Abstract
Acute myeloid leukemia (AML) is a deadly hematologic malignancy characterized by the uncontrolled growth of immature myeloid cells. Over the past several decades, we have learned a tremendous amount regarding the genetic aberrations that govern disease development in AML. Among these are genes that encode noncoding RNAs, including the microRNA (miRNA) family. miRNAs are evolutionarily conserved small noncoding RNAs that display important physiological effects through their posttranscriptional regulation of messenger RNA targets. Over the past decade, studies have identified miRNAs as playing a role in nearly all aspects of AML disease development, including cellular proliferation, survival, and differentiation. These observations have led to the study of miRNAs as biomarkers of disease, and efforts to therapeutically manipulate miRNAs to improve disease outcome in AML are ongoing. Although much has been learned regarding the importance of miRNAs in AML disease initiation and progression, there are many unanswered questions and emerging facets of miRNA biology that add complexity to their roles in AML. Moving forward, answers to these questions will provide a greater level of understanding of miRNA biology and critical insights into the many translational applications for these small regulatory RNAs in AML.
Introduction
MicroRNA (miRNAs) are small noncoding RNAs (∼20-24 nucleotides) that play vital roles in posttranscriptional gene regulation through repression of target messenger RNAs (mRNAs).1 miRNA-encoding genes in the nucleus are transcribed into primary miRNA transcripts, which then undergo a number of processing steps in the nucleus and cytoplasm to generate the mature miRNA molecule. The mature miRNA is loaded into the RNA-induced silencing complex (RISC), and this miRNA-RISC complex targets the 3′ untranslated region (UTR) of specific mRNAs on the basis of sequence complementarity, resulting in reduced protein outputs through mechanisms involving decreased mRNA stability and reduced translation.2
miRNAs are now recognized to play roles in nearly all physiological processes and have been implicated in a number of human diseases including cancer, where miRNAs can act as either oncogenes (oncomiRs) or tumor suppressors.3 Hematologic malignancies are no exception, as dysregulated miRNA expression contributes to blood cancers from many different hematopoietic lineages.4 This review will focus on recent advances in understanding the role of miRNAs in a hematologic malignancy with a particularly high rate of mortality, acute myeloid leukemia (AML).
miRNAs in AML: background
AML is a heterogeneous disease characterized by the increased proliferation and survival of immature myeloid cells and is the result of a number of genetic abnormalities, including mutations and chromosomal rearrangements.5 Early studies characterizing the role of miRNAs in AML focused on identifying AML-specific miRNA expression patterns. Distinctive miRNA profiles were identified for many cytogenetic subtypes of AML,6-8 as well as for several specific mutations in cytogenetically normal AML, including mutations in NPM1, FLT3, and CEBPA.9-13 miRNA expression profiles also correlate with prognosis,12,14 highlighting the potential importance of miRNAs in this disease. However, although miRNAs are enriched in leukemia-associated genomic alterations, only ∼100 are expressed above background level,15 suggesting that only a subset of miRNAs have functional effects in AML.
Beyond dysregulated miRNA expression profiles, it is now well accepted that miRNAs can function as either oncomiRs or tumor suppressors in many subtypes of AML, affecting a broad range of leukemic processes, including proliferation, survival, differentiation, self-renewal, epigenetic regulation, in vivo disease progression, and chemotherapy resistance8,15-57 (Table 1). miRNAs impact leukemic development and progression through collaboration with known oncogenes or tumor suppressors, either by directly targeting them on the mRNA level or by working in concert with these proteins to promote malignancy. To illustrate these concepts, we have summarized findings for selected miRNAs consistently found to play a role in AML in Table 1, and we discuss some of the more novel aspects of miRNA biology in AML below, as a greater understanding of miRNA biology will enable more strategic design of therapies in the future.
Selected miRNAs involved in AML pathogenesis
| miRNA . | Expression change . | Mechanism of dysregulation . | Confirmed targets . | Functional effect of expression . | References . | |
|---|---|---|---|---|---|---|
| OncomiR . | Tumor suppressor . | |||||
| miR-9 | ↑ MLL-rearranged AML, ↓ t(8;21), ↓ EVI1-induced AML | Promoter targeted by MLL-fusion proteins; EVI1 hypermethylates promoter | RHOH, RYBP, HMGA2, LIN28B, FOXO1, FOXO3 | ↑ Proliferation, ↑ survival, ↑ leukemogenesis in mice (MLL rearranged) | ↓ Proliferation, ↑ monocytic differentiation (t(8;21) and EVI1+) | 16-18 |
| miR-17-92 cluster | ↑ In LSCs in MLL-associated AML | Activated by MYC; epigenetically activated by MLL-fusion proteins; genomic amplification in MLL-rearranged AML | P21 | ↑ Proliferation, ↑ survival, ↓ differentiation, ↑ self-renewal, ↑ colony-forming capacity, ↑ leukemogenesis in mice | 19, 20 | |
| miR-22 | ↑ MDS/MDS-derived AML, ↓ de novo AML | Downregulated via TET1/GFI1/EZH2/SIN3A-mediated epigenetic repression and DNA copy-number loss; increased with loss of PU.1 | TET2, CRTC1, FLT3, MYCBP | ↑ Proliferation, ↑ survival, ↓ differentiation, ↑ self-renewal, overexpression leads to myeloid malignancy in mice (MDS/MDS-derived AML) | ↓ AML blast cell growth, ↑ differentiation, ↓ leukemic progression in mice (de novo AML) | 21-23 |
| miR-29b | ↓ Various subtypes of AML | Downregulated via loss of CEBPA; chromosome 7q deletions; MYC represses; NF-kB represses | MCL-1, CXXC6, CDK6, AKT2, CCND2, SP1, DNMT3A, DNMT3B | ↓ Cell growth, ↑ apoptosis, ↓ leukemic progression in vivo, ↓ KIT activation, prevents global DNA hypermethylation | 24-27, 56 | |
| miR-125b | ↑ t(2;11)(p21;q23) AML, ↑ MDS/MDS-derived AML, ↑ pediatric AML | Increased by t(2;11)(p21;q23) translocation | LIN28A, IRF4 | ↑ Proliferation, ↑ production of myeloid progenitors, ↑ self-renewal, overexpression leads to AML in mice | 28-32 | |
| miR-126 | ↑ t(8;21) AML, ↑ in LSCs of CN-AML | Promoter demethylated in t(8;21) AML | PLK2, ADAM9, ILK, GOLPH3, CDK3, TOM1, ERRFI1, SPRED1, FZD7 | ↑ Proliferation, ↑ survival, ↓ differentiation, ↑ colony-forming ability, ↑ LSC maintenance and self-renewal, ↓ cell cycling, ↑ quiescence, ↑ chemotherapy resistance | 8, 33-36 | |
| miR-146a | ↓ Various subtypes of AML, ↓ 5q syndrome MDS/MDS-derived AML | Deletion in del(5q) MDS/MDS-derived AML | TRAF6, IRAK1, TIRAP | ↓ Proliferation, ↓ survival, ↓ NF-kB activation, deletion leads to myeloproliferation in mice | 15, 37-41 | |
| miR-155 | ↑ CN-AML (highest in FLT3-ITD+ AML) | Targeted by STAT5 and NF-kB in FLT3-ITD+ AML; upregulated by MLL-fusion genes via MEIS1 | CEBPB, SHIP1, PU.1 | ↑ Proliferation, ↑ survival, overexpression leads to myeloproliferative neoplasm in mice, confers negative prognosis in CN-AML, no effect in MLL-AF9 mouse model of leukemia | 42-46 | |
| miR-193a | ↓ Various subtypes of AML (lowest in t(8;21) AML) | Epigenetically silenced by AML1/ETO | AML1/ETO, DNMT3A, HDAC3, KIT, CCND1, MDM2 | ↓ Cell growth, ↑ apoptosis, ↑ differentiation, ↓ cell cycling, ↓ KIT expression | 47, 48 | |
| miR-196b | ↑ MLL-associated AML | Activated by MLL-fusion proteins; co-expressed with HOXA9 in MLL-rearranged leukemia | HOXA9, MEIS1, FAS | ↑ Proliferation, ↑ survival, ↓ differentiation, ↓ replating potential, ↑ MLL-AF9–induced leukemic progression in mice | 49, 50 | |
| miR-223 | ↓ t(8;21) AML, ↓ various subtypes of AML | Targeted by transcription factors CEBPA (activates) and E2F1 (represses); epigenetically silenced by AML1/ETO | E2F1, MEF2C, FBXW7 | ↓ Proliferation, ↑ apoptosis, ↑ differentiation/granulopoiesis | 51-54 | |
| miRNA . | Expression change . | Mechanism of dysregulation . | Confirmed targets . | Functional effect of expression . | References . | |
|---|---|---|---|---|---|---|
| OncomiR . | Tumor suppressor . | |||||
| miR-9 | ↑ MLL-rearranged AML, ↓ t(8;21), ↓ EVI1-induced AML | Promoter targeted by MLL-fusion proteins; EVI1 hypermethylates promoter | RHOH, RYBP, HMGA2, LIN28B, FOXO1, FOXO3 | ↑ Proliferation, ↑ survival, ↑ leukemogenesis in mice (MLL rearranged) | ↓ Proliferation, ↑ monocytic differentiation (t(8;21) and EVI1+) | 16-18 |
| miR-17-92 cluster | ↑ In LSCs in MLL-associated AML | Activated by MYC; epigenetically activated by MLL-fusion proteins; genomic amplification in MLL-rearranged AML | P21 | ↑ Proliferation, ↑ survival, ↓ differentiation, ↑ self-renewal, ↑ colony-forming capacity, ↑ leukemogenesis in mice | 19, 20 | |
| miR-22 | ↑ MDS/MDS-derived AML, ↓ de novo AML | Downregulated via TET1/GFI1/EZH2/SIN3A-mediated epigenetic repression and DNA copy-number loss; increased with loss of PU.1 | TET2, CRTC1, FLT3, MYCBP | ↑ Proliferation, ↑ survival, ↓ differentiation, ↑ self-renewal, overexpression leads to myeloid malignancy in mice (MDS/MDS-derived AML) | ↓ AML blast cell growth, ↑ differentiation, ↓ leukemic progression in mice (de novo AML) | 21-23 |
| miR-29b | ↓ Various subtypes of AML | Downregulated via loss of CEBPA; chromosome 7q deletions; MYC represses; NF-kB represses | MCL-1, CXXC6, CDK6, AKT2, CCND2, SP1, DNMT3A, DNMT3B | ↓ Cell growth, ↑ apoptosis, ↓ leukemic progression in vivo, ↓ KIT activation, prevents global DNA hypermethylation | 24-27, 56 | |
| miR-125b | ↑ t(2;11)(p21;q23) AML, ↑ MDS/MDS-derived AML, ↑ pediatric AML | Increased by t(2;11)(p21;q23) translocation | LIN28A, IRF4 | ↑ Proliferation, ↑ production of myeloid progenitors, ↑ self-renewal, overexpression leads to AML in mice | 28-32 | |
| miR-126 | ↑ t(8;21) AML, ↑ in LSCs of CN-AML | Promoter demethylated in t(8;21) AML | PLK2, ADAM9, ILK, GOLPH3, CDK3, TOM1, ERRFI1, SPRED1, FZD7 | ↑ Proliferation, ↑ survival, ↓ differentiation, ↑ colony-forming ability, ↑ LSC maintenance and self-renewal, ↓ cell cycling, ↑ quiescence, ↑ chemotherapy resistance | 8, 33-36 | |
| miR-146a | ↓ Various subtypes of AML, ↓ 5q syndrome MDS/MDS-derived AML | Deletion in del(5q) MDS/MDS-derived AML | TRAF6, IRAK1, TIRAP | ↓ Proliferation, ↓ survival, ↓ NF-kB activation, deletion leads to myeloproliferation in mice | 15, 37-41 | |
| miR-155 | ↑ CN-AML (highest in FLT3-ITD+ AML) | Targeted by STAT5 and NF-kB in FLT3-ITD+ AML; upregulated by MLL-fusion genes via MEIS1 | CEBPB, SHIP1, PU.1 | ↑ Proliferation, ↑ survival, overexpression leads to myeloproliferative neoplasm in mice, confers negative prognosis in CN-AML, no effect in MLL-AF9 mouse model of leukemia | 42-46 | |
| miR-193a | ↓ Various subtypes of AML (lowest in t(8;21) AML) | Epigenetically silenced by AML1/ETO | AML1/ETO, DNMT3A, HDAC3, KIT, CCND1, MDM2 | ↓ Cell growth, ↑ apoptosis, ↑ differentiation, ↓ cell cycling, ↓ KIT expression | 47, 48 | |
| miR-196b | ↑ MLL-associated AML | Activated by MLL-fusion proteins; co-expressed with HOXA9 in MLL-rearranged leukemia | HOXA9, MEIS1, FAS | ↑ Proliferation, ↑ survival, ↓ differentiation, ↓ replating potential, ↑ MLL-AF9–induced leukemic progression in mice | 49, 50 | |
| miR-223 | ↓ t(8;21) AML, ↓ various subtypes of AML | Targeted by transcription factors CEBPA (activates) and E2F1 (represses); epigenetically silenced by AML1/ETO | E2F1, MEF2C, FBXW7 | ↓ Proliferation, ↑ apoptosis, ↑ differentiation/granulopoiesis | 51-54 | |
Table represents selected miRNAs involved in AML disease progression, with a focus on miRNAs with clinical evidence of dysregulation, as well as in vivo and in vitro functional evidence of role in leukemogenesis.
CN, cytogenetically normal; LSC, leukemic stem cell.
Mechanisms of dysregulated miRNA expression in AML
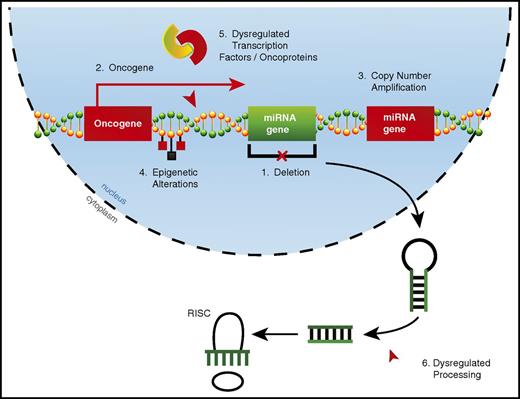
Alterations in miRNA expression can occur through a variety of mechanisms in AML (Figure 1). Copy number alterations (CNAs), which include deletions22,25 and amplifications,20 can drastically alter miRNA expression. However, acquired CNAs specifically targeting miRNAs may be relatively rare in AML. By using a combination of comparative genomic hybridization and whole-genome sequencing, researchers found that 18% of patients had CNAs involving miRNA genes, with a single CNA affecting up to 121 miRNAs.58 However, these CNAs always contained one or more protein-coding genes, suggesting that the miRNA genes involved in these CNAs may be passenger alterations. miRNAs may also be aberrantly expressed when located in oncogenic genomic locations, which occurs through chromosomal translocations28 or overexpression of nearby protein-coding genes.49
Mechanisms of dysregulated miRNA expression in AML. miRNA dysregulation can contribute to the development of AML. Thus far, numerous mechanisms by which miRNAs become dysregulated in AML have been identified, including (1) deletions leading to decreased miRNA expression, (2) improper expression because of close proximity to an oncogenic genomic region created as a result of either a translocation event or overexpression of a neighboring protein-coding gene, (3) copy number amplifications leading to increased miRNA expression, (4) epigenetic alterations affecting miRNA expression, (5) miRNA promoter regions being aberrantly targeted by dysregulated transcription factors or oncoproteins, and (6) dysregulated miRNA processing leading to altered levels of mature miRNAs.
Mechanisms of dysregulated miRNA expression in AML. miRNA dysregulation can contribute to the development of AML. Thus far, numerous mechanisms by which miRNAs become dysregulated in AML have been identified, including (1) deletions leading to decreased miRNA expression, (2) improper expression because of close proximity to an oncogenic genomic region created as a result of either a translocation event or overexpression of a neighboring protein-coding gene, (3) copy number amplifications leading to increased miRNA expression, (4) epigenetic alterations affecting miRNA expression, (5) miRNA promoter regions being aberrantly targeted by dysregulated transcription factors or oncoproteins, and (6) dysregulated miRNA processing leading to altered levels of mature miRNAs.
The most common mechanisms by which miRNA expression becomes dysregulated in AML are epigenetic alterations and via targeting by dysregulated transcription factors or oncogenic fusion proteins. These two mechanisms are not always distinct, as epigenetic alterations to miRNA loci often occur via dysregulated transcription factors or oncoproteins.18,47 There is some evidence that alterations in miRNA expression in cancer can be the result of dysregulated miRNA processing59,60 ; however, it is unclear whether this occurs in AML.
Although mutations in the mature miRNA sequence would likely have no effect on expression levels, these mutations could change mRNA target specificity and dramatically alter phenotypic effects in AML. In perhaps the most thorough AML sequencing effort to date, The Cancer Genome Atlas group reported that miR-142-3p was the only miRNA bearing recurrent somatic single nucleotide variants in its mature strand that could alter binding to targets (4 of 187).13,61 Only 7 miRNA single nucleotide variant mutations were discovered in the 187 samples analyzed, indicating that these are rare events. However, while mutations in the miRNA sequence itself are uncommon, polymorphisms in the mRNA 3′ UTR miRNA binding site may happen more frequently and could predispose patients to AML by altering miRNA regulation of specific genes.62 Taken together, it seems that aberrant miRNA levels that are observed in AML are largely driven by altered transcription of miRNA primary transcripts, which suggests that targeting of key transcription factors or epigenetic regulators may be one way to restore proper miRNA expression in AML.
Translational aspects of miRNA biology
It is now well established that miRNAs play a variety of critical roles in AML, in which they can either promote or inhibit tumor cell biology. However, these advances have yet to make a clinical impact. Here we will highlight the efforts being made toward moving miRNA research in AML to the clinic and focus on the potential for using miRNAs as disease biomarkers, as well as advances in miRNA-targeting therapeutic strategies in AML.
miRNAs as AML biomarkers
Perhaps the most encouraging clinical application of miRNA research to date is the potential use of miRNAs as disease biomarkers in AML.63 When a patient initially presents with leukemia, proper classification is critical to determining the correct treatment. However, a small number of leukemias are difficult to identify as myeloid or lymphoid, thus making treatment decisions challenging. miRNA expression profiling can help classify acute leukemias of ambiguous lineage as either AML or acute lymphoblastic leukemia,64 with 1 group claiming that as few as 2 miRNAs can be used to discriminate between acute lymphoblastic leukemia and AML at an accuracy of >95%.65 As mentioned earlier, specific subtypes and mutant drivers of AML are associated with distinctive miRNA expression profiles, again suggesting that miRNAs could be useful in the initial classification of disease.
Beyond classification, miRNA expression profiles may provide important prognostic information. Several groups have reported that miRNA expression at diagnosis adds relevant prognostic information in patients with AML and can even predict survival in some cases.66-68 It was also recently reported that miRNA expression can predict progression of myelodysplastic syndrome (MDS) to AML.69
A major issue with patients receiving treatment for AML is the persistence of a small number of leukemic blasts in the bone marrow after intensive chemotherapy known as minimal residual disease (MRD), which can eventually give rise to leukemia relapse. Because the appearance of leukemic blasts in circulation often occurs late in the relapse process, a number of highly sensitive polymerase chain reaction– and flow cytometry–based methods for detection of blast nucleic acid or protein products have been developed for monitoring MRD,70 as recognizing MRD before patient relapse could allow for preemptive therapy.71 A number of groups have proposed screening circulating miRNAs as an inexpensive, noninvasive, and sensitive option to monitor for MRD, because the serum expression of miRNAs changes after standard chemotherapy,72 and patients with AML have a distinctive serum miRNA expression profile compared with healthy controls.73,74 These early results are promising, because a specific AML-associated miRNA serum profile could not only be used to track MRD after chemotherapy, but could also potentially provide an important screening tool for early detection of de novo AML in the clinic, as alterations in serum miRNA profiles may precede the entry of leukemic blasts into the periphery. However, there has been a lack of concordance between these individual studies, suggesting that more work is needed on larger AML cohorts with more rigorous study design to validate these initial findings.
Advances in miRNA-based therapeutics
As the list of miRNAs and their mRNA targets that are relevant in AML disease progression continues to grow, therapeutic manipulation of these miRNAs becomes more enticing. It is easy to imagine delivering locked nucleic acid (LNA) oligonucleotide inhibitors to target known oncomiRs in AML or delivery of synthetic miRNA mimics that act as tumor suppressors. These approaches have exciting therapeutic potential, because miRNAs are endogenous molecules that often repress multiple targets, either in the same pathway or by affecting a common biological process. Thus, resistance to miRNA-based therapies through target site mutation would be unlikely.
A good example of the effectiveness of an miRNA-based therapeutic in AML was recently demonstrated with targeted delivery of miR-29b via transferrin-conjugated lipid nanoparticles both in vitro and in mice engrafted with human AML cell lines.75 Delivery of miR-29b led to decreased leukemic cell growth and improved survival in the AML xenograft mouse model, which was attributed to miR-29b downregulating CDK6, SP1, FLT3, DNMTs, and KIT, either directly or indirectly. These target genes affect a variety of cellular processes in AML, and this study highlights the ability of 1 miRNA-based treatment to target many pathways simultaneously. Several studies involving the use of miR-based therapeutics have shown encouraging results in preclinical in vitro and animal models,22,33,75-77 the results of which are summarized in Table 2.
Preclinical studies of miRNA-based therapeutics
| Therapy . | Delivery method . | Targets . | Efficacy of delivery . | In vitro results . | In vivo results . | References . |
|---|---|---|---|---|---|---|
| miR-22 mimic | G7 poly(amidoamine) dendrimer nanoparticles | CRTC1, FLT3, MYCBP | N/A | N/A | ↑ Survival in MV4-11 xenotransplantation mouse models; ↑ Survival (40% cure rate) in primary mouse AML (MLL-AF9 and AE9a) transplant models | 22 |
| miR-29b mimic | Transferrin-conjugated anionic lipid-based nanoparticle | DNMT3A, DNMT3B, DNMT1, SP1, CDK6, FLT3, KIT | >100-fold ↑ in vitro | ↓ Cell proliferation and ↓ colony formation in Kasumi-1, OCI-AML3, and MV4-11 cells | ↑ Survival and ↓ splenomegaly in NSG mice xenografted with MV4-11 cells | 69 |
| 20-fold ↑ in vivo | ↓ Cell viability in primary AML samples; pretreatment ↑ efficacy of decitabine treatment | Pretreatment ↑ efficacy of decitabine treatment | ||||
| miR-21/miR-196b antagomiRs | Naked antagomiR delivered via implanted osmotic pumps | N/A | 60%-80% ↓ of miRNAs in peripheral WBCs in vivo | ↓ Colony formation in HOXA9, Nup98-HOXA9, or MLL-AF9–transduced mouse Lin– BM cells | Cured MLL-AF9 transplanted primary mouse AML; ↑ survival in combination with induction chemotherapy in MLL-AF9 xenotransplantation model | 71 |
| miR-126 antagomiR | Transferrin or CD45.2- conjugated anionic lipid-based nanoparticle | MMP7, CHD7, JAG1 | 80% ↓ in primary AML blasts in vitro; 50% ↓ in BM and spleen in vivo | ↓ Long-term colony formation, ↓ self-renewal capacity, and ↓ quiescence of LSCs | ↑ Survival in NSG mice engrafted with human AML primary blasts and MLL FLT3-ITD mouse model | 33 |
| miR-181a mimic | Transferrin-conjugated anionic lipid-based nanoparticle | KRAS, NRAS, MAPK1 | 2.6-fold ↑ in BM and 35-fold ↑ in spleen in vivo | ↓ Cell proliferation and ↓ colony forming in KG1a, MV4-11, OCI-AML3 cell lines; ↑ apoptosis in primary AML blasts | ↑ Survival and ↓ splenomegaly in NSG mice xenografted with MV4-11 cells | 70 |
| Therapy . | Delivery method . | Targets . | Efficacy of delivery . | In vitro results . | In vivo results . | References . |
|---|---|---|---|---|---|---|
| miR-22 mimic | G7 poly(amidoamine) dendrimer nanoparticles | CRTC1, FLT3, MYCBP | N/A | N/A | ↑ Survival in MV4-11 xenotransplantation mouse models; ↑ Survival (40% cure rate) in primary mouse AML (MLL-AF9 and AE9a) transplant models | 22 |
| miR-29b mimic | Transferrin-conjugated anionic lipid-based nanoparticle | DNMT3A, DNMT3B, DNMT1, SP1, CDK6, FLT3, KIT | >100-fold ↑ in vitro | ↓ Cell proliferation and ↓ colony formation in Kasumi-1, OCI-AML3, and MV4-11 cells | ↑ Survival and ↓ splenomegaly in NSG mice xenografted with MV4-11 cells | 69 |
| 20-fold ↑ in vivo | ↓ Cell viability in primary AML samples; pretreatment ↑ efficacy of decitabine treatment | Pretreatment ↑ efficacy of decitabine treatment | ||||
| miR-21/miR-196b antagomiRs | Naked antagomiR delivered via implanted osmotic pumps | N/A | 60%-80% ↓ of miRNAs in peripheral WBCs in vivo | ↓ Colony formation in HOXA9, Nup98-HOXA9, or MLL-AF9–transduced mouse Lin– BM cells | Cured MLL-AF9 transplanted primary mouse AML; ↑ survival in combination with induction chemotherapy in MLL-AF9 xenotransplantation model | 71 |
| miR-126 antagomiR | Transferrin or CD45.2- conjugated anionic lipid-based nanoparticle | MMP7, CHD7, JAG1 | 80% ↓ in primary AML blasts in vitro; 50% ↓ in BM and spleen in vivo | ↓ Long-term colony formation, ↓ self-renewal capacity, and ↓ quiescence of LSCs | ↑ Survival in NSG mice engrafted with human AML primary blasts and MLL FLT3-ITD mouse model | 33 |
| miR-181a mimic | Transferrin-conjugated anionic lipid-based nanoparticle | KRAS, NRAS, MAPK1 | 2.6-fold ↑ in BM and 35-fold ↑ in spleen in vivo | ↓ Cell proliferation and ↓ colony forming in KG1a, MV4-11, OCI-AML3 cell lines; ↑ apoptosis in primary AML blasts | ↑ Survival and ↓ splenomegaly in NSG mice xenografted with MV4-11 cells | 70 |
This table summarizes the results from selected pre-clinical miRNA-based therapies, including information such as method of delivery, miRNA targets (direct and indirect), efficacy of delivery, and in vivo/in vitro results.
BM, bone marrow; N/A, not applicable; WBC, white blood cell.
Another underexplored area of miRNA-based therapy is the possibility of repurposing existing drugs known to influence miRNA levels by targeting the pathways that regulate miRNA expression. MLN4924 (Pevonedistat), a drug known to reduce nuclear factor κB (NF-κB) activation that is currently being evaluated in clinical trials, was recently shown to decrease the levels of oncogenic miR-155 in FLT3-ITD+ AML cell lines, leading to decreased leukemic phenotypes both in vitro and in vivo.78 miRNA-based therapeutics may also be efficacious when used in combination with existing chemotherapeutics. Manipulation of miRNA expression levels can increase AML responsiveness to standard chemotherapeutic regimens.34,75,79-81
Although several studies have implicated miRNAs and their putative targets as being clinically actionable, the vast majority of these studies have yet to achieve clinical relevance, with the first therapies targeting miRNAs just entering clinical trials within the last few years.82,83 One miRNA-based therapy in clinical trials that shows promise is treatment of hepatitis C virus by miR-122, in which researchers found that patients treated with an LNA inhibitor of mature miR-122 have reductions in hepatitis C virus RNA levels in a dose-dependent manner.84 This study provides proof of principle that should encourage future endeavors of this kind in the cancer arena. Although the list of miRNA-based therapeutics entering clinical trials continues to grow, to the best of our knowledge, no miRNA-based therapies have made their way to clinical trials specifically for the treatment of AML.
A major barrier preventing the development of miRNA-based therapies is the lack of more efficient and specific delivery methods, because synthetic miRNAs or oligonucleotide inhibitors are degraded rapidly in circulation and have limited cellular uptake and specificity. To further complicate matters, delivery of drugs to the bone marrow is difficult, and higher doses are often required to elicit a therapeutic effect. This highlights the importance of developing novel targeting techniques for more effective delivery. Consequently, there are many new approaches being explored for improved delivery of miRNA-based therapies, including liposomes, nanoparticles, LNAs with increased stability, peptide-based inhibitors, and several other creative approaches,85 some of which are highlighted in Table 2. Efficient and specific delivery of miRNA mimics or antagonists to the proper cell types in vivo is a key step toward unlocking the therapeutic potential of manipulating miRNA function to combat AML.
Emerging concepts
Going forward, there remain several aspects of miRNA biology that need further investigation to fully grasp how miRNAs function within AML cells. This includes a continued effort to better answer certain questions that have been raised in the past and work in novel areas that have recently emerged. Several of these areas and how they relate to AML are described below.
Improper regulation of inflammatory pathways leads to AML
A newly appreciated mechanism by which miRNAs promote malignancy is through their impacts on classical inflammatory pathways. It has long been known that there is a strong link between inflammation and cancer; however, the mechanisms governing this association are still largely unclear. NF-κB is critical in initiating inflammatory responses, and dysregulation of this critical transcription factor is heavily integrated into cancer biology because of its role in promoting proliferation and survival.86
Several groups have shown that dysregulation of certain miRNAs can disrupt normal NF-κB signaling that results in cancerous transformation,87,88 including in myeloid malignancies.39 Because miR-146a is a negative regulator of NF-κB signaling, and overactivation of NF-κB is involved in malignant transformation, one could predict that loss of miR-146a might lead to the development of hematopoietic cancers. Indeed, a miR-146a deficiency has been shown to result in the development of both lymphoid and myeloid malignancies in an age-dependent manner.39,41 Chromosome 5q deletions, which are common in MDS progressing to AML, leads to loss of miR-145 and miR-146a because they are both encoded on the long arm of chromosome 5.37 The loss of these miRNAs leads to myeloproliferation and eventual progression to AML in mice as a result of increased NF-κB signaling,37,40 as miR-145 and miR-146a target TIRAP, IRAK1, and TRAF6, known activators of NF-κB. Targeted inhibition of IRAK1 has significant activity against MDS/AML cells in vitro and in xenograft mouse models,89 suggesting that targeting of these traditional innate immune pathways may have clinical efficacy.
Not only does miRNA regulation of NF-κB signaling seem to be important in AML progression, but there is also evidence that NF-κB activates miRNA expression to promote leukemic phenotypes.90 Additionally, there could be some contribution from the bone marrow microenvironment. Activation of inflammatory signaling in mesenchymal cells was recently found to drive development of an MDS preleukemic condition in mice.91 A further understanding of how alterations to these miRNA-regulated classical inflammatory pathways can promote AML progression will be an interesting new area of miRNA research in the future.
miRNAs play context-dependent roles in AML
An interesting aspect of miRNA biology in AML is that a miRNA can have opposing roles, depending on the disease context. For example, miR-9 was identified as being specifically upregulated in MLL-rearranged AML, in which it plays an oncogenic role in promoting leukemogenesis in the presence of MLL-AF9.16 However, other studies have found miR-9 to play a tumor suppressive role in AML, including in pediatric AML with t(8;21), in which miR-9 overexpression reduced leukemic growth and induced monocytic differentiation in human AML cells and in xenotransplantation mouse models,17 as well as in EVI1-induced AML, in which miR-9 is epigenetically silenced leading to decreased apoptosis and myelopoiesis.18
There are many other examples of miRNAs displaying context-dependent roles in AML. miR-155 seems to have no phenotypic effect in MLL-rearranged AML,44 but it is consistently found to play an oncogenic role in FLT3-ITD–driven AML pathogenesis.42,45 miR-126 plays different roles and regulates different targets in normal vs malignant hematopoietic stem cells35 and, interestingly, both overexpression and knockout of miR-126 promote leukemogenesis.34 These studies highlight how the influence of an miRNA in AML can be dependent on the underlying genetic abnormalities that drive disease or cell type of expression.
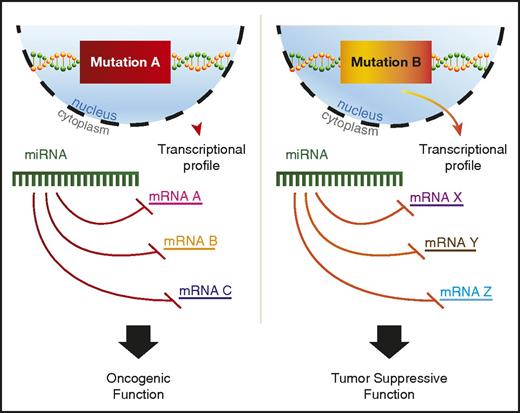
A variety of potential explanations for context-dependent discrepancies have been proposed, including RNA-binding protein regulation of miRNA binding to 3′ UTRs or differential splicing to include or exclude a given 3′ UTR.92 In addition, recent findings suggest that mutant proteins in AML can alter miRNA-mRNA interactions.93 Perhaps the most plausible explanation would be differences in the mRNA target availability for the miRNA, because AML driven by independent mutations would have distinct transcriptional profiles (Figure 2). However, most studies that find context-specific roles for miRNAs in AML do not go on to explore the mechanistic basis underlying this phenomenon, and more work in this area is needed to better understand context-dependent miRNA functions.
miRNAs play context-dependent roles in AML. A model for context-dependent effects of a specific miRNA given different transcriptional backgrounds between 2 distinct AML-driver mutations, mutation A and mutation B. Mutation A leads to the transcription of mRNA A, B, and C, whereas mutation B drives the transcription of mRNA X, Y, and Z. All mRNAs have predicted targeting by the example miRNA. The miRNA depicted in mutation A and mutation B is the same hypothetical miRNA.
miRNAs play context-dependent roles in AML. A model for context-dependent effects of a specific miRNA given different transcriptional backgrounds between 2 distinct AML-driver mutations, mutation A and mutation B. Mutation A leads to the transcription of mRNA A, B, and C, whereas mutation B drives the transcription of mRNA X, Y, and Z. All mRNAs have predicted targeting by the example miRNA. The miRNA depicted in mutation A and mutation B is the same hypothetical miRNA.
Varying levels of miRNA expression may have opposing effects
One potential explanation for the context-dependent effects observed for miRNA dysregulation in AML is that different levels of miRNA expression may have vastly different effects on host-cell phenotypes. miR-125b is one of the more interesting known oncomiRs because it plays a role in promoting both myeloid and lymphoid malignancies. Overexpression in mice has been shown to cause both lymphoproliferative and myeloproliferative disorders and, ultimately, a frank malignancy in these compartments.29,30,94,95 Recently, some light was shed on the dual nature of miR-125b in promoting hematologic malignancy in which miR-125b was found to selectively induce either myeloid or lymphoid leukemia based on the level and time course of miR-125b overexpression.31
miR-155 is another well-studied oncogenic miRNA in AML, and overexpression correlates with a poor prognosis.43 However, evidence has emerged that miR-155 may play a role as a tumor suppressor in certain contexts.96 A recent study examined the role of miR-155 in AML more closely to help resolve these opposing effects. The study found that when overexpressing miR-155 in 3 different murine models of AML (HoxA9/Meis1, MLL-ENL, MLL-AF9) to an intermediate level (∼5- to 10-fold above control), miR-155 displayed oncogenic function, leading to increased proliferation and enhanced colony-forming potential.97 This was in contrast to miR-155 high levels (>10-fold above control), in which miR-155 acted as a tumor suppressor by repressing colony formation and proliferation, establishing a dose-dependent effect of miR-155 in these AML mouse models. The study did confirm that the intermediate miR-155 expression levels were a better representation of what was seen in their pediatric AML data set (increased ∼one- to sevenfold), values that were consistent with miR-155 expression in other AML data sets,12,45 suggesting that miR-155 likely has a predominately oncogenic effect in human AML. The study highlights the importance of considering the level of expression in various model systems when studying miRNAs in AML.
5p vs 3p transcripts
A long-standing question regarding miRNAs is the significance of differential use of 5p vs 3p miRNA transcripts. 5p and 3p miRNAs are encoded by the same genomic region and are both contained within the initial transcript and mature miRNA duplex before 1 of them is chosen as the active or guide strand (miR) and the other as the passenger strand (miR*). The passenger strand is then typically degraded and traditionally thought not to have a functional role. Interestingly, the 2 miRNA strands each have unique seed sequences and therefore do not share the same mRNA target spectrum. This means that the biological processes and pathways being regulated by any given pri-miRNA transcript could be vastly different depending on the selection of either the 5p or 3p strand as the guide. There are several examples of 5p and 3p transcripts from the same duplex having distinct biological functions.98,99
In a small percentage of cases, the passenger strand can be stabilized at the same level as the active strand, and may even exhibit important physiological function in myeloid cells.100 A recent report identified a specific passenger strand, miR-9*, that is not detectable in normal myeloid cells but is expressed in 59% of AML cases, and expression levels correlated with prognosis.101 This group also found that miR-9* expression levels had prognostic value, because patients with high miR-9* expression vs low expression were associated with positive patient outcome. Although the mechanisms behind passenger strand–retained expression are still largely unknown, there is some evidence that posttranscriptional modifications to the RNA duplex and differential expression of various RISC components could play an important role in strand selection.102,103 Whether these processes are dysregulated in AML remains to be determined.
A variety of miRNA sources and noncanonical targeting
Although the traditional dogma of miRNA biology states that miRNAs are encoded from their own genes, go through a distinct processing pathway, and then repress mRNA targets via binding the 3′ UTR, there is mounting evidence that this canonical biogenesis pathway might not be exclusive. It is now understood that the mature miRNA can come from a variety of sources, including from the 5p or 3p transcript, long noncoding RNAs (lncRNAs),104 small nucleolar RNAs,105,106 or spliced from introns107 (Figure 3). Identifying and characterizing miRNAs generated from these nontraditional sources may be challenging, but it is key to comprehensively understanding the breadth of small RNAs in AML.
Alternate miRNA sources and noncanonical targeting. A schematic depicting the varying sources identified for production of mature miRNAs, including the 5′ or 3′ strand of the traditional miRNA hairpin structure, processing of other noncoding RNAs, such as lncRNAs, small nucleolar RNAs (snoRNAs), miRNAs transcribed from the same gene but having different mature miRNA sequences (isomiRs), and miRNA spliced from introns (miRtrons). Nontraditional miRNA targets beyond the 3′ UTR are also depicted, including promoter regions of protein-coding genes, the 5′ UTR, the RNA coding sequence, and protein.
Alternate miRNA sources and noncanonical targeting. A schematic depicting the varying sources identified for production of mature miRNAs, including the 5′ or 3′ strand of the traditional miRNA hairpin structure, processing of other noncoding RNAs, such as lncRNAs, small nucleolar RNAs (snoRNAs), miRNAs transcribed from the same gene but having different mature miRNA sequences (isomiRs), and miRNA spliced from introns (miRtrons). Nontraditional miRNA targets beyond the 3′ UTR are also depicted, including promoter regions of protein-coding genes, the 5′ UTR, the RNA coding sequence, and protein.
Functional effects exhibited by miRNAs are often attributed to a handful of targets predicted by seed sequence complementarity in the 3′ UTR. But miRNAs can also bind and repress targets without predicted binding sites in their 3′ UTRs, referred to as noncanonical targets. Researchers found that when they pulled down the RISC complex to identify mRNA targets loaded in an miR-155–specific manner, ∼40% of the targets identified were noncanonical targets.108 This was explained, in part, by laxity in the seed matching of miRNAs and mRNA 3′UTRs but could also be explained by the concept of isomiRs, which are miRNAs transcribed from the same gene but having different mature miRNA sequences, found extensively in a murine model of leukemia.109 These variants are a result of posttranscriptional modifications, including “errors” in miRNA processing, nucleotide addition to the 3′ end, and nucleotide substitution.110,111 Beyond isomiRs, there is also some evidence that miRNAs can bind to promoter regions of DNA,112 5′ UTRs,113 the mRNA coding sequence,114,115 and even proteins.116 Thus far, the evidence for noncanonical targeting playing a functional role in AML is lacking, but it could be a more prominent mode of miRNA function than we realize.
Transfer of miRNAs in exosomes alters leukemic phenotypes
Recently, miRNAs have been found within extracellular vesicles, including exosomes that are produced by the multivesicular body pathway.117 Both primary and malignant cells can release miRNAs in exosomes, which can be taken up by certain recipient cells where they deliver their miRNA cargo in a functionally relevant manner.118,119 Although evidence for the functional role of exosomally transferred miRNAs in AML is somewhat limited, preliminary studies indicate this could be a paradigm-shifting field of study.
Early work showed that both primary AML cells and AML cell lines do in fact release exosomes containing miRNAs.120 Moreover, these exosomes contain an miRNA population that is compositionally distinct from the miRNA population of the host cell,120 suggesting that there is specificity with the loading of miRNAs into exosomes. Other functional studies have revealed that miRNAs can be transferred in exosomes from AML cells to both stromal and normal hematopoietic cells and alter their function in a manner that promotes leukemic phenotypes.121,122 Interestingly, exosomes from extramedullary tumors have been shown to alter the bone marrow niche, suggesting that miRNA-containing exosomes can home to the bone marrow and alter function independent of cell contact.121 In a recent study,122 authors found that exosomes containing miR-150 and miR-155 released from AML cells suppressed normal hematopoietic stem cell proliferation and differentiation through inhibition of c-MYB, thus perpetuating a malignant phenotype by directly altering hematopoietic stem cell biology.
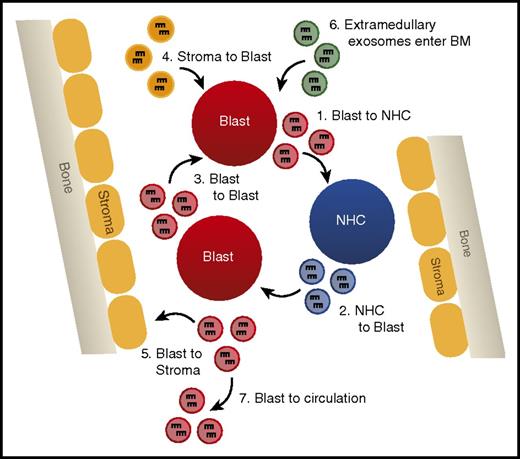
Such studies provide evidence that miRNAs secreted in exosomes are a novel form of intercellular communication that may play vital regulatory roles in suppressing normal hematopoiesis and disrupting the hematopoietic niche to promote leukemic cell outgrowth (Figure 4). Analysis of the miRNA content of exosomes has even been suggested as a novel biomarker for the detection of AML, because blast-derived exosomes can be isolated from circulation before the appearance of circulating blast cells in a xenograft mouse model.74 Further study of the machinery required for specific miRNA loading into exosomes and uptake by recipient cells is critical to manipulating this system in a manner that will better test the relevance of exosomal miRNAs in AML.
Exosomally transferred miRNAs alter leukemic phenotypes. A model depicting the different possibilities of exosomal miRNA transfer in the bone marrow, including (1) transfer from AML blast to normal hematopoietic cells (NHCs), (2) NHC to AML blast, (3) AML blast to AML blast, (4) bone marrow (BM) stromal cells to AML blast, (5) AML blast to BM stromal cells, (6) entrance of extramedullary produced exosomes into the hematopoietic niche, and (7) AML blast-derived exosomes leaving the BM and entering circulation.
Exosomally transferred miRNAs alter leukemic phenotypes. A model depicting the different possibilities of exosomal miRNA transfer in the bone marrow, including (1) transfer from AML blast to normal hematopoietic cells (NHCs), (2) NHC to AML blast, (3) AML blast to AML blast, (4) bone marrow (BM) stromal cells to AML blast, (5) AML blast to BM stromal cells, (6) entrance of extramedullary produced exosomes into the hematopoietic niche, and (7) AML blast-derived exosomes leaving the BM and entering circulation.
lncRNAs can interfere with miRNA function in AML
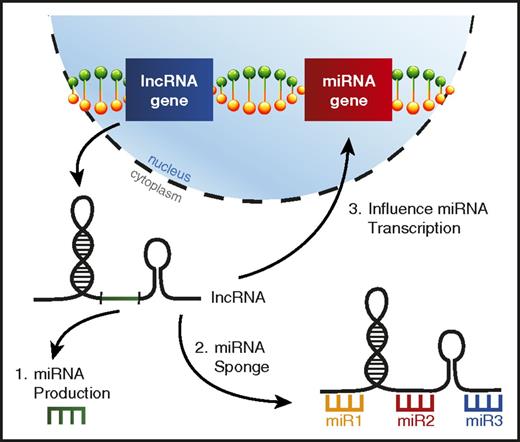
lncRNAs are a distinct class of noncoding RNAs much longer than mature miRNAs (>200 nucleotides) that have been observed to exhibit a variety of functions, but are typically involved in regulating gene expression,123 including miRNA genes.124 It has recently been learned that lncRNA dysregulation in AML can alter the function of specific miRNAs leading to skewed disease phenotypes (Figure 5).
lncRNAs can interfere with miRNA function in AML. A picture depicting the ways lncRNAs can affect miRNA biology in AML, including (1) lncRNAs serving as a source for mature miRNA production, (2) lncRNAs acting as miRNA sponges, binding miRNAs to prevent them from repressing their target mRNAs, and (3) altering miRNA gene transcription.
lncRNAs can interfere with miRNA function in AML. A picture depicting the ways lncRNAs can affect miRNA biology in AML, including (1) lncRNAs serving as a source for mature miRNA production, (2) lncRNAs acting as miRNA sponges, binding miRNAs to prevent them from repressing their target mRNAs, and (3) altering miRNA gene transcription.
HOTAIRM1 is a lncRNA located in the HOXA genomic region that was recently found to impact prognosis in various subtypes of AML and was associated with a distinct miRNA expression profile.125 Additional work on lncRNA HOTAIRM1 revealed that it was acting to sequester autophagy-regulating miRNAs miR-20a, miR-106b, and miR-125b, which affected the degradation of PML-RARA in acute promyelocytic leukemia.126 Another lncRNA, HOTAIR1, was also found to act as a miRNA sponge in AML, competitively binding miR-193a, which leads to increased c-KIT expression and ultimately confers a poor prognosis.127 Other examples of lncRNAs acting to competitively bind miRNAs in myeloid malignancy have started to emerge.128,129
As mentioned above, lncRNAs can provide a nontraditional source for mature miRNA production. It was recently learned that the pri-miR-223 transcript is actually a functional lncRNA in AML that displays tumor suppressive functions by sponging oncogenic miRNAs miR-125a and miR-125b.104 Interestingly, while miR-223 is processed out of this lncRNA, miR-223 and lncRNA-223 are expressed at different levels, and these 2 noncoding RNAs have distinct functions in the myeloid lineage. It is unclear how prevalent this phenomenon is in AML, but there is additional evidence that lncRNAs can serve as precursors for miRNAs in T-cell lymphomas.130,131 A better understanding of the ways in which lncRNAs regulate miRNAs in AML could shed more light on their biology in this setting.
Conclusion
miRNAs are now widely regarded as playing a critical role in AML pathogenesis. Specific miRNA expression profiles can help classify subtype, determine prognosis, and predict response to treatment in AML, but the use of miRNAs as biomarkers is not yet routine practice. Therapies targeting miRNAs in AML have shown promise in preclinical models but have not made the leap to human clinical trials, which will require improvements in our delivery methods.
The continued development of advanced genomic approaches, including CRISPR-Cas9 technology, will allow us to more quickly identify and efficiently study relevant miRNAs and their targets in AML. Indeed, genome-wide CRISPR-Cas9 screening has been used to identify functionally relevant miRNA-mRNA target pairs that regulate AML cell line growth132 and will likely be extended to additional preclinical models of AML.
Many complexities and mysteries of miRNA biology remain, but their solutions will substantially improve our understanding of how miRNAs function in AML. In some cases, novel aspects of miRNA biology have already shed additional light on how AML cells are regulated, whereas in other cases, emerging mechanisms have yet to be explored in AML despite their potential to help us understand how miRNAs influence this deadly leukemia. Addressing long-standing questions and exploring emerging concepts of miRNA biology will provide important insights into how miRNAs function in AML, and only through an improved understanding of these mechanisms can we better exploit miRNAs therapeutically to improve disease outcomes in the clinic.
Acknowledgments
The authors acknowledge Rachel Merrill for assistance with figure preparation.
This work was supported by the National Institutes of Health (NIH) under the Ruth L. Kirschstein National Research Service Award NIH 5T32DK007115 from the National Institute of Diabetes and Digestive and Kidney Diseases (J.A.W.); the NIH National Cancer Institute under Award Number F30CA217027 (J.A.W.); the NIH New Innovator Award DP2GM111099-01 from the National Institute of General Medical Sciences (R.M.O.); Gabrielle's Angel Foundation Award (R.M.O.); and the Leukemia & Lymphoma Society Scholar Award (R.M.O.).
Authorship
Contribution: J.A.W. and R.M.O. reviewed the literature and wrote the review.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ryan M. O’Connell, Department of Pathology, University of Utah, 15 North Medical Dr East, JMRB 4280B, Salt Lake City, UT 84112; e-mail: ryan.oconnell@path.utah.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal