hope to clarify findings that appeared discordant from previous studies. We believe the apparent disagreement among the studies is likely a result of differences in analytical approaches, as well as sampling issues inherent in studying relatively small sample cohorts.
The driver genes we have identified segregate into 14 functional pathways. These include cell cycle regulators, chromatin-modifying genes, DNA damage response genes, an RNA splicing gene, a Hippo signaling gene, JAK/STAT signaling genes, MAPK signaling genes, NF-κB signaling genes, PI-3-kinase signaling genes, immune evasion genes, and cytoskeleton remodeling genes, as well as genes involved in T-cell signaling, migration, and differentiation. These signaling pathways are recurrently mutated, likely because of their fundamental importance to CTCL lymphomagenesis. In other words, cells with functionally similar mutations arise in multiple patients because these mutations confer a selective advantage in vivo. We predict that a deeper understanding of how these genes and pathways contribute to disease will lead to the development of novel therapeutic strategies. Importantly, a subset of these pathways, the chromatin modification,40 the MAPK,41 the JAK/STAT,42 the NF-κB,43 and immune evasion pathways,44 may be directly targetable with therapies that are either currently approved by the US Food and Drug Administration or in clinical development for other cancer types. These data collectively highlight the importance of genotyping patients before choosing therapies, particularly for future clinical trials.
The genomics highlight shared features and important differences between CTCL and other T-cell lymphoma types. At this time, we do not have an explanation for the disease-specific distribution of mutations in RHOA. One possibility may be that CTCLs have a preference for RHOA (p.N117I) because this mutation has a selective effect on skin-homing CD4+ T cells; that is, the presumed cells of origin of CTCLs.
Last, we have identified and functionally validated a novel gain of function mutation in RLTPR (p.Q575E). In unstimulated cells, RLTPR (p.Q575E) potentiates interactions with CARMA1, an NF-κB signaling component. This interaction is not sufficient to alter TCR-dependent transcriptional programs. However, in the presence of TCR activation, the mutation selectively upregulates the NF-κB signaling pathway and confers the most dramatic effect on TCR signaling flux of any CTCL mutation studied to date.5,8-10 Mechanistically, these data suggest a 2-hit model. The mutation enables preassembly of signaling complexes in unstimulated cells. However, it transduces mutation-specific signals only in the presence of the second hit; that is, activation of the T-cell receptor signaling pathways.
Importantly, the RLTPR mutation highlights key concepts about CTCL pathogenesis. To date, all the gain-of-function mutations in TCR signaling genes (CARD11,8 CD28,45 PLCG1,46 and RLTPR; Figure 6C-F) have been shown to activate the NF-κB pathway. The convergence of diverse mutations on this pathway highlights its fundamental importance to CTCL lymphomagenesis and support revisiting NF-κB inhibitors as a core therapeutic strategy for this disease.47
Moreover, we postulate that the relationship between RLTPR mutations and TCR signaling provides important insights into the CTCL tumor microenvironment. Because this mutation has no effect without TCR signals, it suggests that positive selection for this mutation could only have occurred if the CTCL cells were receiving repeated chronic or tonic TCR stimulation. Therefore, these data underscore the fundamental importance of the interactions between CTCL cells and antigen-presenting cells in the skin and elsewhere. This interaction may be critical for the CTCLs to receive the TCR and costimulatory signals apparently critical for disease pathogenesis.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the patients who have contributed to this study, the Northwestern Skin Disease Research Center (DNA/RNA Delivery Core), the Robert H. Lurie Comprehensive Cancer Center (Flow Cytometry Core), the Yale Center for Genome Analysis, and the Northwestern University Research Computing Services for their invaluable contribution for providing effective solutions for the storage and analysis of data.
This study was supported in part by the National Institutes of Health, National Cancer Institute (grant K08-CA191019-01A1), the American Cancer Society (grant ACS-IRG-15-173-21), the Skin Cancer Foundation Research Grant, and the Leukemia Research Foundation New Investigator Grant (J.C.). J.C. is a Doris Duke–Damon Runyon Clinical Investigator supported in part by the Damon Runyon Cancer Research Foundation (DRCRF# CI-84-16) and by the Doris Duke Charitable Foundation (DDCF# 2016095). J.P. was supported by an award from the Translational Bridge Program of the Robert H. Lurie Cancer Center and the Northwestern University Clinical and Translational Sciences Institute.
Authorship
Contribution: J.P. and J.C. designed the project and wrote the manuscript; J.P., J.K., B.P., E.M.-E., J.G., M.L.M., J.N.S., N.A., T.J.B., and J.C. reviewed the data and the manuscript; J.P. and A.R. performed all the functional experiments in the manuscript; J.P., J.Y., A.T.W., W.J.L., and J.C.D. performed all of the bioinformatics analysis in the manuscript; and T.J.B. performed all the protein 3D structure analysis of the mutations in CK1α, RARA, RHOA, and RLTPR.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jaehyuk Choi, Robert H. Lurie Comprehensive Cancer Center, Northwestern University, 303 E. Superior St, Room 5-115, Chicago, IL 60611; e-mail: jaehyuk.choi@northwestern.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal