Key Points
Increased FOXO1 is oncogenic in human CD34+ cells and promotes preleukemia transition.
FOXO1 is required by AE preleukemia cells for the activation of a stem cell molecular program.
Abstract
Understanding and blocking the self-renewal pathway of preleukemia stem cells could prevent acute myeloid leukemia (AML) relapse. In this study, we show that increased FOXO1 represents a critical mechanism driving aberrant self-renewal in preleukemic cells expressing the t(8;21)-associated oncogene AML1-ETO (AE). Although generally considered as a tumor suppressor, FOXO1 is consistently upregulated in t(8;21) AML. Expression of FOXO1 in human CD34+ cells promotes a preleukemic state with enhanced self-renewal and dysregulated differentiation. The DNA binding domain of FOXO1 is essential for these functions. FOXO1 activates a stem cell molecular signature that is also present in AE preleukemia cells and preserved in t(8;21) patient samples. Genome-wide binding studies show that AE and FOXO1 share the majority of their binding sites, whereby FOXO1 binds to multiple crucial self-renewal genes and is required for their activation. In agreement with this observation, genetic and pharmacological ablation of FOXO1 inhibited the long-term proliferation and clonogenicity of AE cells and t(8;21) AML cell lines. Targeting of FOXO1 therefore provides a potential therapeutic strategy for elimination of stem cells at both preleukemic and leukemic stages.
Introduction
Acute myeloid leukemia (AML) is an aggressive hematopoietic malignancy. Leukemogenesis is a hierarchical process, whereby an initiating mutation drives the development of preleukemic stem cells that evolve over time to overt disease through additional cooperating mutations.1 Preleukemic stem cells can survive chemotherapy and serve as a potential reservoir of disease relapse.2,3 The translocation at chromosome t(8;21) generating the AML1-ETO (RUNX1-RUNX1T1; AE) fusion protein is one of the most frequent initiating mutations, accounting for ∼10% of total AML.4 Although t(8;21) AML has a comparatively good prognosis and most patients enter remission, approximately half of the patients relapse, and only a 60% overall survival is achieved after 5 years.5 Preleukemic stem cells are evident in this AML subtype, as cells positive for AE can be detected long before disease onset or after complete remission.6-8 Thus, disrupting the self-renewal program of malignant stem cells will provide therapeutic opportunities for preventing disease relapse.
We and others have shown that expression of AE in human CD34+ hematopoietic stem and progenitor cells (HSPCs) causes dysregulated differentiation and increased self-renewal of cells but without inducing AML,9,10 serving as an ideal model to study the preleukemic stage of t(8;21) AML (AE cells). Until now, the mechanism by which AE programs progenitor cells into preleukemia with aberrant self-renewal has not been completely understood.
The FOXO transcription factors include FOXO1, FOXO3, FOXO4, and FOXO6. Except for FOXO6, which is expressed predominantly in the brain, all other FOXOs are ubiquitously expressed and act as essential regulators controlling oxidative stress and metabolic homeostasis,11 which is critical in hematopoietic stem cell (HSC) maintenance.12-14 Because of their capacity to arrest the cell cycle and activate apoptosis, FOXOs are well-known tumor suppressors.15 However, the precise function of FOXOs in AML is unclear. The inactivation of FOXO3 in AML was reported, and restoring its activity impaired cell growth.16 On the other hand, in line with its critical role in HSC maintenance,17 FOXO3 was found to be important for maintaining AML stem cells, although the molecular mechanism was not defined.18 It is therefore unclear whether FOXOs serve as tumor suppressors, as oncogenes or as stem cell maintenance genes.
Different FOXO family members may have nonredundant functions.19 For instance, an HSC defect has been observed only in Foxo3- but not Foxo1-deficient mice.12 However, the role of FOXO1 in preleukemia stem cells has not been explored. Here, we show FOXO1 can function as an oncogene whose upregulation promotes self-renewal and blocks the differentiation of human CD34+ HSPCs. In AE preleukemic cells, this oncogenic function is required for the activation of a self-renewal program. Our results demonstrate a new function of FOXO1 in AML pathogenesis, strengthen our understanding of the mechanisms that mediate the aberrant self-renewal of preleukemia stem cells, and reveal therapeutic strategies for their elimination.
Methods
Plasmids and reagents
MSCV-IRES-GFP/THY.1 (MIG/MIT) and MSCV-AE-IRES-GFP/THY.1 retroviral vectors were as described.10,20 pSIN-TREtight-DsRED-IRES and MSCV-GFP-IRES-tTA retroviral vectors were generously provided by Johannes Zuber. Insertion of AE was subcloned into pSIN-TREtight-DsRED-IRES to build an AE-tet-off system. pSG5L-FOXO1 wild-type (WT) and pBabe-puro-FOXO1-DB (DNA binding domain mutant) vectors were obtained from Addgene (#10693 and #10695). Insertions of FOXO1 WT and FOXO1 DB were then subcloned into MIG. Lentiviral vector MISSION pLKO.1- short hairpin RNA (shRNA)–puro constructs targeting human FOXO1 (TRCN0000039578, sh-1) and nontarget shRNA control (shNT-1) were obtained from Sigma; the puromycin-resistant gene in the constructs was replaced with Venus marker. GIPZ lentiviral shRNA targeting FOXO1 (V3LHS_405827, sh-2) and nonsilencing control (shNT-2) were obtained from Open Biosystems; the shRNA insertions were subcloned into SF-LV-shRNA-EGFP lentiviral vector, a gift from K. Lenhard Rudolph. FOXO1 inhibitor AS1842856 was from Millipore (344355), dissolved in dimethyl sulfoxide (DMSO).
Cells and culture
Human umbilical cord blood cells were obtained by the Translational Trials Support Laboratory at Cincinnati Children's Hospital Medical Center (CCHMC) under institution-approved protocol. CD34+ cells were enriched using CD34+ selection kit (Miltenyi). AE preleukemic cells were established as described.10 AE cells and MIG/FOXO1-transduced CD34+ cells were cultured in Iscove modified Dulbecco medium with 20% BIT Serum Substitute (Stemcell Technologies #9500), supplemented with 10 ng/mL stem cell factor (SCF), thrombopoietin, FLT3 ligand, interleukin-3 (IL-3), and IL-6. Kasumi-1 and SKNO-1 cells were obtained from Deutsche Sammlung von Mikroorganismen und Zelljulturen, and cultured in RPMI 1640 with 20% fetal bovine serum or RPMI 1640 with 10% fetal bovine serum plus 10 ng/mL granulocyte-macrophage colony-stimulating factor, respectively.
Methylcellulose colony-forming assays
Assays were performed in MethoCult H4100 medium (Stemcell Technologies) supplemented with 20% BIT, 50 μM β-mercaptoethanol, 2 mM l-glutamine, 100 U/mL penicillin/streptomycin, and the human cytokines erythropoietin (6 U/mL), granulocyte colony-stimulating factor (10 ng/mL), IL-6 (20 ng/mL), IL-3 (20 ng/mL), and SCF (20 ng/mL). Colonies were scored at day 14, and then the cells were collected for replating. Some experiments were performed by using MethoCult Express medium (Stemcell Technologies); colonies were scored at day 7 to 10.
Xenograft transplantation
Transplantation was performed on NOD/SCID/IL2RG−/− immunodeficient mice with transgenic expression of human SCF, granulocyte-macrophage colony-stimulating factor, and IL-3 (NSGS). Six- to 8-week-old mice were conditioned with 30 mg/kg busulfan (Sigma) through intraperitoneal injection 24 h before transplantation. For shRNA knockdown experiment, 5 × 105 transduced cells were transplanted to each mouse via tail vein. For the FOXO1 overexpression experiment, 1 × 105 cells were transplanted through intrafemoral injection immediately after transduction. Bone marrow was aspirated for flow cytometry at the indicated time. All experiments were performed in accordance with CCHMC institutional guidelines.
RNA sequencing (RNA-seq)
Three different units of CD34+ cord blood cells were transduced with MIG, AE, FOXO1 WT, and FOXO1 DB. RNA was collected from sorted GFP+CD11b− cells 5 days after transduction using RNeasy Mini Kit (Qiagen). The integrity of RNA was analyzed by Bioanalyzer (Agilent). One microgram total RNA was used for poly(A) RNA selection, followed by complementary DNA synthesis using PrepX messenger RNA (mRNA) Library kit (WaferGen) and Apollo 324 NGS automatic library prep system. Sample-specific index was added to the adaptor-ligated complementary DNA by polymerase chain reaction (PCR) with index-specific primers for 13 cycles. The cluster generation of indexed libraries was carried out on cBot system (Illumina) using Illumina’s TruSeq SR Cluster kit v3, and then sequenced on Illumina HiSeq system using TruSeq SBS kit to generate single-end 50 cycle reads. Reads (20-50 million) were generated for each sample. More details about the data analyses can be found in supplemental Methods (available on the Blood Web site).
Chromatin immunoprecipitation
Chromatin immunoprecipitation–sequencing (ChIP-seq) assays in AE cells were performed as previously described.21 More details about the protocol and the data analyses can be found in supplemental Methods.
Data access
RNA-seq and ChIP-seq data have been deposited in Gene Expression Omnibus (accession numbers GSE81084 and GSE80773, respectively).
Results
FOXO1 is upregulated in AE leukemia and preleukemia cells
To identify critical regulators of self-renewal in AE AML, we examined an AML patient data set published by Ross et al22 for stem cell–related genes that are significantly upregulated in t(8;21) AML cells compared with other AML subtypes. FOXO1 was one such gene (Figure 1A; supplemental Figure 1A), a result that was further validated in additional data sets (supplemental Figure 1B). Expression of other FOXO family members did not differ significantly between t(8;21) and other AML subtypes (supplemental Figure 1C). We confirmed the upregulation of FOXO1 protein in t(8;21) AML primary patient samples compared with cytogenetically normal AML samples (Figure 1B). Increased FOXO1 transcript and protein levels were also evident in AE preleukemia cells compared with control vector (MIT)–transduced CD34+ HSPCs (Figures 1C-D), at levels similar to those in t(8;21) cell lines (Kasumi-1 and SKNO-1) and primary patient samples (supplemental Figure 1D-E). In contrast, AE did not elicit upregulation of other FOXOs in the same cells (supplemental Figure 1F). The transcriptional activity of FOXOs is dependent on their nuclear localization, as they can be inactivated in AML cells by nuclear export.16 Cellular fractionation and immunoblot analysis showed that FOXO1 had a primarily nuclear distribution in AE cells and Kasumi-1 cells (Figure 1E; supplemental Figure 1G). Immunostaining confirmed this finding, suggesting that FOXO1 is active in AE preleukemic and leukemic cells (Figure 1F).
FOXO1 is upregulated in AE leukemia and preleukemia cells. (A) Microarray analysis of FOXO1 transcript levels in t(8;21)-positive compared with t(8;21)-negative AML patient samples from Ross et al.22 (B) Immunoblot showing FOXO1 protein levels in t(8;21) and cytogenetically normal [non-t(8;21)] AML patient samples. FOXO1 mRNA levels determined by quantitative PCR (qPCR) (C; n = 4, error bars represent standard deviation [SD]) and protein levels (D) in AE- or empty vector (MIT)–transduced CD34+ HSPCs. Cellular fractionation immunoblot (E) and immunofluorescence analysis (F) to determine the subcellular localization of FOXO1 in AE cells. The images were obtained by Leica DMI6000 fluorescent microscope with ×40 objective. Bar represents 20 μm. All P values were calculated by unpaired 2-tailed Student t test.
FOXO1 is upregulated in AE leukemia and preleukemia cells. (A) Microarray analysis of FOXO1 transcript levels in t(8;21)-positive compared with t(8;21)-negative AML patient samples from Ross et al.22 (B) Immunoblot showing FOXO1 protein levels in t(8;21) and cytogenetically normal [non-t(8;21)] AML patient samples. FOXO1 mRNA levels determined by quantitative PCR (qPCR) (C; n = 4, error bars represent standard deviation [SD]) and protein levels (D) in AE- or empty vector (MIT)–transduced CD34+ HSPCs. Cellular fractionation immunoblot (E) and immunofluorescence analysis (F) to determine the subcellular localization of FOXO1 in AE cells. The images were obtained by Leica DMI6000 fluorescent microscope with ×40 objective. Bar represents 20 μm. All P values were calculated by unpaired 2-tailed Student t test.
To test whether FOXO1 is a downstream target of AE we developed a conditional (tet-off) AE expression system, and found that the expression of FOXO1 was highly dependent on continued expression of AE (Figure 2A-B). Similarly, when analyzing our previous RNA-seq study,21,23 we found that depletion of AE in Kasumi-1 cells led to a decrease of FOXO1 transcripts (Figure 2C-D). ChIP-seq analysis in Kasumi-1 cells21,23 showed that AE bound to the FOXO1 gene locus, where multiple RUNX1 binding motifs were identified (Figure 2E). We validated this binding using the anti-HA antibody to pull down the HA-tagged AE protein in AE cells (Figure 2F). These results suggest that FOXO1 could be a direct transcriptional target of AE.
Upregulation of FOXO1 depends on the presence of AE. qPCR and immunoblot analysis to determine FOXO1 mRNA (A; n = 3, error bars represent SD) and protein (B) in AE-tet-off system following doxycycline (DOX) addition. (C and D) RNA-seq analysis of FOXO1 expression of Kasumi-1 cells transected with control siRNA (siMM) or AE siRNA (siAE). RNA-seq tracks (C) and relative fragments per kilobase of transcript per million mapped reads (FPKM) values (D) are shown. n = 3; error bars represent SD. P value was calculated by unpaired 2-tailed Student t test. (E) ChIP-seq analysis showed AE binding patterns at FOXO1 locus in Kasumi-1 cells. The sequence of the binding peak is shown with RUNX1 binding motifs highlighted. (F) ChIP-qPCR analysis of chromatin occupancy of AE (HA-tagged) at FOXO1 locus in AE cells, with primers indicated in panel E by arrow. RPL30 locus was used as negative control. n = 3; error bars represent SD. IgG, immunoglobulin G.
Upregulation of FOXO1 depends on the presence of AE. qPCR and immunoblot analysis to determine FOXO1 mRNA (A; n = 3, error bars represent SD) and protein (B) in AE-tet-off system following doxycycline (DOX) addition. (C and D) RNA-seq analysis of FOXO1 expression of Kasumi-1 cells transected with control siRNA (siMM) or AE siRNA (siAE). RNA-seq tracks (C) and relative fragments per kilobase of transcript per million mapped reads (FPKM) values (D) are shown. n = 3; error bars represent SD. P value was calculated by unpaired 2-tailed Student t test. (E) ChIP-seq analysis showed AE binding patterns at FOXO1 locus in Kasumi-1 cells. The sequence of the binding peak is shown with RUNX1 binding motifs highlighted. (F) ChIP-qPCR analysis of chromatin occupancy of AE (HA-tagged) at FOXO1 locus in AE cells, with primers indicated in panel E by arrow. RPL30 locus was used as negative control. n = 3; error bars represent SD. IgG, immunoglobulin G.
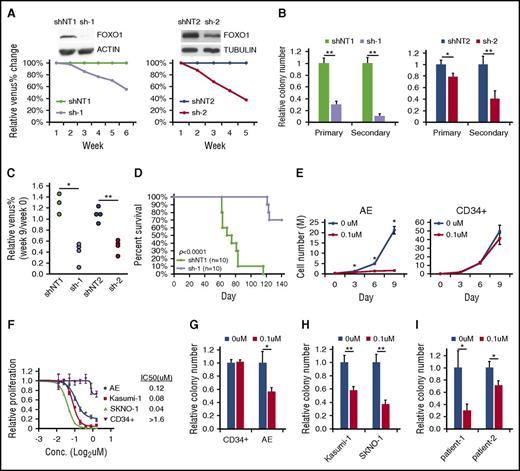
FOXO1 is required to sustain the growth of AE cells
To investigate the function of FOXO1 in AE preleukemia cells, we knocked down FOXO1 using 2 independent pairs of shRNAs. AE cells with FOXO1 depletion displayed slightly increased apoptosis and reduced cell cycle progression compared with nontargeting shRNA (shNT)–transduced cells (supplemental Figure 2A-B), although the differentiation status of the cells was not altered as indicated by CD11b expression (data not shown). FOXO1 knockdown impaired growth in liquid culture and clonogenicity of AE cells in a methylcellulose assay (Figure 3A-B). Additionally, FOXO1-depleted AE cells showed a reduced long-term engraftment capacity in immunodeficient mice, as the Venus+ fraction in engrafted human cells was reduced at week 9 posttransplantation and continuously decreased at later time points (Figure 3C; supplemental Figure 2C). These results suggest that FOXO1 does not act as a tumor suppressor; instead, FOXO1 ablation impairs the self-renewal of AE preleukemia cells both in vitro and in vivo. In addition, depletion of FOXO1 inhibited cell growth and colony formation of t(8;21) AML cell lines (supplemental Figure 2D-E), indicating that FOXO1 is also critical for leukemia maintenance. To further test this notion, we transduced shRNAs into an AE overt leukemic clone that spontaneously evolved from preleukemia cells24 and evaluated its leukemogenicity in immunodeficient mice. The majority of mice receiving shNT-transduced cells died of disease between 60 and 80 days after transplantation as expected. In contrast, recipients of FOXO1-depleted AE leukemia cells survived much longer (Figure 3D). Bone marrow examination showed that the cells with FOXO1 knockdown displayed a much lower engraftment level compared with shNT-transduced cells at a time point when the latter were already showing overt disease (supplemental Figure 2F).
FOXO1 inhibition impairs growth of AE cells. (A) AE cells were transduced with 2 sets of shNT and FOXO1 shRNA (sh-1 and sh-2) vectors that coexpressed Venus. Knockdown efficiency was confirmed by immunoblot. Growth of AE cells in liquid culture was measured as change of percentage of Venus+ cells relative to shNT by flow cytometry. One representative experiment of 3 replicates is shown. (B) Colony-forming unit (CFU) assay of sorted shRNA-transduced AE cells. n = 3; results represent mean ± SD. *P < .05;**P < .01. (C) shRNA-transduced AE cells were transplanted into immunodeficient mice. Venus+ percentage of human engrafted cells in bone marrow was examined 9 weeks later and normalized to those before the transplantation. *P < .005; **P < .001. One representative experiment of 2 replicates is shown. (D) Survival curve of immunodeficient mice received fully transformed AE cells expressing shNT or FOXO1 shRNA. Two independent experiments were included; P value was calculated by log-rank test. (E) CD34+ HSPCs and AE cells were treated with the FOXO1 inhibitor AS1842856 or DMSO; cell numbers were monitored over time. n = 3; results represent mean ± SD. *P < .01, paired 2-tailed Student t test. (F) Proliferation rate of cells treated with FOXO1 inhibitor at indicated concentration for 6 days, which is determined by WST-1 assay. Data are normalized to DMSO control (0 μM) and are shown as mean ± SD (n = 3). The 50% inhibitory concentration values for each type of cells are indicated. CFU assay of CD34+ HSPCs, AE cells (G), t(8;21) cell lines (H), and primary patient cells (I) treated with FOXO1 inhibitor. Data are normalized to DMSO-treated cells (0 μM). n = 3; results represent mean ± SD. *P < .05; **P < .005. All P values were calculated by unpaired 2-tailed Student t test unless noted. Conc., concentration.
FOXO1 inhibition impairs growth of AE cells. (A) AE cells were transduced with 2 sets of shNT and FOXO1 shRNA (sh-1 and sh-2) vectors that coexpressed Venus. Knockdown efficiency was confirmed by immunoblot. Growth of AE cells in liquid culture was measured as change of percentage of Venus+ cells relative to shNT by flow cytometry. One representative experiment of 3 replicates is shown. (B) Colony-forming unit (CFU) assay of sorted shRNA-transduced AE cells. n = 3; results represent mean ± SD. *P < .05;**P < .01. (C) shRNA-transduced AE cells were transplanted into immunodeficient mice. Venus+ percentage of human engrafted cells in bone marrow was examined 9 weeks later and normalized to those before the transplantation. *P < .005; **P < .001. One representative experiment of 2 replicates is shown. (D) Survival curve of immunodeficient mice received fully transformed AE cells expressing shNT or FOXO1 shRNA. Two independent experiments were included; P value was calculated by log-rank test. (E) CD34+ HSPCs and AE cells were treated with the FOXO1 inhibitor AS1842856 or DMSO; cell numbers were monitored over time. n = 3; results represent mean ± SD. *P < .01, paired 2-tailed Student t test. (F) Proliferation rate of cells treated with FOXO1 inhibitor at indicated concentration for 6 days, which is determined by WST-1 assay. Data are normalized to DMSO control (0 μM) and are shown as mean ± SD (n = 3). The 50% inhibitory concentration values for each type of cells are indicated. CFU assay of CD34+ HSPCs, AE cells (G), t(8;21) cell lines (H), and primary patient cells (I) treated with FOXO1 inhibitor. Data are normalized to DMSO-treated cells (0 μM). n = 3; results represent mean ± SD. *P < .05; **P < .005. All P values were calculated by unpaired 2-tailed Student t test unless noted. Conc., concentration.
We also evaluated FOXO1’s function using a FOXO1-specific inhibitor.25 Inhibitor treatment dramatically reduced the growth of AE preleukemia cells as well as of t(8;21) cell lines (Figure 3E; supplemental Figure 2G), partially by inducing apoptosis and blocking the cell cycle (supplemental Figure 2H-I). By contrast, the inhibitor showed only minimal effects on the growth of normal human CD34+ HSPCs, with an 50% inhibitory concentration 10 times higher than those of AE-expressed cells (Figure 3E-F). Consistent with the observation in liquid culture, the colony-forming potential of normal CD34+ HSPCs was unaffected by the inhibitor. However, the clonogenic activity of AE preleukemia cells and AML cell lines was impaired upon FOXO inhibition (Figure 3G-H). Importantly, the inhibitor also displayed efficacy on primary cells from t(8;21) AML patients, impeding their colony formation as well (Figure 3I). These findings are in accordance with data showing that Foxo1 knockout mice do not present with hematopoietic defects,12 demonstrating that FOXO1 is specifically required in AE preleukemia and leukemia cells.
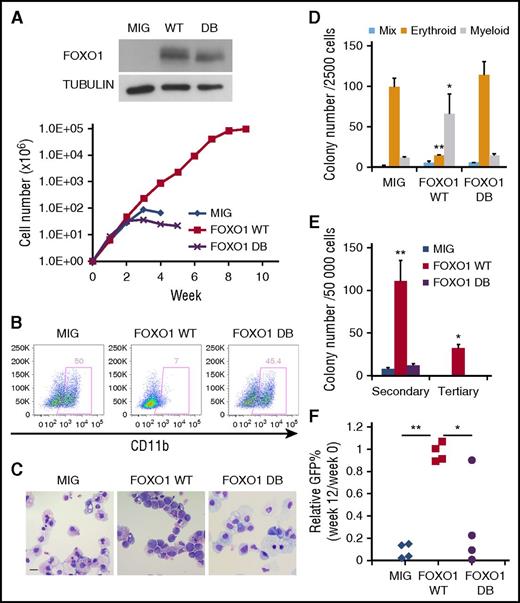
Increased FOXO1 has oncogenic activity in human CD34+ cells
The increase in FOXO1 upon AE expression in normal CD34+ HSPCs suggested that FOXO1 could play a direct role in AE pathogenesis. To test this hypothesis, we transduced CD34+ HSPCs with a retrovirus expressing FOXO1 WT or only GFP (MIG) as control. To determine whether the transforming ability of FOXO1 relied on its DNA-binding activity, we also used a FOXO1 mutant unable to bind DNA (FOXO1 DB). FOXO1 WT promoted long-term proliferation of HSPCs in liquid culture, whereas control and FOXO1 DB cells grew for only a limited period (Figure 4A). Immunophenotyping and morphological analyses showed that FOXO1 WT cells retained an immature morphology with very little CD11b expression, whereas control and FOXO DB cells underwent terminal myeloid differentiation (Figures 4B-C). In methylcellulose clonogenic assays, FOXO1 WT cells gave rise to significantly more myeloid colonies with a dramatic loss of erythroid colonies when compared with MIG and FOXO1 DB cells (Figure 4D). In addition, FOXO1 WT cells generated substantially more colonies in both secondary and tertiary replatings, in striking contrast to control cells, where replating activity was limited (Figure 4E). All these effects are similar to the phenotype elicited upon AE expression.10
Increased FOXO1 promotes a preleukemic state in human CD34+HSPCs . (A) Weekly cell count of liquid culture of CD34+ HSPCs transduced with FOXO1 WT, DB, or control (MIG) retroviral vectors. FOXO1 expression was confirmed by immunoblot. One representative experiment of 3 replicates is shown. (B) Flow cytometry analysis of the myeloid differentiation marker CD11b on cells from a week-4 culture. (C) Wright-Giemsa staining of week-4 cultured cells. The images were obtained using Motic BA310 microscope with ×40 objective. Bar represents 20 μm. (D and E) CFU assay with transduced cells. Results represent mean ± SD of colony counts of first round (D; *P < .05, **P < .001) and second and third rounds (E; *P < .01, **P < .005); n = 3. (F) Transduced cells were transplanted into immunodeficient mice. GFP+ percentage of human engrafted cells in bone marrow was examined 12 weeks later and normalized to those before the transplantation. *P < .05; **P < 1 × 10−5. One representative experiment of 2 replicates is shown. All P values were calculated by unpaired 2-tailed Student t test.
Increased FOXO1 promotes a preleukemic state in human CD34+HSPCs . (A) Weekly cell count of liquid culture of CD34+ HSPCs transduced with FOXO1 WT, DB, or control (MIG) retroviral vectors. FOXO1 expression was confirmed by immunoblot. One representative experiment of 3 replicates is shown. (B) Flow cytometry analysis of the myeloid differentiation marker CD11b on cells from a week-4 culture. (C) Wright-Giemsa staining of week-4 cultured cells. The images were obtained using Motic BA310 microscope with ×40 objective. Bar represents 20 μm. (D and E) CFU assay with transduced cells. Results represent mean ± SD of colony counts of first round (D; *P < .05, **P < .001) and second and third rounds (E; *P < .01, **P < .005); n = 3. (F) Transduced cells were transplanted into immunodeficient mice. GFP+ percentage of human engrafted cells in bone marrow was examined 12 weeks later and normalized to those before the transplantation. *P < .05; **P < 1 × 10−5. One representative experiment of 2 replicates is shown. All P values were calculated by unpaired 2-tailed Student t test.
To investigate the effect of FOXO1 expression in vivo, we transplanted transduced HSPCs into immunodeficient mice. Twelve weeks after transplantation, the proportion of GFP+ cells within human engraftment was measured. Consistent with our previous observation,26 control MIG cells did not maintain long-term engraftment in vivo, and thus the percentage of engrafted human cells expressing GFP decreased over time. FOXO DB cells showed a similar loss in vivo, but with FOXO1 WT cells, the fraction of transduced cells was maintained (Figure 4F). However, as seen for AE cells, FOXO1 WT cells were unable to initiate AML. Taken together, these data indicate that enforced expression of FOXO1 in normal CD34+ HSPCs enhances stem cell function and disrupts differentiation, thus initiating a preleukemic phenotype that resembles that of AE cells and highlighting an oncogenic function of FOXO1.
Increased FOXO1 elicits a gene expression signature shared with AE-expressing cells
The transcriptional targets of FOXOs in normal HSCs and leukemia stem cells are poorly explored. To gain a better understanding of the underlying gene network accounting for FOXO1’s oncogenic role, we performed RNA-seq analysis on CD34+ HSPCs transduced with AE, FOXO1 WT, FOXO1 DB, or MIG on day 5 posttransduction. Hierarchical clustering analysis showed that gene expression patterns from FOXO1 DB and MIG cells clustered together, indicating that the expression of FOXO1 DB does not have a major impact on gene expression and thus was nonfunctional in this context. In contrast, gene expression patterns from AE and FOXO1 WT cells clustered separately from the control cells (Figure 5A; supplemental Table 1). More strikingly, a significant portion of the AE gene expression signature was also present in FOXO1 cells (Figure 5A; “Core gene” in supplemental Table 1). Pathway enrichment analysis on genes activated by FOXO1 compared with control cells showed a significant enrichment of published gene signatures characterizing AE preleukemia and leukemia. In addition, HSC signature genes were also significantly enriched (Figure 5B). These results suggest that FOXO1 regulates a core network associating with HSC function that is critical for AE-mediated preleukemia stem cell programming. Accordingly, the FOXO1-activating gene signature was overrepresented in t(8;21) patient samples (supplemental Table 2), and the average expression level of FOXO1-activated genes was significantly higher in t(8;21) AML compared with other subtypes of AML (Figure 5C-D; supplemental Figure 3). Therefore, upregulation of the FOXO1-driven gene regulatory network was retained as a molecular feature in the acute leukemia stage of AE AML.
FOXO1 gene signature is overrepresented in AE preleukemia and leukemia cells. (A) RNA-seq analysis was performed on human CD34+ HSPCs transduced with indicated genes. Heat map depicting hierarchical clustering of 1000 significantly differentially expressed genes of AE, FOXO1 WT, and FOXO1 DB cells compared with MIG cells. The detailed gene list can be found in supplemental Table 1. (B) Pathway enrichment analysis of FOXO1-activating target genes showed enrichment of AE and HSC signatures. (C) Heat map showing the expression level of FOXO1-activating gene signatures in different subtypes of AML patient samples. The data set was from Ross et al.22 (D) Bean plot depicting the average of the mean-centered log2 expression values of FOXO1-activating genes in different subtypes of AML patient samples. The data set was from Ross et al.22 Black lines show the means of subgroups; each white line represents the value of individual samples; polygons represent the estimated density of the data. P values were calculated by 2-tailed Student t test.
FOXO1 gene signature is overrepresented in AE preleukemia and leukemia cells. (A) RNA-seq analysis was performed on human CD34+ HSPCs transduced with indicated genes. Heat map depicting hierarchical clustering of 1000 significantly differentially expressed genes of AE, FOXO1 WT, and FOXO1 DB cells compared with MIG cells. The detailed gene list can be found in supplemental Table 1. (B) Pathway enrichment analysis of FOXO1-activating target genes showed enrichment of AE and HSC signatures. (C) Heat map showing the expression level of FOXO1-activating gene signatures in different subtypes of AML patient samples. The data set was from Ross et al.22 (D) Bean plot depicting the average of the mean-centered log2 expression values of FOXO1-activating genes in different subtypes of AML patient samples. The data set was from Ross et al.22 Black lines show the means of subgroups; each white line represents the value of individual samples; polygons represent the estimated density of the data. P values were calculated by 2-tailed Student t test.
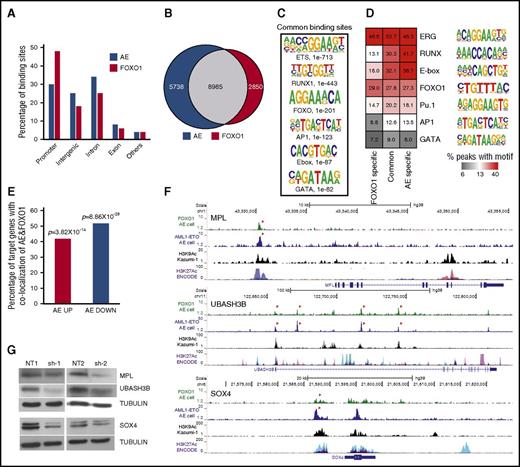
FOXO1 and AE share genome-wide localization, and FOXO1 maintains activation of key AE target genes
To identify the genomic targets of FOXO1 and AE we performed ChIP-seq analysis in AE preleukemia cells. In total, 14 723 AE- and 11 835 FOXO1-bound loci were identified. Peak distributions differed slightly, with FOXO1 showing a greater localization to promoter regions than AE (Figure 6A). In agreement with our previous report showing FOXO binding motif enrichment in AE bound loci mapped in Kasumi-1 cells,21 we found 61% of AE and 76% of FOXO1 bound loci were in common (Figure 6B). Unbiased motif enrichment analysis of joint peaks showed that both RUNX1 and FOXO motifs were significantly enriched, together with ERG/ETS and E-Box motifs, consistent with our previous report (Figures 6C-D).21 Taken together, these results suggest that the FOXO1 and AE molecular networks are broadly interconnected.
FOXO1 regulates AE key target genes. (A) Genomic location of AE and FOXO1 binding sites. (B) Venn diagram showing the overlap between AE and FOXO1 ChIP-seq peaks. (C) Enriched transcription factor binding motifs in jointly bound AE and FOXO1 ChIP-seq peaks. (D) Percentage of peaks containing predicted enriched motifs. (E) Percentage of AE-regulated genes associated with both AE and FOXO1 binding. (F) ChIP-seq tracks at SOX4, MPL, and UBASH3B loci showing binding patterns of AE and FOXO1 in AE cells, H3K9Ac in Kasumi-1 cells, and layered H3K27Ac peaks of 7 cell lines from ENCODE. Arrows indicate jointly bound AE and FOXO1 peaks. (G) Immunoblot to determine protein levels of MPL, UBASH3B, and SOX4 upon FOXO1 knockdown in AE cells.
FOXO1 regulates AE key target genes. (A) Genomic location of AE and FOXO1 binding sites. (B) Venn diagram showing the overlap between AE and FOXO1 ChIP-seq peaks. (C) Enriched transcription factor binding motifs in jointly bound AE and FOXO1 ChIP-seq peaks. (D) Percentage of peaks containing predicted enriched motifs. (E) Percentage of AE-regulated genes associated with both AE and FOXO1 binding. (F) ChIP-seq tracks at SOX4, MPL, and UBASH3B loci showing binding patterns of AE and FOXO1 in AE cells, H3K9Ac in Kasumi-1 cells, and layered H3K27Ac peaks of 7 cell lines from ENCODE. Arrows indicate jointly bound AE and FOXO1 peaks. (G) Immunoblot to determine protein levels of MPL, UBASH3B, and SOX4 upon FOXO1 knockdown in AE cells.
Cross analysis with RNA-seq data showed that ∼50% of AE-regulated genes have both AE and FOXO1 bound in close proximity to the gene body (Figure 6E). The shared binding of AE and FOXO1 at several target genes was validated in t(8;21) cell lines as well (supplemental Figure 4A). The genome-wide overlap of FOXO1 and AE binding sites compelled us to test whether FOXO1 was required for AE binding to its loci. Although we could detect a weak interaction between FOXO1 and AE in AE cells (supplemental Figure 4B), knockdown of FOXO1 did not affect the target binding of AE (supplemental Figure 4C), suggesting that the shared localization of the 2 proteins in chromatin does not require physical interaction. We identified MPL, UBASH3B, and SOX4 as genomic targets for both AE and FOXO1. It has been reported that AE leukemic and preleukemic cells depend on MPL and UBASH3B for self-renewal and long-term proliferation.26-28 In addition, SOX4 is an AML oncogene that can efficiently transform mouse HSPCs.29 Histone H3 lysine 9 acetylation (H3K9Ac) and lysine 27 acetylation (H3K27Ac) are epigenetic markers associated with active promoters and enhancers.30,31 The AE and FOXO1 binding peaks at these gene loci overlapped with our own published H3K9Ac-enriched regions in Kasumi-1 cells as well as with H3K27Ac-enriched regions identified in the Encyclopedia of DNA Elements (ENCODE) database,23,30 suggesting that AE and FOXO1 bind at bona fide gene regulatory elements (Figure 6F). Interestingly, FOXO1 knockdown in AE cells led to the downregulation of all 3 genes (Figure 6G), which may account for the impaired growth of AE cells. Similar results were observed when FOXO1 was depleted in Kasumi-1 cells, although MPL was barely expressed in these cells, possibly because of them being cytokine-independent and not relying on MPL signaling (supplemental Figure 4D). These data suggest that the activation of a substantial set of AE targets and preleukemic stem cell programming cannot be fulfilled by AE itself and requires the upregulation of FOXO1.
Discussion
Understanding the mechanisms through which leukemic oncogenes reprogram transcriptional networks and lead to enhanced self-renewal is an important prerequisite for efforts to specifically target leukemic and preleukemic stem cells and thus preventing relapse. The key mediators for leukemic programming of several subtypes of AML have been identified, including HOXA9/MEIS1 for MLL-translocation–related AML, SOX4 for AML associated with CEBPA mutations, and GATA2 for t(3;11) leukemia.29,32,33 Yet how AE promotes self-renewal and whether a key mediator exists in t(8;21) AML has been less clear. In this study, we identified FOXO1 as a critical regulator of the self-renewal program in AE preleukemia cells that is necessary and sufficient to initiate a preleukemic phenotype. We show that FOXO1 is required for full activation of a set of self-renewal genes and the long-term proliferation and clonogenicity of AE cells. Importantly, enforced expression of FOXO1 in normal CD34+ HSPCs can partially recapitulate the cellular phenotype and gene signature of AE cells, revealing increased FOXO1 activity as a new molecular mechanism by which preleukemia stem cells establish an aberrant self-renewal program.
FOXOs have complex functions, and their role in leukemia is still unclear. In AML, it has been well documented that FOXO3 acts as tumor suppressor. It was shown that active IκB kinase sustains AML proliferation by phosphorylating FOXO3 and thus preventing its nuclear entry, conversely, restoring nuclear localization of FOXO3 impaired AML cell growth.16 Similarly, it was demonstrated that FLT3-ITD, one frequent mutant in AML associated with poor disease outcome, can promote FOXO3 phosphorylation and thus prevent FOXO3-mediated apoptosis induction.34 Accordingly, a study of 511 patients showed that high levels of phosphorylated FOXO3 indicating low FOXO3 activity was an independent adverse prognostic factor in AML.35 In addition, FOXO3 was shown to localize in the cytoplasm in PML-RARα-expressing AML cells and becoming activated during all-trans retinoic acid treatment. Knockdown of FOXO3 blocked all-trans retinoic acid–induced granulocytic differentiation and apoptosis.36 In contrast, FOXO3 was found to be important for maintaining AML stem cells in a mouse MLL-AF9 AML model. FOXO3 localized to the nucleus of AML stem cells, and depletion of FOXO3 promoted myeloid differentiation and cell death, leading to a reduction of AML stem cell frequency.18 However, the molecular mechanism by which FOXO3 is required for AML stem cells was not defined, and it is therefore unclear whether FOXO3 preserves leukemia stem cell activity by coordinating metabolic balance, similar to its role in normal HSCs. In the current study, we show that FOXOs can also act as oncogenic factors and actively transform normal human HSPCs to AML upon increased expression, highlighting a new functional aspect of FOXOs in AML. Mechanistically, our data suggest that the oncogenic function of FOXO1 in AML is through the activation of an HSC program, which is critical for AE-mediated preleukemia stem cell programming.
Upregulation of FOXO1 is not restricted to t(8;21) AML. Interestingly, the FOXO1 core signature is also relatively enriched in inv(16) AML, correlating with high expression of FOXO1 in these samples (Figures 5C-D; supplemental Figures 1A and 3). The inv(16) mutations disrupt the CBFB gene, generating the CBFB-MYH11 fusion. CBFB interacts with AML1 (RUNX1) to form the core binding factor transcription complex, a critical regulator for normal hematopoiesis.37 In addition, FOXO1 upregulation was also revealed in cytogenetically normal AML with AML1 mutations.38 It is therefore possible that FOXO1 is important for AML with core binding factor gene–related mutations in general and could play a more widespread role in development of other subtypes of AML as well.
By virtue of selecting and regulating a set of AE target genes, FOXO1 reinforces the AE molecular network. Our analysis showed that target binding of AE did not depend on FOXO1. Together with the fact that increased FOXO1 was sufficient to elicit part of the AE gene signature in the absence of AE, this suggests that the genomic binding of FOXO1 and AE is independent. It is possible the genomic arrangement of binding motifs could serve as the basis for the coordination of AE and FOXO1. A significant number of genes across the genome have adjacent FOXO and RUNX binding motifs.39,40 It was also shown that FOXO1 and AML1 had functional interactions in breast cancer cell lines by jointly regulating a common set of genes.39 Because AE also recognizes RUNX motifs for chromatin binding, AE could hijack the network of FOXO1 and AML1 and thereby cooperate with FOXO1 for promoting a preleukemia program. Alternatively, other cobinding factors may be involved in orchestrating the genomic binding of FOXO1 and AE. ETS transcription factors ERG and FLI1 have been reported to direct AE to specific binding regions.41 Genomic colocalization of ETS factors and FOXO in nonhematopoietic tissues has been suggested.42 As evidenced by our finding that ETS motifs were significantly enriched in FOXO1 bound loci, it is possible that ERG/FLI1 binding sites also demarcate targets for FOXO1, thus leading to the coselection of targets by FOXO1 and AE.
With the understanding that FOXOs are tumor suppressors, restoration of FOXO activity is being considered as a potential cancer therapy.15 Given the various functions of FOXOs in AML, caution has to be taken in applying this strategy until we fully understand the AML subtype specific role of FOXOs and evaluate the possible oncogenic effects on normal HSPCs. Considering the selective toxicity of the FOXO1 inhibitor on AE preleukemic and leukemic cells, and the demonstration that an HSC defect was not seen in Foxo1-deleted mice,12 FOXO1 may be an effective therapeutic target for specifically eliminating malignant stem cells in AE and other FOXO1-overexpressing AMLs.
The data reported in this article have been deposited in the Gene Expression Omnibus database (accession numbers GSE81084 and GSE80773).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank members of the Mulloy laboratory for experimental assistance and Stephen Kissane for support with the running of ChIP-seq libraries.
This work was supported by an Institutional Clinical and Translational Science Award, National Institute of General Medical Sciences, National Institutes of Health (grant 1UL1RR026314-01); Translational Trials Development and Support Laboratory award, US Public Health Service (grant MO1 RR 08084); a Center of Excellence in Molecular Hematology P30 award, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health (grant DK090971); a CancerFree KIDS Research Award (S.L.); a Leukemia and Lymphoma Society Scholar award (J.C.M.); the assistance of the CCHMC Research Flow Cytometry Core, Translational Core Laboratory, Pathology Core, and Comprehensive Mouse and Cancer Core; and the Genomics, Epigenomics, and Sequencing Core (National Institute of Environmental Health Sciences, National Institutes of Health grant P30-ES006096) and Statistical Genomics and Systems Biology Core at the University of Cincinnati. Leukemia Research in the Bonifer laboratory is supported by a program grant from Bloodwise, United Kingdom, as well as by the Kay Kendall Leukaemia Fund.
Authorship
Contribution: S.L. performed experiments; S.L. and J.C.M. conceived and designed the study, analyzed data, and together with C.B. wrote the manuscript; A.P., S.A.A., P.S.C., and M.R.I. performed experiments and analyzed data; M.S. performed experiments; and X.C., B.J.A., J.Z., M.T.W., and C.B. analyzed data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: James C. Mulloy, Cincinnati Children's Hospital Medical Center, 3333 Burnet Ave, ML 7013, Cincinnati, OH 45229; e-mail: james.mulloy@cchmc.org.

![Figure 1. FOXO1 is upregulated in AE leukemia and preleukemia cells. (A) Microarray analysis of FOXO1 transcript levels in t(8;21)-positive compared with t(8;21)-negative AML patient samples from Ross et al.22 (B) Immunoblot showing FOXO1 protein levels in t(8;21) and cytogenetically normal [non-t(8;21)] AML patient samples. FOXO1 mRNA levels determined by quantitative PCR (qPCR) (C; n = 4, error bars represent standard deviation [SD]) and protein levels (D) in AE- or empty vector (MIT)–transduced CD34+ HSPCs. Cellular fractionation immunoblot (E) and immunofluorescence analysis (F) to determine the subcellular localization of FOXO1 in AE cells. The images were obtained by Leica DMI6000 fluorescent microscope with ×40 objective. Bar represents 20 μm. All P values were calculated by unpaired 2-tailed Student t test.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/130/10/10.1182_blood-2016-11-750976/4/m_blood750976f1.jpeg?Expires=1765920590&Signature=IYZOq14yUzlpsfpBKOuaicP79d-nnsYY3MDWb5QMxAKybe~jdwRQwWCOUSUcPfGjjzEJjMp40Sj9DPEOzbgMxe1pPx67fJkM6HFCoE5lpa51NrkC3ZfQx7GqsfusCSIsUL1zYcf-T5aYlHH-ScWY-jj~692fjU62j~dpldVZ~lVDC8W79NRr0JfPNxtvMpSHQLdqKo5GxMZEFQFnoU-ECFycoNbQILReIbr3LfjWzGVeK3rgM75ju1XRxcFGterUsrGv8MPiIwCvALRoN3jg-CQ8dmaE0FUJCYTB6Oz~Rfm0KVQhcA16qiq7cqw2H-sdoKuiqhy458FToN~yaYfFoA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal