Key Points
PIM kinases are ubiquitously expressed in RS cells of cHL.
PIM inhibition decreases NFκB and STAT3/5 activity, cell viability, and expression of immunoregulatory proteins PD-L1/2 and galectin-1.
Abstract
Reed-Sternberg (RS) cells of classical Hodgkin lymphoma (cHL) express multiple immunoregulatory proteins that shape the cHL microenvironment and allow tumor cells to evade immune surveillance. Expression of certain immunoregulatory proteins is modulated by prosurvival transcription factors, such as NFκB and STATs. Because these factors also induce expression of the oncogenic PIM1/2/3 serine/threonine kinases, and as PIMs modulate transcriptional activity of NFκB and STATs, we hypothesized that these kinases support RS cell survival and foster their immune privilege. Here, we investigated PIM1/2/3 expression in cHL and assessed their role in developing RS cell immune privilege and survival. PIM1/2/3 were ubiquitously expressed in primary and cultured RS cells, and their expression was driven by JAK-STAT and NFκB activity. Genetic or chemical PIM inhibition with a newly developed pan-PIM inhibitor, SEL24-B489, induced RS cell apoptosis. PIM inhibition decreased cap-dependent protein translation, blocked JAK-STAT signaling, and markedly attenuated NFκB-dependent gene expression. In a cHL xenograft model, SEL24-B489 delayed tumor growth by 95.8% (P = .0002). Furthermore, SEL24-B489 decreased the expression of multiple molecules engaged in developing the immunosuppressive microenvironment, including galectin-1 and PD-L1/2. In coculture experiments, T cells incubated with SEL24-B489-treated RS cells exhibited higher expression of activation markers than T cells coincubated with control RS cells. Taken together, our data indicate that PIM kinases in cHL exhibit pleiotropic effects, orchestrating tumor immune escape and supporting RS cell survival. Inhibition of PIM kinases decreases RS cell viability and disrupts signaling circuits that link these cells with their niches. Thus, PIM kinases are promising therapeutic targets in cHL.
Introduction
Classical Hodgkin lymphoma (cHL) is characterized by the presence of the rare malignant Hodgkin/Reed Sternberg (RS) cells, surrounded by an abundant reactive infiltrate of T and B lymphocytes, granulocytes, macrophages, plasma cells, eosinophils, mast cells, and fibroblasts.1 This peculiar infiltrate exhibits profound local immunosuppressive properties and an extensive network of bilateral connections with RS cells. Paracrine signals and surface ligands provided by the infiltrating cells contribute to aberrant activation of certain pathways in RS cells, of which NFκB, JAK-STAT, and MAPK/AP-1 seem to play a pivotal role in cHL pathogenesis.2-10 These activated transcription factors induce expression of molecules critical for RS cell survival and proliferation, but also increase the expression of multiple cytokines, chemokines, and immunomodulatory proteins, such as PD-1 ligands (PD-L1/2) or galectin-1 (Gal-1), that directly facilitate RS cell escape from host immune surveillance.4,11-14 Consistent with these pathogenetic dependencies, recent studies demonstrate that targeting immune checkpoints in cHL with anti-PD1 antibodies is a highly promising therapeutic strategy.15
The family of PIM serine/threonine kinases comprises 3 highly conserved oncogenes (PIM1, PIM2, and PIM3), overexpressed in multiple solid cancers and hematologic malignancies. PIMs are unique among other kinases, as they lack a regulatory domain and thus, once transcribed, become constitutively active.16 PIMs have been shown to promote oncogenesis by modulating the activity of proteins involved in regulation of translation, transcription, cell cycle, and survival.17-21 PIMs also modulate the activity of STATs and NFκB, key transcription factors supporting RS cell survival and responsible for the cross-talk with the inflammatory infiltrate.22-24 Because expression of PIM kinases is controlled by mitogens, growth factors, and cytokines via STATs and NFκB transcription factors, MAPK, and PI3K-AKT pathways, we hypothesized that PIMs might be expressed in RS cells, support RS cell survival, and foster their immune privilege.16,25-29 Here, we demonstrate that PIM1, PIM2, and PIM3 are induced by JAK-STAT and NFκB pathways and are expressed in primary and cultured RS cells. We show that genetic or chemical PIM inhibition decreases proliferation and viability of the malignant cells in vitro and in vivo. In addition, inhibition of PIM kinases downregulated expression of multiple factors engaged in developing the immunosuppressive microenvironment in cHL, including Gal-1 and PD-L1, and increased activation of T cells cocultured with RS cells.
Methods
Cell lines, culture conditions, and chemicals
Human cHL/RS cell lines L540, L428, L1236, HDLM-2, SUP-HD1 KM-H2, and Jurkat cells were maintained as described.12,30 The pan-PIM inhibitor SEL24-B489 had been developed and provided by Selvita S.A. (Kraków, Poland). JAK inhibitors CYT387 and fedratinib were purchased from Selleckchem; all chemicals were dissolved in dimethyl sulfoxide (DMSO). In vehicle-control experiments, final DMSO concentrations were 0.5% (control for 5- and 10-µM dose of SEL24-B489) or 0.3% (for 3 µM dose).
Immunoblotting
Protein lysates were resolved by sodium dodecyl sulfate/polyacrylamide gel electrophoresis, transferred to polyvinylidene difluoride membranes (Millipore), and immunoblotted with primary and appropriate horseradish peroxidase-labeled secondary antibodies (listed in supplemental Table 1, available on the Blood Web site). Signals were developed by enhanced luminescence, using enhanced chemiluminescence reagent (Perkin Elmer) and a digital image acquisition system (G:Box, Syngene). To reprobe with another antibody, blots were incubated in stripping buffer at 50°C for 30 minutes and analyzed as described earlier. Densitometric quantifications of band intensities were performed using ImageJ software (https://imagej.nih.gov/ij/).
Patient samples and immunohistochemistry
A retrospective group of 67 patients diagnosed according to the 2008 World Health Organization classification with cHL (42 nodular sclerosis, 23 mixed cellularity, 2 lymphocyte-rich) and 4 patients with follicular hyperplasia were enrolled for the study. Immunohistochemistry on formalin-fixed paraffin embedded sections was performed using an automated stainer (Dako Denmark A/S) and indicated antibodies (supplemental Table 1). EnVision Detection System (Dako Denmark A/S) was used for signal detection. Stained cHL sections were independently reviewed for expression of PIMs by 2 hematopathologists (M.P.-S. and A.S.-C.). Staining controls and the PIM1/2/3 scoring system are described in the supplemental Data. Expression of Galectin-1 in RS cells was assessed and scored by 2 hematopathologists (M.P.-S. and O.S.-G.), as previously described.31 All microphotographs were taken by a microscope DP72 Olympus BX63 camera (Olympus, Japan).
Real-time, quantitative polymerase chain reaction
RNA was extracted using a GeneMATRIX Universal RNA purification kit (EURx) and reverse-transcribed using a Transcriptor First strand cDNA synthesis kit (Roche), according to the manufacturer’s instructions. Gene expression levels were measured in triplicate with the 7500 Fast RT-PCR system (Applied Biosystems), using the SYBR Green PCR Master Mix (Life Technologies) and the gene-specific primers (sequences are given in supplemental Table 2). Obtained CT values for target genes and housekeeping control (glyceraldehyde-3-phosphate dehydrogenase; GAPDH) were used to calculate relative transcript abundance, using the 2−ΔΔCT method.32 Raw ΔCT (CT gene of interest − CT GAPDH) are also presented as indicated.
Vectors and retroviral transductions
An IκBα super-repressor construct in pMSCV-IRES-GFP (pMIG-SR-IkBα) vector was used to inhibit the canonical NFκB pathway in SUP-HD1 and HDLM-2 cells as described.33 Virus production and infections were performed as described.34 Twenty-four hours after infection, the GFP-positive cells were isolated by fluorescence-activated cell sorting (FACS Aria III; Beckton Dickinson).
RNAi-mediated silencing of gene expression
SMARTpool siRNAs targeting PIM1, PIM2, PIM3, RELA, REL, RELB, and a negative (nontargeting) control were purchased from GE Healthcare (catalog numbers are listed in supplemental Table 3). For siRNA transduction, cells were suspended at 5 × 105 cells/mL in Accell siRNA delivery media (Dharmacon/GE Healthcare) supplemented with 2% fetal bovine serum; individual siRNA pools were added to cells at a final concertation of 1 µmol/L. Cells were then incubated at 37°C with 5% CO2 for 96 hours before further experiments.
Proliferation and apoptosis assays
Cellular proliferation was measured by the CellTiter96 AQueous Nonradioactive cell proliferation assay (Promega). Fifty percent effective concentration (EC50) values were calculated using GraphPad Prism Version 6 (GraphPad software). For apoptosis, cells were washed in PBS, suspended in 1× AnnexinV binding buffer (Becton Dickinson; BD), stained with AnnexinV-PE and 7-AAD (BD), and analyzed using a FACS Canto flow cytometer (BD).
NFκB-p65 DNA binding assay
DNA binding activities of NFκB complexes were assessed using a colorimetric NFκB binding assay (TransAM NFκB/p65 kit, Active Motif, cat. 40096). After 24 hours of incubation with 5 µmol/L SEL24-B489 or DMSO, 20 µg protein extracts were added to microwells. Signals were developed according to manufacturer’s instructions and quantified using a Multiskan GO plate reader (Thermo Scientific).
Flow cytometry
PD-L1, PD-L2, CD69, and CD25 surface expression was evaluated by flow cytometry as previously described,35 with antibodies listed in supplemental Table 1. Cells were analyzed using FACS Canto flow cytometer (BD).
T-cell activation assay
HDLM-2 and SUP-HD1 RS cells were treated with DMSO alone or SEL24-B489 (2 µM) for 120 hours. Peripheral mononuclear cells were obtained from normal blood donors by Histopaque gradient centrifugation (Sigma-Aldrich). Naive T cells were purified from peripheral mononuclear cells using a Naive Pan T-cell isolation kit (Miltenyi Biotech). T-cell purity (≥90%) was confirmed by flow cytometry using CD3-FITC antibody (BD Biosciences). After purification, peripheral T cells were stimulated for 24 hours, using human T-activator CD3/CD28 Dynabeads (Thermo Scientific) and mixed with pretreated RS cells at 1:4 ratio (T cells to RS cells). After 16 hours, expression of CD69 and CD25 on T-cell surface was assessed in cocultures by flow cytometry, using APC-CD69, PE-CD25, and FITC-CD3 antibodies. CD3 gating was used to discriminate T cells from RS cells (T cells: CD3-positive; RS cells: CD3-negative).
Statistical analyses
Differences between continuous variables (gene expression, apoptosis, NFκB DNA binding activity) were compared by the Gosset’s 2-sided t-test, using GraphPad QuickCals/Prism 6 software; error bars represent SD as indicated.
Results
PIM1, PIM2, and PIM3 are ubiquitously expressed in primary and cultured RS cells
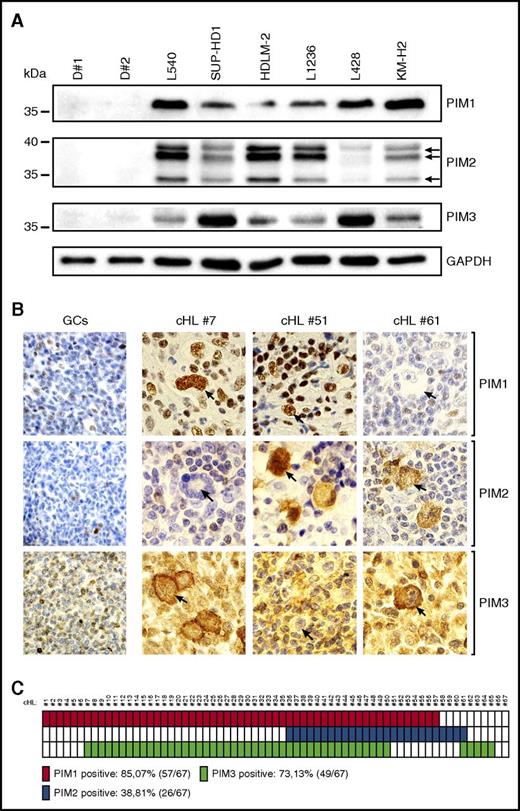
To assess the expression of PIM kinases in cHL RS cells, and to compare it with normal B cells, we first performed an immunoblot analysis with protein extracts of 6 RS cell lines and peripheral B lymphocytes obtained from 2 healthy donors. All tested RS cell lines expressed at least 2 PIM isoforms; in contrast, normal B cells were PIM-1/2/3-negative (Figure 1A). These data suggest that neoplastic RS cells overexpress oncogenic PIM kinases. To further explore this possibility, we assessed PIM expression in primary cHL and reactive lymph node sections by IHC. In a series of 67 diagnostic cHL tumor biopsies, 57 (85.07%) were PIM1+, 26 (38.81%) were PIM2+, and 49 (73.13%) were PIM3+ (Figure 1B-C). There were 15 (22.39%) triple-positive cases, 37 (55.22%) double-positive cases, and only 2 PIM1/2/3− cases (2.98%). PIM1 exhibited exclusively nuclear staining, whereas PIM2 was present in the nucleus and in the cytosol, and PIM3 only in the cytosol of RS cells. PIM expression was abundant in RS cells, but was also present in certain infiltrating cells (eg, macrophages). In marked contrast, in a reactive lymph node germinal center, centrocytes and centroblasts were negative or weakly positive, with few macrophages and mantle zone small lymphocytes that revealed PIM1/2/3 expression (Figure 1B; supplemental Figure 1).Taken together, these data indicate that PIM kinases are abundantly expressed in the vast majority of RS cells of cHL.
Primary and cultured RS cells express PIM1, PIM2, and PIM3 kinases. (A) Western blot analysis of PIM1, PIM2, and PIM3 expression in peripheral B cells isolated from healthy donors (D#1, D#2) and a panel of 6 cHL-derived RS cell lines. GAPDH served as a loading control. (B) Immunohistochemical analysis of PIM expression in reactive germinal centers and cHL samples. Shown are representative germinal centers (GCs) originating from follicular hyperplasia lymph node samples and cHL cases, double positive for PIM1 and PIM3 (#7), PIM1 and PIM2 (#51), and PIM2 and PIM3 (#61). RS cells are indicated with arrows. Original magnification was ×600. (C) Summary of the immunohistochemical analysis of PIM expression performed in a panel of 67 patients with cHL. Solid blocks, positive cases; open blocks, negative cases.
Primary and cultured RS cells express PIM1, PIM2, and PIM3 kinases. (A) Western blot analysis of PIM1, PIM2, and PIM3 expression in peripheral B cells isolated from healthy donors (D#1, D#2) and a panel of 6 cHL-derived RS cell lines. GAPDH served as a loading control. (B) Immunohistochemical analysis of PIM expression in reactive germinal centers and cHL samples. Shown are representative germinal centers (GCs) originating from follicular hyperplasia lymph node samples and cHL cases, double positive for PIM1 and PIM3 (#7), PIM1 and PIM2 (#51), and PIM2 and PIM3 (#61). RS cells are indicated with arrows. Original magnification was ×600. (C) Summary of the immunohistochemical analysis of PIM expression performed in a panel of 67 patients with cHL. Solid blocks, positive cases; open blocks, negative cases.
NFκB and STAT transcription factors drive expression of PIM kinases in RS cells
Because aberrant activity of the NFκB and JAK-STAT signaling pathways is a typical feature of RS cells, and both cascades have been shown to induce PIM expression,5-8,25-28 we hypothesized that overexpression of PIM kinases in RS cells in cHL is driven by NFκB and/or STAT transcription factors. To test this hypothesis, we transduced RS cells with a nondegradable super-repressor form of inhibitor of κBα (SR-IκBα), in which serines 32 and 36 were replaced with alanines (SR-IκBα).36 SR-IκBα, which cannot be phosphorylated by IκK, remains complexed to the NFκB heterodimer, inhibiting NFκB translocation and activation of canonical NFκB activity.33 In SR-IκBα-transduced SUP-HD1 and HDLM-2 cells, expression of a direct NFκB target, IκBα, decreased by 63% and 86%, respectively, confirming reduced activity of the canonical NFκB pathway37 (Figure 2A; supplemental Figures 2 and 3). Compared with empty vector controls, SUP-HD1 cells transduced with SR-IκBα exhibited reduced expression of PIM2 (−57%) and PIM3 (−49%), but not PIM1 (Figure 2A). In HDLM-2 cells, SR-IκBα-mediated inhibition of the canonical NFκB pathway caused marked decrease in PIM1 (−31%) and PIM3 (−80%) levels, and only minor decrease in PIM2 (supplemental Figure 3). To further confirm the role of NFκB dimers in regulation of PIM kinases in SUP-HD1 cells, we knocked down expression of transcriptional activators of the canonical and noncanonical NFκB pathway: NFκB-p65/RELA, REL (c-Rel), and RELB. This approach decreased RELA and RELB expression by 88% to 98% but had only a minor effect on the already low REL expression level (supplemental Figure 4). RELB knock-down had a stronger effect on PIM2 and PIM3 expression than RELA (Figure 2B), and concurrent RELA and RELB silencing led to significant downregulation of PIM2 and PIM3, but not PIM1 (Figure 2B; supplemental Figure 5). Collectively, these results indicate that both canonical and noncanonical NFκB pathways regulate PIM1, PIM2, and PIM3 expression in RS cells, but also suggest cell line-specific contribution of NFκB to expression of PIM1 and PIM2 isoforms and/or an additional role of another pathway or pathways.
PIM expression in RS cells is regulated by NFκB and JAK-STAT activity. (A) Expression of PIM1, PIM2, and PIM3 after inhibition of the canonical NFκB signaling by expression of an IκBα super-repressor (SR-IκBα) in SUP-HD1 RS cells. Gene expression was assessed by qPCR and presented as relative changes using the 2−ΔΔCT method. IκB, a direct target of canonical NFκB pathway, was used as a control. Raw ΔCT (CT gene of interest − CT GAPDH) values are shown in supplemental Figure 2. P values were determined using 2-sided Gosset’s t-test. *P < .05; **P < .01; ***P < .001. (B) Expression of PIM1, PIM2, and PIM3 after siRNA-mediated RELA and RELB knock-down in SUP-HD1 cells. Cells were transfected with a nontargeting siRNAs pool (control, Ctrl) or siRNAs targeting RELA, RELB, or RELA and RELB (2×Rel). Cells were cultured for 4 days and assessed for changes in PIM1, PIM2, and PIM3 gene expression, using qPCR. Raw ΔCT values are shown in supplemental Figure 5. P values were determined using 2-sided Gosset’s t-test: *P < .05; **P < .01; ***P < .001. (C) Expression of PIM1, PIM2, and PIM3 after JAK inhibition. Cell lines were incubated with 1-3 µM CYT387 or DMSO alone for 24 hours, harvested and analyzed by western blot, using indicated antibodies. GAPDH was used as a loading control. Densitometric quantifications of band intensities are shown in supplemental Table 5. Data in A-C are representative of 3 independent experiments.
PIM expression in RS cells is regulated by NFκB and JAK-STAT activity. (A) Expression of PIM1, PIM2, and PIM3 after inhibition of the canonical NFκB signaling by expression of an IκBα super-repressor (SR-IκBα) in SUP-HD1 RS cells. Gene expression was assessed by qPCR and presented as relative changes using the 2−ΔΔCT method. IκB, a direct target of canonical NFκB pathway, was used as a control. Raw ΔCT (CT gene of interest − CT GAPDH) values are shown in supplemental Figure 2. P values were determined using 2-sided Gosset’s t-test. *P < .05; **P < .01; ***P < .001. (B) Expression of PIM1, PIM2, and PIM3 after siRNA-mediated RELA and RELB knock-down in SUP-HD1 cells. Cells were transfected with a nontargeting siRNAs pool (control, Ctrl) or siRNAs targeting RELA, RELB, or RELA and RELB (2×Rel). Cells were cultured for 4 days and assessed for changes in PIM1, PIM2, and PIM3 gene expression, using qPCR. Raw ΔCT values are shown in supplemental Figure 5. P values were determined using 2-sided Gosset’s t-test: *P < .05; **P < .01; ***P < .001. (C) Expression of PIM1, PIM2, and PIM3 after JAK inhibition. Cell lines were incubated with 1-3 µM CYT387 or DMSO alone for 24 hours, harvested and analyzed by western blot, using indicated antibodies. GAPDH was used as a loading control. Densitometric quantifications of band intensities are shown in supplemental Table 5. Data in A-C are representative of 3 independent experiments.
Because transcription of PIM kinases can also be induced by cytokine signaling,25,26,38,39 which primarily activates the JAK-STAT axis, we determined its role in PIM expression. In RS cells incubated with a pan-JAK inhibitor CYT387, activities of STAT1, STAT3, and STAT5 were markedly suppressed and were accompanied by decreases in PIM2 protein levels, whereas PIM1 and PIM3 expression decreased in 5 of the 6 tested cell lines (Figure 2C). This pattern of decrease in PIM1, PIM2, and PIM3 was closely reproduced in all investigated RS cell lines on treatment with a specific JAK2 inhibitor fedratinib (supplemental Figure 6). Taken together, these results demonstrate that both NFκB and STAT transcription factors are important inducers of PIM1, PIM2, and PIM3 expression in RS cells.
Prosurvival role of PIM kinases in RS cells
Because oncogenic PIM kinases support cell viability and proliferation in multiple tumor types, we next asked whether their activity plays a similar role in RS cells. To inhibit PIM activity, we transiently silenced PIM1, PIM2, and PIM3 expression in cHL RS cells using siRNA. After confirming successful knockdowns by quantitative polymerase chain reaction (qPCR) and western blotting (Figure 3A-B; supplemental Figure 7), cellular apoptosis was assessed by AnnexinV/7AAD staining. Knockdowns of individual PIM members were associated with increased expression of remaining isoforms and did not significantly induce apoptosis of HDLM-2 cells (Figure 3C). In contrast, simultaneous downregulation of all 3 PIMs markedly increased cellular apoptosis. To confirm these observations, we knocked down all PIMs in an additional cell line, SUP-HD1, which similarly led to increased apoptosis (Figure 3B-C). Given the redundant role of PIMs, for subsequent studies, we used a newly developed pan-PIM inhibitor, SEL24-B48940 (supplemental Table 4). Because SEL24-B489 also targets FLT3,40 to ensure the inhibitor’s specificity, we first confirmed complete absence of the FLT3 expression in a panel of RS cell lines (supplemental Figure 8). Next, we assessed the inhibitor’s effect on cellular proliferation and apoptosis. SEL24-B489 markedly decreased proliferation in a dose-dependent manner in all tested cell lines (Figure 3D), with EC50 ranging from 3 to 5.3 μM. We next determined whether SEL24-B489 was cytotoxic to RS cells, using the 5 μM dose, derived from the EC50 analysis. RS cells were cultured with the inhibitor or vehicle alone (DMSO) for 96 hours and assessed for apoptosis by AnnexinV/7-AAD staining. SEL24-B489 induced apoptosis in all cell lines, increasing the fraction of AnnexinV/7-AAD positive cells by 22.7% to 69% (Figure 3E; supplemental Figure 9). In marked contrast, exposure of normal B cells to 5 and 10 µM of SEL24-B489 did not induce cell death (supplemental Figure 10). We further assessed the antitumor efficacy of SEL24-B489 in vivo, in a murine cHL xenograft model. For this purpose, HDLM-2 cells were inoculated subcutaneously into NOD SCID IL2γnull mice and allowed to develop tumors. Subsequently, cHL xenograft-bearing mice were treated once daily with 50 mg/kg SEL24-B489 or vehicle (H2O) alone (control animals). In SEL24-B489-treated mice, tumor growth was inhibited by 95.82% (2-way ANOVA, P = .00002; supplemental Figure 11A). Taken together, these results indicate that PIM kinases are prosurvival factors in RS cells and that the pan-PIM inhibitor SEL24-B489 exhibits cytotoxic activity selectively in PIM-positive tumor cells.
PIM1, PIM2, and PIM3 kinases support RS cell survival. (A) SiRNA-mediated PIM1, PIM2, and PIM3 silencing in HDLM-2 cell line. Cells were transfected with a nontargeting siRNA (control, Ctrl), siRNAs targeting individual PIM isoforms, or with a siRNA cocktail targeting all 3 PIM kinases (3×PIM) and cultured for 4 days. Relative changes in transcript abundance are calculated using the 2−ΔΔCT method. Raw ΔCT values are shown in supplemental Figure 7. P values were determined using 2-sided Gosset’s t-test: *P < .05; **P < .01; ***P < .001). (B) SiRNA-mediated PIM1, PIM2, and PIM3 protein knock-down in HDLM-2 and SUP-HD1 cells. Cells were transfected with a nontargeting siRNA (control, Ctrl) or with a siRNA cocktail targeting all 3 PIM kinases (3×PIM). Thereafter, cells were cultured for 4 days, and PIM1, PIM2, and PIM3 expression was assessed by immunoblotting. Densitometric quantifications of band intensities (normalized to GAPDH) are indicated above the lanes. (C) Induction of apoptosis in HDLM-2 and SUP-HD1 RS cells after silencing PIM1, PIM2, and PIM3 expression individually or in combination. Cells were transduced with siRNAs as in A. Apoptosis was assessed by AnnexinV/7AAD staining. Bar graphs represent mean values derived from 3 independent experiments. P values were determined using 2-sided Gosset’s t-test: *P < .05. (D) EC50 dose-response curves for SEL24-B489 in cHL-derived cell lines. Cellular proliferation was determined by 3-(4,5 dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl-2-(4-sulfophenyl)-2H-tetrazolium reduction assay after 72 hours of incubation. The individual data points on dose-response curves represent the mean ± SD. Calculated EC50 values are given below the plots. (E) Cellular apoptosis in SEL24-B489-treated RS cells. After 96 hours of incubation with 5 µM SEL24-B489, apoptosis was measured by AnnexinV/7AAD staining, followed by FACS analysis (summary and statistics of 3 independent experiments is shown in supplemental Figure 9). Data in A-E are representative of 3 independent experiments
PIM1, PIM2, and PIM3 kinases support RS cell survival. (A) SiRNA-mediated PIM1, PIM2, and PIM3 silencing in HDLM-2 cell line. Cells were transfected with a nontargeting siRNA (control, Ctrl), siRNAs targeting individual PIM isoforms, or with a siRNA cocktail targeting all 3 PIM kinases (3×PIM) and cultured for 4 days. Relative changes in transcript abundance are calculated using the 2−ΔΔCT method. Raw ΔCT values are shown in supplemental Figure 7. P values were determined using 2-sided Gosset’s t-test: *P < .05; **P < .01; ***P < .001). (B) SiRNA-mediated PIM1, PIM2, and PIM3 protein knock-down in HDLM-2 and SUP-HD1 cells. Cells were transfected with a nontargeting siRNA (control, Ctrl) or with a siRNA cocktail targeting all 3 PIM kinases (3×PIM). Thereafter, cells were cultured for 4 days, and PIM1, PIM2, and PIM3 expression was assessed by immunoblotting. Densitometric quantifications of band intensities (normalized to GAPDH) are indicated above the lanes. (C) Induction of apoptosis in HDLM-2 and SUP-HD1 RS cells after silencing PIM1, PIM2, and PIM3 expression individually or in combination. Cells were transduced with siRNAs as in A. Apoptosis was assessed by AnnexinV/7AAD staining. Bar graphs represent mean values derived from 3 independent experiments. P values were determined using 2-sided Gosset’s t-test: *P < .05. (D) EC50 dose-response curves for SEL24-B489 in cHL-derived cell lines. Cellular proliferation was determined by 3-(4,5 dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl-2-(4-sulfophenyl)-2H-tetrazolium reduction assay after 72 hours of incubation. The individual data points on dose-response curves represent the mean ± SD. Calculated EC50 values are given below the plots. (E) Cellular apoptosis in SEL24-B489-treated RS cells. After 96 hours of incubation with 5 µM SEL24-B489, apoptosis was measured by AnnexinV/7AAD staining, followed by FACS analysis (summary and statistics of 3 independent experiments is shown in supplemental Figure 9). Data in A-E are representative of 3 independent experiments
Inhibition of PIM kinases modulates cap-dependent translation in RS cells
The cytotoxic activity of SEL24-B489 in RS cells prompted us to investigate molecular mechanisms associated with PIM inhibition. Because PIM kinases are involved in regulation of 4EBP1 and the ribosomal protein S6, and thus control cap-dependent translation, we assessed the phosphorylation status of these substrates in RS cells after PIM inhibition, using siRNA and after SEL24-B489 treatment. PIM knock-down decreased 4EBP1 phosphorylation at the PIM-specific S65 residue and S6 phosphorylation at S235/236 in 2 RS cell lines (Figure 4A). Consistently, all RS cell lines exposed to SEL24-B489 exhibited rapid and dose-dependent decrease in 4EBP1 and S6 phosphorylations (Figure 4B). In addition, SEL24-B489 markedly downregulated p-S6 S235/236 levels in tumor sections from PIM inhibitor-treated animals (supplemental Figure 11B-C).
PIM inhibition blocks cap-dependent translation in RS cells. (A) PIM depletion decreases activity of 4E-BP1 and S6. Cells were transfected with a nontargeting siRNA (control, Ctrl) or with a siRNA cocktail targeting all 3 PIM kinases (3×PIM). After 4 days, p-4EBP1 (S65) and p-S6 (S235/236) protein expression was assessed by immunoblotting. GAPDH served as a loading control. Densitometric quantifications of band intensities (p-4EBP1 and p-S6, normalized to 4EBP1 and S6, respectively) are indicated above the lanes. (B) SEL24-B489 downregulates PIM-specific 4EBP1 and S6 phosphorylation. Cells were incubated for 4 hours with DMSO alone or with increasing doses (0.1-10 µM) of SEL24-B489, harvested, and lysed. Expression of p-4EBP1 (S65) and p-S6 (S235/236) were analyzed by immunoblotting. GAPDH served as a loading control. Densitometric quantifications of band intensities are shown in supplemental Table 6. Data in A-B are representative of 3 independent experiments.
PIM inhibition blocks cap-dependent translation in RS cells. (A) PIM depletion decreases activity of 4E-BP1 and S6. Cells were transfected with a nontargeting siRNA (control, Ctrl) or with a siRNA cocktail targeting all 3 PIM kinases (3×PIM). After 4 days, p-4EBP1 (S65) and p-S6 (S235/236) protein expression was assessed by immunoblotting. GAPDH served as a loading control. Densitometric quantifications of band intensities (p-4EBP1 and p-S6, normalized to 4EBP1 and S6, respectively) are indicated above the lanes. (B) SEL24-B489 downregulates PIM-specific 4EBP1 and S6 phosphorylation. Cells were incubated for 4 hours with DMSO alone or with increasing doses (0.1-10 µM) of SEL24-B489, harvested, and lysed. Expression of p-4EBP1 (S65) and p-S6 (S235/236) were analyzed by immunoblotting. GAPDH served as a loading control. Densitometric quantifications of band intensities are shown in supplemental Table 6. Data in A-B are representative of 3 independent experiments.
PIM kinases modulate NFκB and STAT3/5 activity in RS cells
Substantial induction of apoptosis in RS cells, caused by PIM inhibition, led us to suspect an additional PIM-dependent mechanism or mechanisms of SEL24-B489 toxicity. RELA (p65) dimers are known to play a crucial pathogenetic role in RS cells by stimulating proliferation and conferring resistance to apoptosis.5,6,41,42 RS cells also exhibit constitutive activity of the JAK/STAT signaling pathway.4,43 Because PIM kinases directly modulate the activity of STAT3 and STAT5 factors,23,24 we first assessed the phosphorylation levels of these STATs in RS cells after PIM knock-down and SEL24-B489 treatment. As expected, genetic (siRNA) PIM inhibition was accompanied by a decrease in phosphorylation of STAT3 and STAT5 (Figure 5A). Chemical PIM kinase inhibition in RS cells similarly decreased p-STAT3 and p-STAT5 levels (Figure 5B). Because PIM1 directly and indirectly modulates NFκB function,22,44 we assessed the activity of this transcription factor in RS cells after genetic and chemical PIM inhibition. PIMs knock-down and SEL24-B489 decreased PIM-specific RELA-S276 phosphorylation (Figure 5C-D). Furthermore, SEL24-B489 decreased RELA binding to its consensus DNA sequences by 17.8% to 54.81% (Figure 5E) and decreased the mRNA expression of BCL2A1 (Bfl-1) and RELB, being the 2 direct NFκB target genes implicated in cHL pathogenesis and RS cell survival45,46 (Figure 5F; supplemental Figure 12).
PIM inhibition downregulates NFκB and STAT3/5 activity in cHL-derived cell lines. (A) PIM1/2/3 depletion decreases STAT3/5 activity in RS cells. Cells were transfected with a nontargeting siRNA (control, Ctrl) or with a siRNA cocktail targeting all 3 PIM kinases (3×PIM). Thereafter, cells were cultured for 4 days, and phospho-STAT5 (Y694) and phospho-STAT3 (Y705) protein levels were assessed by immunoblotting. Densitometric quantifications of band intensities (p-STAT5 and p-STAT3 normalized to STAT5 and STAT3, respectively) are indicated above the lanes. (B) SEL24-B489 decreases STAT3/5 activity in RS cells. Cells were incubated for 24 hours with 3-5 µM SEL24-B489 or DMSO alone, harvested, and lysed. Phospho-STAT5 (Y694) phospho-STAT3 (Y705) levels were assessed by immunoblotting. Densitometric quantifications of band intensities are shown in supplemental Table 7. (C) PIM1/2/3 depletion decreases PIM-specific RELA/p65-S276 phosphorylation in RS cells. Cells were transfected with a nontargeting siRNA (control, Ctrl) or with a siRNA cocktail targeting all 3 PIM kinases (3×PIM). Thereafter, cells were cultured for 4 days, and phospho-RELA/p65 (S276) levels were assessed by immunoblotting. Densitometric quantifications of band intensities (p-RELA/p65 normalized to RELA/p65) are indicated above the lanes. (D) SEL24-B489 decreases RELA/p65 S276 phosphorylation in RS cells. Cells were incubated for 4 hours with DMSO alone or with 0.1-10 µM SEL24-B489, harvested, and lysed. The abundance of phospho-RELA/p65 (S276) was assessed by immunoblotting. GAPDH served as a loading control. Densitometric quantifications of band intensities are shown in supplemental Table 8. (E) SEL24-B489 reduces DNA binding activity of the RELA-containing NFκB complexes. RS cells were incubated either with DMSO alone or SEL24-B489 (5 µM) for 24 hours, lysed, and subjected to TransAM DNA binding assay, using the RELA-specific antibody. (F) PIM inhibition in cHL cell lines decreased NFκB-dependent gene expression. Cells were incubated with DMSO alone or with SEL24-B489 (3 µM) for 24 hours. Thereafter, expression of BCL2A1 and RELB was assessed by qPCR. Raw ΔCT values are shown in supplemental Figure 12. P values were determined using the 2-sided Gosset’s t-test: *P < .05; **P < .01; ***P < .001. Data in A-F are representative of 3 independent experiments.
PIM inhibition downregulates NFκB and STAT3/5 activity in cHL-derived cell lines. (A) PIM1/2/3 depletion decreases STAT3/5 activity in RS cells. Cells were transfected with a nontargeting siRNA (control, Ctrl) or with a siRNA cocktail targeting all 3 PIM kinases (3×PIM). Thereafter, cells were cultured for 4 days, and phospho-STAT5 (Y694) and phospho-STAT3 (Y705) protein levels were assessed by immunoblotting. Densitometric quantifications of band intensities (p-STAT5 and p-STAT3 normalized to STAT5 and STAT3, respectively) are indicated above the lanes. (B) SEL24-B489 decreases STAT3/5 activity in RS cells. Cells were incubated for 24 hours with 3-5 µM SEL24-B489 or DMSO alone, harvested, and lysed. Phospho-STAT5 (Y694) phospho-STAT3 (Y705) levels were assessed by immunoblotting. Densitometric quantifications of band intensities are shown in supplemental Table 7. (C) PIM1/2/3 depletion decreases PIM-specific RELA/p65-S276 phosphorylation in RS cells. Cells were transfected with a nontargeting siRNA (control, Ctrl) or with a siRNA cocktail targeting all 3 PIM kinases (3×PIM). Thereafter, cells were cultured for 4 days, and phospho-RELA/p65 (S276) levels were assessed by immunoblotting. Densitometric quantifications of band intensities (p-RELA/p65 normalized to RELA/p65) are indicated above the lanes. (D) SEL24-B489 decreases RELA/p65 S276 phosphorylation in RS cells. Cells were incubated for 4 hours with DMSO alone or with 0.1-10 µM SEL24-B489, harvested, and lysed. The abundance of phospho-RELA/p65 (S276) was assessed by immunoblotting. GAPDH served as a loading control. Densitometric quantifications of band intensities are shown in supplemental Table 8. (E) SEL24-B489 reduces DNA binding activity of the RELA-containing NFκB complexes. RS cells were incubated either with DMSO alone or SEL24-B489 (5 µM) for 24 hours, lysed, and subjected to TransAM DNA binding assay, using the RELA-specific antibody. (F) PIM inhibition in cHL cell lines decreased NFκB-dependent gene expression. Cells were incubated with DMSO alone or with SEL24-B489 (3 µM) for 24 hours. Thereafter, expression of BCL2A1 and RELB was assessed by qPCR. Raw ΔCT values are shown in supplemental Figure 12. P values were determined using the 2-sided Gosset’s t-test: *P < .05; **P < .01; ***P < .001. Data in A-F are representative of 3 independent experiments.
PIM inhibition in RS cells impairs expression of cytokines, chemokines, and immunomodulatory molecules and leads to increased T-cell activation
In addition to the pro-survival genes, NFκB and STATs in RS cells induce the expression of multiple molecules that affect the microenvironment’s cellular composition and activity of infiltrating cells. We thus investigated the transcript abundance of direct NFκB targets, tumor necrosis factor (TNF) α, CXCL8/interleukin 8 (IL-8), CCL5/RANTES, IL-13, and CD40, in RS cells on PIM inhibition.42,47-50 As expected, SEL24-B489 significantly decreased mRNA levels of these genes (Figure 6A; supplemental Figure 13).
PIM inhibition attenuates expression of NFκB-regulated cytokines, chemokines, and immunoregulatory molecules PD-L1/2 and Gal-1 in RS cells. (A) PIM inhibition in cHL cell lines decreased NFκB-dependent gene expression. Cells were incubated with DMSO alone or with SEL24-B489 (3 µM) for 24 hours. Thereafter, expression of TNF, IL-8, CCL5, IL-13, and CD40 was assessed by qPCR. Raw ΔCT values are shown in supplemental Figure 13. P values were determined using the 2-sided Gosset’s t-test: *P < .05; **P < .01; ***P < .001. (B) PIM depletion decreases Gal-1 expression in RS cells. Cells were transfected with a nontargeting siRNA (control, Ctrl) or with a siRNA cocktail targeting all 3 PIM kinases (3×PIM). Thereafter, cells were cultured for 4 days, and Gal-1 protein abundance was assessed by immunoblotting. GAPDH served as a loading control. Densitometric quantifications of band intensities (normalized to GAPDH) are indicated above the lanes. (C) Inhibition of PIM kinases attenuates Gal-1 protein expression in RS cells. CHL cells were incubated for 24 hours with 3-5 µM SEL24-B489 or DMSO alone, harvested, and lysed. Expression of Gal-1 was assessed by immunoblotting. GAPDH served as a loading control. Densitometric quantifications of band intensities (normalized to GAPDH) are indicated above the lanes. (D) PIM knock-down decreases surface PD-L1 and PD-L2 expression. HDLM-2 and SUP-HD1 RS cells were transfected with a nontargeting siRNA (control, Ctrl) or with a siRNA cocktail targeting all 3 PIM kinases (3×PIM). Thereafter, cells were cultured for 5 days, and cell surface expression of PD-L1 and PD-L2 was assessed by flow cytometry. (E) PIM inhibition decreases surface PD-L1 and PD-L2 expression. Cells were incubated with DMSO alone or with SEL24-B489 (2 µM) for 120 hours. Thereafter, cell surface expression of PD-L1 and PD-L2 was assessed by flow cytometry. Data in A-E are representative of 3 independent experiments.
PIM inhibition attenuates expression of NFκB-regulated cytokines, chemokines, and immunoregulatory molecules PD-L1/2 and Gal-1 in RS cells. (A) PIM inhibition in cHL cell lines decreased NFκB-dependent gene expression. Cells were incubated with DMSO alone or with SEL24-B489 (3 µM) for 24 hours. Thereafter, expression of TNF, IL-8, CCL5, IL-13, and CD40 was assessed by qPCR. Raw ΔCT values are shown in supplemental Figure 13. P values were determined using the 2-sided Gosset’s t-test: *P < .05; **P < .01; ***P < .001. (B) PIM depletion decreases Gal-1 expression in RS cells. Cells were transfected with a nontargeting siRNA (control, Ctrl) or with a siRNA cocktail targeting all 3 PIM kinases (3×PIM). Thereafter, cells were cultured for 4 days, and Gal-1 protein abundance was assessed by immunoblotting. GAPDH served as a loading control. Densitometric quantifications of band intensities (normalized to GAPDH) are indicated above the lanes. (C) Inhibition of PIM kinases attenuates Gal-1 protein expression in RS cells. CHL cells were incubated for 24 hours with 3-5 µM SEL24-B489 or DMSO alone, harvested, and lysed. Expression of Gal-1 was assessed by immunoblotting. GAPDH served as a loading control. Densitometric quantifications of band intensities (normalized to GAPDH) are indicated above the lanes. (D) PIM knock-down decreases surface PD-L1 and PD-L2 expression. HDLM-2 and SUP-HD1 RS cells were transfected with a nontargeting siRNA (control, Ctrl) or with a siRNA cocktail targeting all 3 PIM kinases (3×PIM). Thereafter, cells were cultured for 5 days, and cell surface expression of PD-L1 and PD-L2 was assessed by flow cytometry. (E) PIM inhibition decreases surface PD-L1 and PD-L2 expression. Cells were incubated with DMSO alone or with SEL24-B489 (2 µM) for 120 hours. Thereafter, cell surface expression of PD-L1 and PD-L2 was assessed by flow cytometry. Data in A-E are representative of 3 independent experiments.
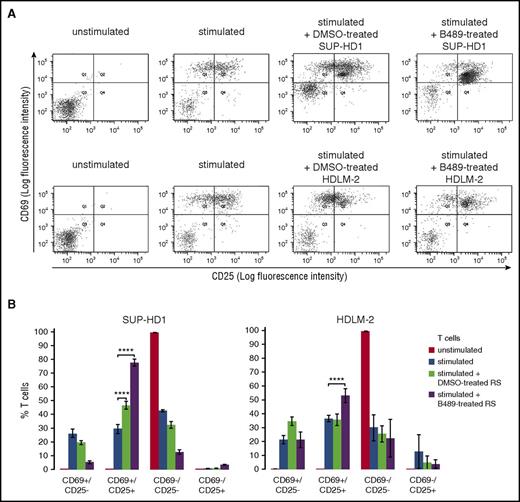
RS cells also express potent immunomodulatory proteins facilitating tumor immune escape, Gal-1, PD-L1, and PD-L2, at least in part via increased activity of STAT and NFκB transcription factors.12-14,35 We thus hypothesized that PIM kinases might contribute to expression of these proteins in cHL. Consistent with this hypothesis, genetic or chemical PIM inhibition in RS cells markedly downregulated Gal-1 protein (Figure 6B-C) and decreased surface abundance of PD-L1 and PD-L2 (Figure 6D-E). Of note, SEL24-B489 markedly reduced PD-L1 and Gal-1 protein levels in xenografted cHL tumors in mice exposed to the inhibitor (supplemental Figure 11D). In line with these data, we also found that Gal-1 expression was lower in the primary cHL samples expressing single PIM isoform at low levels compared with cases highly positive for all 3 PIM kinases (supplemental Figure 14). These data indicate that PIM inhibition modulates expression of multiple key immunoregulatory molecules involved in creating an immunosuppressive tumor microenvironment in cHL. To determine whether PIM inhibition in RS cells would influence their interactions with T cells, we cocultured normal blood-derived T lymphocytes or Jurkat cells with DMSO- or SEL24-B489-treated RS cells and assessed surface expression of 2 T-cell activation markers, CD69 and CD25. Compared with T cells incubated with DMSO-treated RS cells, T lymphocytes cocultured with inhibitor-treated RS cells exhibited increased expression of both CD69 and CD25 (Figure 7; supplemental Figure 15). These results demonstrate that PIM inhibition in the neoplastic cells of cHL increases activation of interacting T cells.
Inhibition of PIM kinases in RS cells increases T-cell activation. (A) SUP-HD1 and HDLM-2 RS cells were treated for 120 hours with DMSO alone or B489 (2 µM). Normal blood-derived naive T cells were stimulated with human T-activator CD3/CD28 Dynabeads and mixed with pretreated RS cells at 1:4 ratio (T cells to RS cells). After 16 hours of coculture, CD69 and CD25 expression on T-cell surface was measured by flow cytometry. (B) Relative changes in T- cell activation, presented as changes in CD69+/CD25− (Q1 in Figure 7A), CD69+/CD25+ (Q2 in Figure 7A), CD69−/CD25− (Q3 in Figure 7A), and CD69−/CD25+ (Q4 in Figure 7A) staining in T cell, averaged from 3 independent experiments. P values were determined using the 2-sided Gosset’s t-test: ****P < .0001.
Inhibition of PIM kinases in RS cells increases T-cell activation. (A) SUP-HD1 and HDLM-2 RS cells were treated for 120 hours with DMSO alone or B489 (2 µM). Normal blood-derived naive T cells were stimulated with human T-activator CD3/CD28 Dynabeads and mixed with pretreated RS cells at 1:4 ratio (T cells to RS cells). After 16 hours of coculture, CD69 and CD25 expression on T-cell surface was measured by flow cytometry. (B) Relative changes in T- cell activation, presented as changes in CD69+/CD25− (Q1 in Figure 7A), CD69+/CD25+ (Q2 in Figure 7A), CD69−/CD25− (Q3 in Figure 7A), and CD69−/CD25+ (Q4 in Figure 7A) staining in T cell, averaged from 3 independent experiments. P values were determined using the 2-sided Gosset’s t-test: ****P < .0001.
Discussion
In this study, we document the expression of PIM kinases in cHL and characterize their prosurvival functions in RS cells of cHL. We show that PIM kinases regulate fundamental cellular functions in this tumor, such as cap-dependent protein translation, but also support NFκB and JAK/STAT signaling. We also show that PIMs modulate the RS-specific expression of cytokines, chemokines, and immunomodulatory proteins that shape the protumoral, immune privileged microenvironment. Thus, their inhibition exhibits pleiotropic effects that combine a direct cytotoxic effect with disruption of signaling circuits that link RS cells to their niches.
PIM kinase genes display characteristics of primary response genes that are induced by the activation of transcription factors downstream of growth factor signaling pathways, such as the JAK/STAT, PI3K-AKT, MAPK, and NFκB pathways. As the constitutive activity of these pathways is a hallmark of cHL, PIM kinases are also almost uniformly expressed by RS cells. Inhibition of canonical and alternative NFκB pathway or inhibition of JAK1/2 led to decreased expression of PIM genes in RS cells, indicating that these pathways are functionally implicated in PIM kinase expression.
PIM isoforms are highly homologous, and thus share an overlapping spectrum of substrates and exhibit partial functional redundancy in vitro and in vivo.51,52 Consistent with these data, we found that inhibition of a single PIM isoform was not sufficient to trigger cellular apoptosis, and only simultaneous downregulation of all 3 PIMs led to RS cell death. In addition, knockdown of a specific PIM isoform in RS cells led to a marked compensatory induction of remaining isoforms. These results indicate that expression of PIM kinases in RS cells is an important homeostatic mechanism, required for cell survival. These data also indicate that therapeutic effects of PIM kinase inhibition can be only achieved with a compound targeting all 3 PIM family members simultaneously. In this study, we used a newly developed, chemical pan-PIM inhibitor with promising characteristics and safety profile, approved for an ongoing phase 1/2 study in acute myeloid leukemia (AML; www.clinicaltrials.gov: NCT03008187).40,53 Our studies demonstrate that SEL24-B489 rapidly and efficiently blocked phosphorylation of crucial PIM substrates in vitro and in vivo, including ribosomal protein p-S6. This feature was also consistently observed in other tumor types, including AML,24,53,54 indicating that in the setting of clinical trial, p-S6 inhibition might be used as a target engagement biomarker.
PIM kinases are important hubs consolidating cytokine and growth factor-induced activity of multiple transcription factors, including NFκB subunit p65/RELA and STAT3/5. In RS cells of cHL, constitutive activity of these factors is induced by a number of functional and structural mechanisms. For example, NFκB is activated in RS cells by paracrine stimulation of TNF family receptors, expression of the EBV-encoded protein LMP-1, genomic amplifications of REL, or inactivating mutations of NFKBIA and TNFAIP3.4,55-63 Likewise, increased activity of JAK/STATs in these cells is caused by paracrine cytokine signaling and RS-cell intrinsic 9p24/JAK2 copy gains, SOCS, or PTPN11 mutations.11,35,64 Inhibition of these pathways decreases the viability of RS cells, highlighting the therapeutic potential of such approaches.43 However, unlike JAK/STAT signaling, targeted inhibition of NFκB activity is difficult and clinically not available. Here, we demonstrate that PIM kinases support NFκB activity in RS cells through modulating activity of the canonical NFκB subunit, RELA/p65. The mechanism by which decreased S276 phosphorylation regulates RELA/p65 transcriptional activity may involve several independent pathways. For example, inhibition of S276 phosphorylation leads to decreased RELA stability,22 but also triggers conformational changes in p65 that block its interaction with CBP/p300 coactivator thus reducing NFκB-dependent transcription.44 Therefore, these observations explain the nonlinear relationship between decreased NFκB DNA binding and expression of NFκB target genes in RS cell lines (Figure 5E-F). The relative contribution of these different transcriptional and epigenetic mechanisms to NFκB target gene expression requires further studies.
NFκB activity in RS cells orchestrates the expression of multiple cytokines/chemokines, immunomodulators, and surface receptors involved in RS cell cross-talk with the microenvironment. Consistent with attenuation of NFκB activity, PIM kinase inhibition decreased the expression of NFκB-dependent cytokines and chemokines. Thus, in the in vivo setting, inhibition of PIM kinases will likely interfere with RS-induced shaping of the microenvironment and disrupt certain autocrine prosurvival loops (eg, IL-13/IL13R). Importantly, PIM inhibition in vitro and in vivo decreased the expression of key immunomodulatory molecules, PD-L1 and Gal-1, which are expressed by RS cells, at least in part, in a JAK2/NFκB-dependent manner.13,14,35 These observations suggest PIM kinases contribute to establishing the immunosuppressive microenvironment in this disease, although they are unlikely to be key drivers of PD-L1/2 induction on the RS cell surface. As we previously demonstrated, disease-specific amplification of the 9p24.1 region that harbors PD-L1/2 and JAK2 loci increases gene dosage of PD-1 ligands and their induction by the JAK2-STAT pathway.35 Thus, cells with high-level 9p24.1 amplifications (such as SUP-HD1 and HDLM-2) exhibit the highest expression of PD-1 ligands, and the relative magnitude of PD-L1/2 reductions after PIM inhibition is also the highest. Because of the vast majority of patients with cHL exhibit structural alterations of 9p24.1 locus,65 our observations from in vitro studies are likely clinically and biologically relevant.
Of note, PIM inhibition was sufficient to produce functional consequences on T-cell activation. Given the PIM’s pleiotropic effect on the expression of various immunomodulatory molecules, it is difficult to judge which of them plays the leading role in increasing activation of interacting T cells. More likely, PIM kinase inhibition produces immunomodulatory effects by affecting multiple, nonredundant mechanisms leading to decreased or skewed T-cell responses.
Taken together, our work demonstrates that RS-specific expression of PIM kinases plays an important biological role in cHL. We show that PIM kinases support RS cell viability and facilitate their immune evasion. These results indicate that PIM kinases are promising therapeutic targets in cHL.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgment
This work was supported by a grant from the Polish National Science Centre (DEC-2011/03/B/NZ4/02020).
Authorship
Contribution: M.S. designed and performed the research, analyzed results, and wrote the manuscript; M.P.-S., A.S.-C., E.D., and O.S.-G. performed immunohistochemical stainings and interpreted IHC data; G.H. and D.W. performed in vivo studies; W.C., M.G., R.W., K.B., and J.M.Z. contributed vital reagents and materials; E.J., E.B., T.S., A.P., and K.W. contributed to the design of the experiments, developed vectors, and performed research; and P.J. designed and supervised research, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: P.J. is a member of the Scientific Advisory Board at Selvita S.A. and served as a consultant for Selvita S.A. W.C., M.G., R.W., and K.B. are Selvita S.A. employees. The remaining authors declare no competing financial interests.
Correspondence: Przemysław Juszczyński, Institute of Hematology and Transfusion Medicine, Department of Experimental Hematology, Indiry Gandhi 14, Warsaw, 02-776, Poland; e-mail: pjuszczynski@ihit.waw.pl.






This feature is available to Subscribers Only
Sign In or Create an Account Close Modal