Key Points
IMiDs inhibit TrxR-mediated intracellular decomposition of H2O2 and caused oxidative stress in MM cells.
MM cells with lower antioxidative capacity were more vulnerable to lenalidomide-induced H2O2 accumulation and its associated cytotoxicity.
Abstract
Lenalidomide is an immunomodulatory drug (IMiDs) with clinical efficacy in multiple myeloma (MM) and other late B-cell neoplasms. Although cereblon (CRBN) is an essential requirement for IMiD action, the complete molecular and biochemical mechanisms responsible for lenalidomide-mediated sensitivity or resistance remain unknown. Here, we report that IMiDs work primarily via inhibition of peroxidase-mediated intracellular H2O2 decomposition in MM cells. MM cells with lower H2O2-decomposition capacity were more vulnerable to lenalidomide-induced H2O2 accumulation and associated cytotoxicity. CRBN-dependent degradation of IKZF1 and IKZF3 was a consequence of H2O2-mediated oxidative stress. Lenalidomide increased intracellular H2O2 levels by inhibiting thioredoxin reductase (TrxR) in cells expressing CRBN, causing accumulation of immunoglobulin light-chain dimers, significantly increasing endoplasmic reticulum stress and inducing cytotoxicity by activation of BH3-only protein Bim in MM. Other direct inhibitors of TrxR and thioredoxin (Trx) caused similar cytotoxicity, but in a CRBN-independent fashion. Our findings could help identify patients most likely to benefit from IMiDs and suggest direct TrxR or Trx inhibitors for MM therapy.
Introduction
Multiple myeloma (MM) is a plasma cell neoplasm that produces and secretes monoclonal immunoglobulins.1 Immunoglobulin synthesis is associated with the formation of disulfide bonds, a reaction catalyzed by the Ero1 flavoprotein in the endoplasmic reticulum (ER), that consumes oxygen and generates H2O2.2,3 Excessive H2O2 is deleterious to cells because it activates the oxidative stress response, leading to apoptosis.4 Although a cellular antioxidant defense system exists to combat increasing concentrations of reactive oxygen species, such as H2O2, this system has a finite capacity, and antioxidant defenses can be overwhelmed by increased production of intracellular H2O2.5,6
Lenalidomide, a thalidomide analog, is an immunomodulatory drug (IMiD) widely used to treat MM, often in combination with other agents.7-9 Although cereblon (CRBN) is required for IMiD anti-MM cytotoxicity,10 the exact mechanism of action of these drugs remains elusive. Recent work has shown that lenalidomide binds to CRBN and promotes degradation of B-cell transcription factors IKZF1 and IKZF3 via ubiquitination and proteasomal degradation, thereby inducing cell death.11,12 Another recent study identified a ubiquitin-independent physiological chaperone-like function of CRBN that promotes maturation of basigin (BSG; also known as CD147) and solute carrier family 16 member 1 (SLC16A1; also known as MCT1) proteins. IMiDs outcompete CRBN for binding to CD147 and MCT1, leading to destabilization of the CD147-MCT1 complex and mediating cytotoxicity in MM.13 However, these studies still fail to explain why MM can be resistant to lenalidomide despite expression of CRBN.11-13 Even though previous reports indicate that thalidomide teratogenicity is due to oxidative stress14,15 and that thalidomide analogs induce oxidative killing of leukemic cells,16 the complete molecular and biochemical mechanisms responsible for lenalidomide-mediated anti-MM activity remain unknown. Therefore, we aimed to analyze whether the IMiD, particularly lenalidomide, given its extensive clinical application, interferes with the antioxidative capacity of MM cells to exert its anti-MM activity.
Methods
Cell culture and MTT assay
All human MM cell lines (HMCLs) were maintained in RPMI-1640, supplemented with 10% fetal bovine serum, 100 U/mL penicillin, 100 μg/mL streptomycin, and 2 mM glutamine. All HMCLs were grown at 37°C in a 5% CO2 incubator. Cell viability was assessed by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) colorimetric assay.
Western blot analysis
Whole-cell lysates were prepared from cell pellets using cell lysis buffer (Cell Signaling Technology). Equal amounts of protein extracts were resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis. Most gels were run under reducing conditions (by adding dithiothreitol [DTT]), but analysis of immunoglobulin light chain (IgL) dimers required nonreducing conditions (without DTT). Other procedures and antibodies used for this study are listed in supplemental Methods, available on the Blood Web site.
Novel quantitative measurement of determining cellular antioxidative capacity
HMCL cells (106 cells/mL in phosphate-buffered saline [PBS]) were or were not treated with H2O2 (100 µM) and immediately analyzed for autofluorescence of flavin adenine dinucleotide (FAD; fluorescein isothiocyanate [FITC]−A channel) and reduced nicotinamide adenine dinucleotide and nicotinamide adenine dinucleotide phosphate [NAD(P)H; UV Blue–A channel] with multicolor flow cytometry (LSRFortessa; BD Biosciences). Histogram normalization (FlowJo flow cytometry analysis software v.7.6.5; Tree Star, Inc.) was used to overlay untreated vs treated samples. CD138+ patient samples were treated with or without H2O2 (100 µM or control) for 30 minutes. Fluorescence readings were obtained for FAD at 450/535- and NAD(P)H at 350/460-nm excitation and emission wavelengths, respectively. All individuals provided written informed consent for the collection and use of samples for research purposes under Institutional Review Board approval and according to the Declaration of Helsinki.
CRBN, IgL-λ, and Bim knockdown
CRBN knockdown cells were the same as previously published.10 For Bim and IgL-λ knockdown, lentiviral constructs expressing nontargeting (shCtrl) and Bim or IgL-λ short hairpin RNA (shRNAs; Sigma-Aldrich) were used.
Augmented ectopic expression of CRBN in OCIMY5
Human CRBN complementary DNA was purchased from Thermo Scientific and subcloned into a lentiviral expression vector, pCDH-CMV-MCS-EF1-copGFP (System Bioscience). Lentivirus harboring control vector and CRBN complementary DNA constructs were prepared and used to infect the MM cell line OCIMY5. Infection efficiency was measured by FACScan analysis of GFP expression 3 days after infection. The cells were sorted for GFP expression 14 days after infection. CRBN overexpression was confirmed by immunoblotting.
Knockout of CRBN using CRISPR-associated protein 9 technology
Results
MM cells with lower antioxidative capacity are more vulnerable to lenalidomide-mediated cytotoxicity
MM cells with similar levels of CRBN expression, but variable IKZF1 and IKZF3 status, can have differential sensitivity to lenalidomide,19 suggesting other mechanisms for IMiD-mediated cytotoxicity (Figure 1A-B). We hypothesized that a differential capacity to dispose of H2O2 might contribute to cellular sensitivity to IMiDs, and we therefore analyzed cellular antioxidative capacity as a predictor of lenalidomide sensitivity.
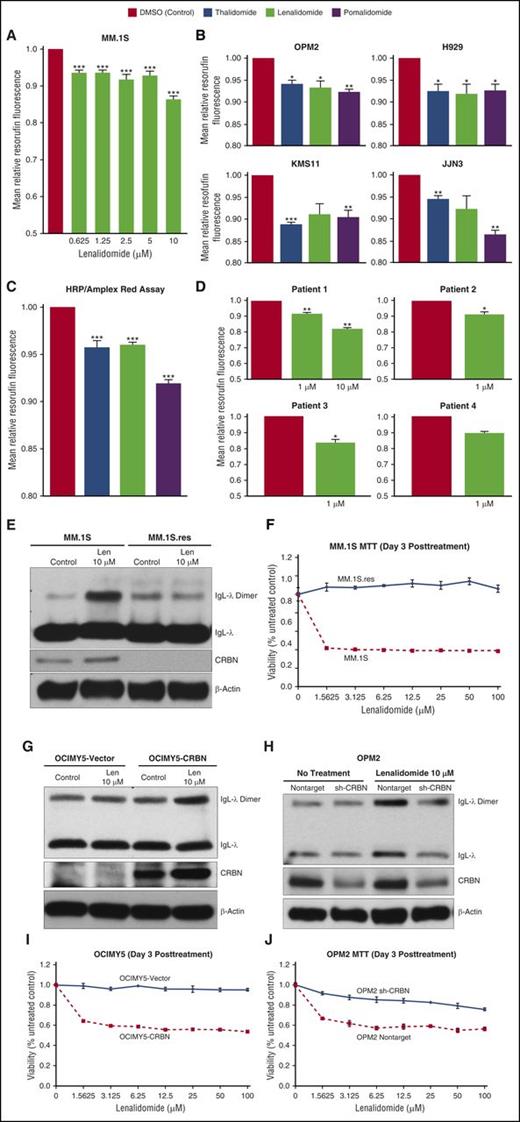
MM cells with lower antioxidative capacity are vulnerable to lenalidomide-mediated cytotoxicity. (A) RPMI-8226, JJN3, MM.1S, and KMS11 cell lysates were prepared, separated by electrophoresis, and immunoblotted with the indicated antibodies: IKZF1, IKZF3, CRBN, and β-actin. β-Actin was used as a loading control. (B) MTT assay of MM cell lines showed that MM.1S was lenalidomide hypersensitive, KMS11 was sensitive, and RPMI-8226 and JJN3 were lenalidomide resistant. Each experimental condition was performed in triplicate and repeated at least twice. (C) Bubble assay depicts release of oxygen from H2O2 by MM cells, with higher antioxidative capacity being associated with lenalidomide resistance. An equal number (1 × 106) of cells in PBS from various cell lines was incubated with H2O2, and oxygen release was qualitatively measured. (D) H2O2-mediated intracellular oxidation of FADH2 and NAD(P)H quantitatively determined antioxidative capacity in MM cells. Flow cytometry of RPMI-8226, JJN3, MM.1S, and KMS11 cells in PBS, with or without H2O2 treatment (100 μM) (gated for 10 000 events), showed significantly increased FAD autofluorescence (FITC-A) and significantly decreased NAD(P)H autofluorescence in RPMI-8226 and JJN3 with H2O2; these changes were associated with greater antioxidative capacity and resistance to lenalidomide compared with MM.1S and KMS11 cells, which have lower antioxidative capacity and greater sensitivity to lenalidomide. (E-F) Mean fluorescence intensity from 3 independent experiments for FAD and NAD(P)H autofluorescence, respectively. Data are shown as mean ± standard error of the mean (SEM). (G) Antioxidative capacity and prediction of lenalidomide sensitivity for samples from patients with primary MM. CD138+ patient samples were treated with or without H2O2 (100 µM or control) for 30 minutes. Fluorescence readings were obtained for FAD at 450/535- and NAD(P)H at 350/460-nm excitation and emission wavelengths, respectively. Data are shown as mean ± SEM of 4 biological repeats. **P < .01 compared with control; ***P < .001 compared with control.
MM cells with lower antioxidative capacity are vulnerable to lenalidomide-mediated cytotoxicity. (A) RPMI-8226, JJN3, MM.1S, and KMS11 cell lysates were prepared, separated by electrophoresis, and immunoblotted with the indicated antibodies: IKZF1, IKZF3, CRBN, and β-actin. β-Actin was used as a loading control. (B) MTT assay of MM cell lines showed that MM.1S was lenalidomide hypersensitive, KMS11 was sensitive, and RPMI-8226 and JJN3 were lenalidomide resistant. Each experimental condition was performed in triplicate and repeated at least twice. (C) Bubble assay depicts release of oxygen from H2O2 by MM cells, with higher antioxidative capacity being associated with lenalidomide resistance. An equal number (1 × 106) of cells in PBS from various cell lines was incubated with H2O2, and oxygen release was qualitatively measured. (D) H2O2-mediated intracellular oxidation of FADH2 and NAD(P)H quantitatively determined antioxidative capacity in MM cells. Flow cytometry of RPMI-8226, JJN3, MM.1S, and KMS11 cells in PBS, with or without H2O2 treatment (100 μM) (gated for 10 000 events), showed significantly increased FAD autofluorescence (FITC-A) and significantly decreased NAD(P)H autofluorescence in RPMI-8226 and JJN3 with H2O2; these changes were associated with greater antioxidative capacity and resistance to lenalidomide compared with MM.1S and KMS11 cells, which have lower antioxidative capacity and greater sensitivity to lenalidomide. (E-F) Mean fluorescence intensity from 3 independent experiments for FAD and NAD(P)H autofluorescence, respectively. Data are shown as mean ± standard error of the mean (SEM). (G) Antioxidative capacity and prediction of lenalidomide sensitivity for samples from patients with primary MM. CD138+ patient samples were treated with or without H2O2 (100 µM or control) for 30 minutes. Fluorescence readings were obtained for FAD at 450/535- and NAD(P)H at 350/460-nm excitation and emission wavelengths, respectively. Data are shown as mean ± SEM of 4 biological repeats. **P < .01 compared with control; ***P < .001 compared with control.
We measured the capacity of MM cells to decompose H2O2 to oxygen and water by using a simple biochemical test that qualitatively measures the amount of oxygen released in vitro after H2O2 exposure. We tested several HMCLs that express wild-type CRBN and with different sensitivity to lenalidomide, including the resistant cells RPMI-8226 and JJN3, lenalidomide-hypersensitive MM.1S, and moderately sensitive KMS11. An equal number of cells were treated with an equal amount of H2O2, and the release of oxygen was observed and subjectively quantified. This analysis showed that lenalidomide-resistant MM cells had the highest capacity to decompose H2O2 compared with lenalidomide-sensitive cells (Figure 1C; supplemental Figure 1A). This differential H2O2 decomposition capacity appears to correspond well with external H2O2-mediated cytotoxicity; MM.1S was hypersensitive, and RPMI-8226 was resistant to H2O2 (supplemental Figure 1B-C).
To develop a quantitative assay of cellular antioxidative capacity, we applied a new strategy that measured total cellular oxidation of reduced flavin adenine dinucleotide (FADH2) and NAD(P)H after H2O2 treatment. Cells with high antioxidative capacity generate more oxidized FAD and NAD(P) after H2O2 treatment, and cells with lower antioxidative capacity (ie, those already in a highly oxidized state) show less oxidation of FADH2 and NAD(P)H with H2O2 treatment (Figure 1D; supplemental Figure 1D-E). The autofluorescent properties of oxidized FAD and reduced NAD(P)H allowed us to determine whether H2O2 treatment increased oxidized FAD and NAD(P) through increased and decreased autofluorescence, respectively. Cells with greater antioxidative capacity, and therefore resistant to lenalidomide (RPMI-8226 and JJN3), exhibited greater increases in FAD autofluorescence and greater decreases in NAD(P)H autofluorescence than lenalidomide-sensitive cells (MM.1S and KMS11) after treatment with 100 μM H2O2 (Figure 1E-F). These results are consistent with the hypothesis that antioxidative capacity determines lenalidomide sensitivity in HMCLs, despite similar CRBN protein expression levels (Figure 1A-F; supplemental Figure 1D-E).
Next, we were interested in determining whether our findings could be clinically useful in predicting which patients are sensitive to lenalidomide. Using a small number of primary patient samples, we found that the antioxidative capacity correlates with lenalidomide sensitivity (Figure 1G; supplemental Figure 1F).
Lenalidomide inhibits intracellular H2O2 decomposition in MM cells
As MM cells’ capacity to decompose H2O2 was correlated with lenalidomide sensitivity, we tested whether lenalidomide could inhibit H2O2 decomposition in HMCLs. Cells were pretreated with the highly specific peroxidase substrate Amplex Red, followed by exposure to increasing concentrations of lenalidomide or vehicle control. Lenalidomide-exposed MM.1S cells showed decreased resorufin (oxidized fluorescent product), indicating that lenalidomide inhibited intracellular H2O2 decomposition (Figure 2A). Because lenalidomide was able to inhibit H2O2 decomposition, we were interested in evaluating whether other IMiDs (ie, thalidomide and pomalidomide) also inhibit intracellular H2O2 decomposition in different HMCLs (Figure 2B). To confirm IMiD-mediated inhibition of H2O2 decomposition, we treated different HMCLs with external H2O2 and observed (1) increased basal resorufin fluorescence and (2) IMiD-mediated inhibition of external H2O2 decomposition by intracellular peroxidases (supplemental Figure 2A). We also showed that all IMiDs analyzed had the same mechanism of action because in vitro decomposition of H2O2 by horseradish peroxidase (HRP) (in vitro HRP assay) was similarly inhibited by thalidomide, lenalidomide, and pomalidomide (Figure 2C). Lenalidomide-mediated inhibition of HRP was not concentration dependent (supplemental Figure 2B) and inhibited H2O2 decomposition in clinically achievable concentrations. Pomalidomide was the most potent inhibitor, consistent with its perceived clinical potency20 (Figure 2B-C; supplemental Figure 2A). Moreover, we also confirmed that lenalidomide inhibited intracellular H2O2 decomposition in samples from MM patients (CD138+) at a clinically achievable concentration (Figure 2D).
Lenalidomide inhibits intracellular H2O2 decomposition in MM cells. (A) MM.1S cell line was pretreated with Amplex Red (50 μM) and incubated with increasing concentrations of lenalidomide (0.625-10 μM) or dimethyl sulfoxide (DMSO; control) for 90 minutes before fluorescence intensity was measured. (B) IMiDs inhibit intracellular H2O2 decomposition in HMCLs. Cells were pretreated with Amplex Red (50 µM) and incubated with thalidomide (20 µM), lenalidomide (20 µM), pomalidomide (10 µM), or DMSO (control) for 40 to 90 minutes. All IMiDs inhibited intracellular H2O2 decomposition in HMCLs. (C) IMiDs inhibit HRP-mediated decomposition of H2O2 in an in vitro assay. Thalidomide, lenalidomide, or pomalidomide (10 µM for all) or DMSO (control) was incubated with HRP, Amplex Red, and H2O2 (5 µM) for 30 minutes before fluorescence intensity was measured. (D) Lenalidomide inhibits intracellular H2O2 decomposition in samples from patients with primary MM. CD138+ samples were pretreated with Amplex Red (50 μM) and incubated with lenalidomide (1 or 10 μM) or DMSO (control) for 90 minutes. Data are shown as mean ± SEM, n = 4 biological repeats. (E) Lenalidomide-sensitive (MM.1S) and lenalidomide-resistant (MM.1S.res) cell lines were treated with lenalidomide for 3 days. Cell lysates were prepared under nonreducing conditions; protein lysates were separated by electrophoresis under nonreducing conditions (without DTT) and immunoblotted as indicated. (F) MM.1S and MM.1S.res cells were seeded and incubated with lenalidomide at the indicated concentration for 3 days, and MTT assays were performed. Each experimental condition was performed in triplicate and repeated at least twice. (G-H) CRBN-overexpressing OCIMY5 and vector control cells and CRBN-knockdown OPM2 cell lines were treated with lenalidomide for 3 days. Cell lysates were both prepared and separated by electrophoresis under nonreducing conditions (without DTT) to detect IgL dimerization. Immunoblots were performed as indicated. Blots are representative of 3 independent experiments. (I-J) CRBN-positive cells were more sensitive to lenalidomide than CRBN-negative cells, as measured by the MTT assay. Each experimental condition was performed in triplicate and repeated at least twice. All data are shown as mean ± SEM for a minimum of 3 independent experiments. *P < .05 compared with DMSO (control); **P < .01 compared with control; ***P < .001 compared with control.
Lenalidomide inhibits intracellular H2O2 decomposition in MM cells. (A) MM.1S cell line was pretreated with Amplex Red (50 μM) and incubated with increasing concentrations of lenalidomide (0.625-10 μM) or dimethyl sulfoxide (DMSO; control) for 90 minutes before fluorescence intensity was measured. (B) IMiDs inhibit intracellular H2O2 decomposition in HMCLs. Cells were pretreated with Amplex Red (50 µM) and incubated with thalidomide (20 µM), lenalidomide (20 µM), pomalidomide (10 µM), or DMSO (control) for 40 to 90 minutes. All IMiDs inhibited intracellular H2O2 decomposition in HMCLs. (C) IMiDs inhibit HRP-mediated decomposition of H2O2 in an in vitro assay. Thalidomide, lenalidomide, or pomalidomide (10 µM for all) or DMSO (control) was incubated with HRP, Amplex Red, and H2O2 (5 µM) for 30 minutes before fluorescence intensity was measured. (D) Lenalidomide inhibits intracellular H2O2 decomposition in samples from patients with primary MM. CD138+ samples were pretreated with Amplex Red (50 μM) and incubated with lenalidomide (1 or 10 μM) or DMSO (control) for 90 minutes. Data are shown as mean ± SEM, n = 4 biological repeats. (E) Lenalidomide-sensitive (MM.1S) and lenalidomide-resistant (MM.1S.res) cell lines were treated with lenalidomide for 3 days. Cell lysates were prepared under nonreducing conditions; protein lysates were separated by electrophoresis under nonreducing conditions (without DTT) and immunoblotted as indicated. (F) MM.1S and MM.1S.res cells were seeded and incubated with lenalidomide at the indicated concentration for 3 days, and MTT assays were performed. Each experimental condition was performed in triplicate and repeated at least twice. (G-H) CRBN-overexpressing OCIMY5 and vector control cells and CRBN-knockdown OPM2 cell lines were treated with lenalidomide for 3 days. Cell lysates were both prepared and separated by electrophoresis under nonreducing conditions (without DTT) to detect IgL dimerization. Immunoblots were performed as indicated. Blots are representative of 3 independent experiments. (I-J) CRBN-positive cells were more sensitive to lenalidomide than CRBN-negative cells, as measured by the MTT assay. Each experimental condition was performed in triplicate and repeated at least twice. All data are shown as mean ± SEM for a minimum of 3 independent experiments. *P < .05 compared with DMSO (control); **P < .01 compared with control; ***P < .001 compared with control.
After showing that lenalidomide inhibited intracellular H2O2 decomposition, we sought a biological marker for lenalidomide-mediated, elevated intracellular H2O2 in MM. Increased intermolecular disulfide bond formation (protein disulfide dimers) is a consequence and hallmark of elevated intracellular H2O2 due to cysteine oxidation.21,22 Using MM.1S, we observed increased formation of IgL-λ dimers after 3 hours of treatment with lenalidomide or H2O2 (supplemental Figure 2C). Because the degree of dimerization was somewhat low after 3 hours of lenalidomide, we subsequently prolonged lenalidomide exposure to enhance dimerization. At the same time, we analyzed the requirement of CRBN for lenalidomide-mediated elevation of intracellular H2O2 to induce IgL-λ dimers. We repeated this analysis with lenalidomide-resistant MM.1Sres (this cell line was generated by culturing MM.1S in gradually increasing concentrations of lenalidomide).23 Our laboratory previously reported that CRBN expression in MM.1Sres is diminished as lenalidomide resistance increases.10 We treated MM.1S and MM.1Sres cells with lenalidomide for 3 days and observed IgL-λ dimers only in MM.1S (Figure 2E), the lenalidomide-sensitive cell line (Figure 2F). By using other isogenic cell lines, positive and negative for CRBN expression, we showed that lenalidomide treatment caused accumulation of IgL-λ dimers only in CRBN-positive cells (Figure 2G-H; supplemental Figure 2D), further confirming that CRBN-bound lenalidomide is required to inhibit intracellular H2O2 decomposition and that the H2O2 had to be high to induce IgL dimerization (Figure 2E,G,H; supplemental Figure 2C-D). Dimerization was associated with sensitivity to lenalidomide (Figure 2F,I-J; supplemental Figure 2E). This dimerization was completely eliminated after electrophoresis with DTT (supplemental Figure 2 F), suggesting that lenalidomide acts as a static drug as it has almost the same biological effect (IgL dimerization) at very low and high concentrations (supplemental Figure 2G-H).
H2O2 effectively degrades IKZF1 and IKZF3 in MM cells expressing CRBN
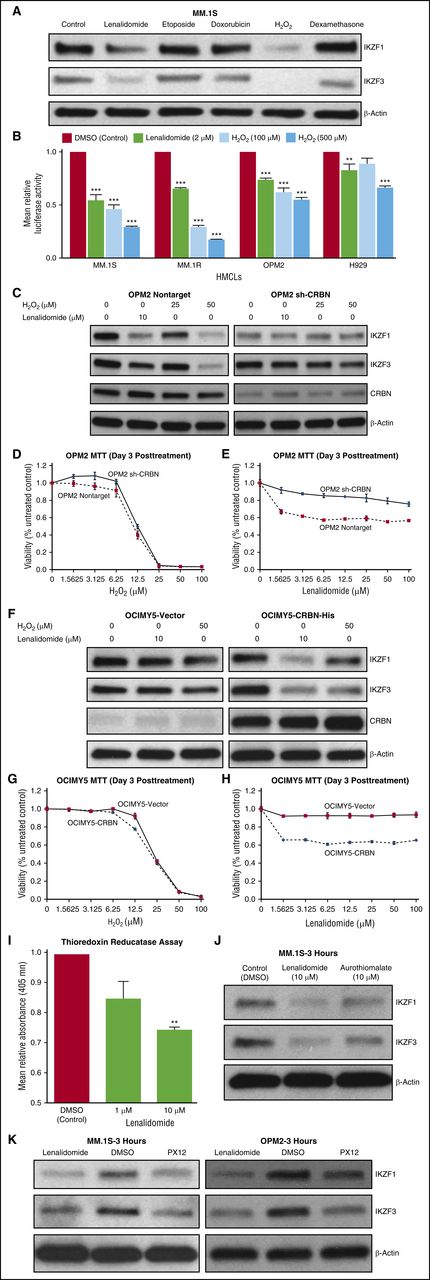
Lenalidomide-bound CRBN has been shown to induce proteasomal degradation of IKZF1 and IKZF3.11,12,24 We hypothesized that lenalidomide-induced IKZF1 and IKZF3 degradation was mediated via H2O2-induced protein oxidation cascade. We found that both lenalidomide and H2O2 treatment induced IKZF1 protein dimerization (supplemental Figure 3A-D). To confirm that elevated intracellular H2O2 caused IKZF1 and IKZF3 degradation, we treated MM.1S cells with H2O2 or other drugs also reported to induce oxidative stress (etoposide, doxorubicin, and dexamethasone). Only H2O2 and lenalidomide degraded IKZF1 and IKZF3 (degradation evident within 3 hours of treatment; Figure 3A). Moreover, H2O2 and lenalidomide-induced IKZF1 and IKZF3 degradation was rescued by proteasomal inhibition with bortezomib (supplemental Figure 3E-G). This finding further supports our hypothesis that lenalidomide and exogenous H2O2 degrade IKZF1 and IKZF3 by a common mechanism.
H2O2 leads to preferential degradation of IKZF1 and IKZF3 in CRBN-positive cells. (A) Degradation of IKZF1 and IKZF3 was observed only with lenalidomide and H2O2. MM.1S cell line was treated with lenalidomide (10 μM), etoposide (1 μM), doxorubicin (0.5 μM), H2O2 (100 μM), and dexamethasone (20 μM) for 3 hours. Cell lysates were prepared, separated by electrophoresis, and immunoblotted with the indicated antibodies. (B) Different HMCLs were infected with AdIKLuc or the control vector (AdLuc), and sufficient luciferase activity was confirmed. Cell lines were treated with increasing concentrations of H2O2 (100 or 500 µM) or lenalidomide (2 µM) for 1 hour, and luciferase activity was measured. Data are shown as mean ± SEM, n = 4 biological repeats. (C) To evaluate the role of CRBN on IKZF1 and IKZF3 degradation, OPM2-nontarget (control) and OPM2-shCRBN (silenced) cells were treated with lenalidomide (10 μM) or increasing concentrations of H2O2 (25 or 50 μM) for 3 hours. Protein lysates were prepared, separated by electrophoresis, and immunoblotted as indicated. (D-E) To differentially evaluate the effect of H2O2 and lenalidomide on cell viability based on the level of CRBN expression, OPM2-NT and OPM2-shCRBN were treated with increasing concentrations of H2O2 and lenalidomide for 3 days. MTT assays were performed, and cell survival was plotted. Each experimental condition was performed in triplicate and repeated at least twice. (F) OCIMY-5-vector cells and an isogenic CRBN-overexpressing cell line were treated with lenalidomide (10 μM) or H2O2 (50 μM) for 3 hours. Protein lysates were prepared, separated by electrophoresis, and immunoblotted as indicated. (G-H) OCIMY-5 cells overexpressing CRBN and control vector were treated with increasing concentrations of H2O2 and lenalidomide for 3 days. MTT assays were performed, and cell survival was plotted. Each experimental condition was performed in triplicate and repeated at least twice. (I) Lenalidomide inhibits TrxR activity in vitro. Rat liver TrxR was treated with lenalidomide (1 µM and 10 µM) or DMSO control, and absorbance was measured at 405 nM. Lenalidomide-treated samples show decreased absorbance compared with control (DMSO). Data are shown as mean ± SEM, n = 3 biological repeats. (J) MM.1S cell line treated with lenalidomide (10 µM), aurothiomalate (10 µM), or DMSO (control) for 3 hours. Protein lysates were prepared and blotted with indicated antibodies. (K) MM.1S and OPM2 cells were treated with lenalidomide (10 µM), PX12 (20 µM), or DMSO (control) for 3 hours. Protein lysates were prepared, separated by electrophoresis, and immunoblotted as indicated. Blots are representative of 3 independent experiments. All data are shown as mean ± SEM for a minimum of 3 independent experiments. **P < .01 compared with control; ***P < .001 compared with control.
H2O2 leads to preferential degradation of IKZF1 and IKZF3 in CRBN-positive cells. (A) Degradation of IKZF1 and IKZF3 was observed only with lenalidomide and H2O2. MM.1S cell line was treated with lenalidomide (10 μM), etoposide (1 μM), doxorubicin (0.5 μM), H2O2 (100 μM), and dexamethasone (20 μM) for 3 hours. Cell lysates were prepared, separated by electrophoresis, and immunoblotted with the indicated antibodies. (B) Different HMCLs were infected with AdIKLuc or the control vector (AdLuc), and sufficient luciferase activity was confirmed. Cell lines were treated with increasing concentrations of H2O2 (100 or 500 µM) or lenalidomide (2 µM) for 1 hour, and luciferase activity was measured. Data are shown as mean ± SEM, n = 4 biological repeats. (C) To evaluate the role of CRBN on IKZF1 and IKZF3 degradation, OPM2-nontarget (control) and OPM2-shCRBN (silenced) cells were treated with lenalidomide (10 μM) or increasing concentrations of H2O2 (25 or 50 μM) for 3 hours. Protein lysates were prepared, separated by electrophoresis, and immunoblotted as indicated. (D-E) To differentially evaluate the effect of H2O2 and lenalidomide on cell viability based on the level of CRBN expression, OPM2-NT and OPM2-shCRBN were treated with increasing concentrations of H2O2 and lenalidomide for 3 days. MTT assays were performed, and cell survival was plotted. Each experimental condition was performed in triplicate and repeated at least twice. (F) OCIMY-5-vector cells and an isogenic CRBN-overexpressing cell line were treated with lenalidomide (10 μM) or H2O2 (50 μM) for 3 hours. Protein lysates were prepared, separated by electrophoresis, and immunoblotted as indicated. (G-H) OCIMY-5 cells overexpressing CRBN and control vector were treated with increasing concentrations of H2O2 and lenalidomide for 3 days. MTT assays were performed, and cell survival was plotted. Each experimental condition was performed in triplicate and repeated at least twice. (I) Lenalidomide inhibits TrxR activity in vitro. Rat liver TrxR was treated with lenalidomide (1 µM and 10 µM) or DMSO control, and absorbance was measured at 405 nM. Lenalidomide-treated samples show decreased absorbance compared with control (DMSO). Data are shown as mean ± SEM, n = 3 biological repeats. (J) MM.1S cell line treated with lenalidomide (10 µM), aurothiomalate (10 µM), or DMSO (control) for 3 hours. Protein lysates were prepared and blotted with indicated antibodies. (K) MM.1S and OPM2 cells were treated with lenalidomide (10 µM), PX12 (20 µM), or DMSO (control) for 3 hours. Protein lysates were prepared, separated by electrophoresis, and immunoblotted as indicated. Blots are representative of 3 independent experiments. All data are shown as mean ± SEM for a minimum of 3 independent experiments. **P < .01 compared with control; ***P < .001 compared with control.
To confirm that external H2O2-induced IKZF1 degradation was a rapid process at the protein level, we used an adenoviral vector expressing IKZF1 fused to luciferase (AdIKLuc) or control vector (AdLuc)25 to infect HMCLs. Cells were treated with increasing concentrations of H2O2 or lenalidomide for 1 hour, and luciferase activity was measured. We observed a marked decrease only in IKZF1-conjugated luciferase (AdIKLuc) activity within 1 hour of H2O2 treatment without affecting control luciferase activity (AdLuc) in MM.1S, MM.1R (resistant to dexamethasone), OPM2, and H929 (Figure 3B), as well as in other HMCLs (supplemental Figure 3H).
To confirm the central role of CRBN in IKZF1 and IKZF3 degradation by the H2O2-induced oxidative cascade, we next examined OPM2 isogeneic cells with CRBN wild-type and CRBN-knockdown as well as CRBN-overexpressing OCIMY5 (transfected with wild-type CRBN). We treated OPM2-NT (nontarget shRNA control) and OPM2-shCRBN with lenalidomide and 2 concentrations of H2O2 (25 and 50 μM) for 3 hours, and we showed that H2O2 mediates IKZF1 and IKZF3 degradation in a CRBN-dependent fashion (Figure 3C). Next, we tested the viability of OPM2-NT and OPM2-shCRBN with increasing concentrations of lenalidomide and H2O2 for 3 days. Lenalidomide-induced cytotoxicity was CRBN-dependent, but H2O2 was not (Figure 3D-E). OCIMY5 cells overexpressing CRBN also showed enhanced IKZF1 and IKZF3 degradation within 3 hours of lenalidomide or H2O2 (50 µM) treatment (Figure 3F). Again, lenalidomide, but not H2O2 cytotoxicity, was CRBN dependent (Figure 3G-H), confirming that CRBN isogenic cells exhibit similar H2O2 decomposition capacity and sensitivity to external H2O2 treatment (Figure 3D,G). However, CRBN is required for lenalidomide to elevate intracellular H2O2 (Figure 3E,H). Moreover, IKZF1 and IKZF3 degradation was a consequence of elevated intracellular H2O2 through a CRBN-dependent pathway (Figure 3A-H).
To demonstrate that antioxidative capacity determined the degree of IKZF1 degradation in CRBN-positive cells, we used RPMI-8226 and MM.1S, which have high and low antioxidative capacity, respectively (supplemental Figure 1A-C). After 3 hours of treatment with lenalidomide or H2O2, IKZF1 degradation was prominent in MM.1S but not in RPMI-8226, even though both cell lines expressed CRBN protein, further indicating that MM antioxidative capacity determined lenalidomide- and H2O2-induced IKZF1 degradation (supplemental Figure 3I). It was evident that high H2O2 concentration was required to degrade IKZF1 in RPMI-8226 compared with MM.1S due to its higher antioxidative capacity (supplemental Figure 3H).
Next, we sought to identify which H2O2-decomposing enzymes could be inhibited by lenalidomide. We performed in vitro assays with glutathione peroxidase and catalase but did not observe any inhibition with lenalidomide (supplemental Figure 3J). Surprisingly, lenalidomide inhibited thioredoxin reductase (TrxR) activity in vitro (Figure 3I). Concomitantly, we asked whether inhibition of TrxR with aurothiomalate (specific inhibitor of TrxR)26 (Figure 3J) or thioredoxin (Trx) inhibition with PX1227 (a major substrate of TrxR for maintaining intracellular H2O2 homeostasis; Figure 3K) could induce specific degradation of IKZF1 and IKZF3 in MM cells. We treated MM.1S and OPM2 cells with aurothiomalate and PX12 and showed that, like lenalidomide, aurothiomalate and PX12 also induced IKZF1 and IKZF3 degradation within 3 hours of treatment (Figure 3J-K). These findings clearly indicate that lenalidomide-mediated inhibition of TrxR leads to elevated intracellular H2O2 and also impairs the cells' capacity to reduce protein disulfide adducts28 (protein disulfide dimers) in MM.
Lenalidomide-mediated intracellular H2O2 elevation induces ER stress
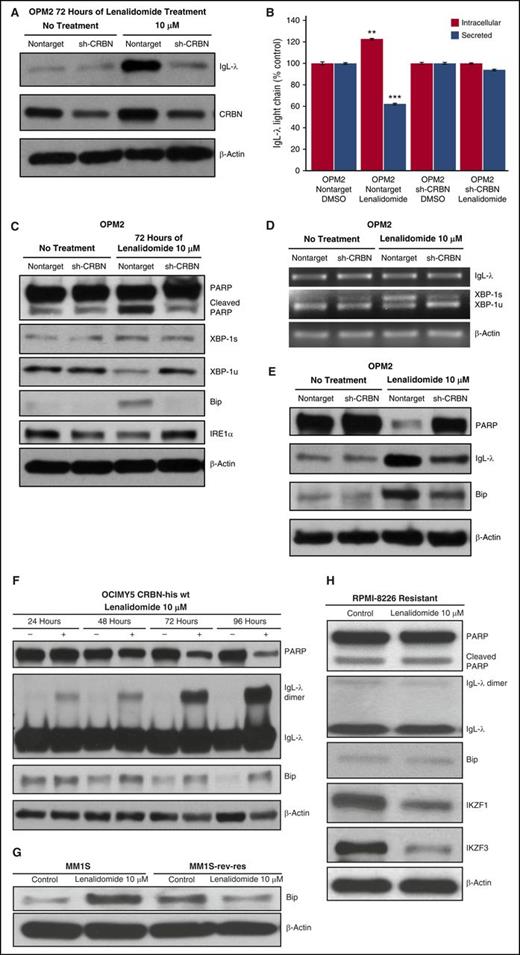
Lenalidomide-induced IgG-λ dimerization led to decreased secretion and consequent intracellular accumulation of IgG-λ, as evidenced by unchanged IgG-λ messenger RNA (mRNA) expression (supplemental Figure 4A), increased total intracellular IgL protein (Figure 4A), and decreased secretion of IgL-λ (Figure 4B; supplemental Figure 4B).
Lenalidomide-mediated intracellular H2O2 elevation induces ER stress. (A) OPM2-NT and shCRBN cells were treated for 72 hours with lenalidomide (10 μM) or DMSO (control). Cell lysates were prepared, separated by electrophoresis, and immunoblotted with the indicated antibodies. (B) OPM2-NT and OPM2-shCRBN knockdown cells were incubated with lenalidomide (10 μM) for 48 hours. Secreted and intracellular IgL-λ levels were determined via enzyme-linked immunosorbent assay. Data are expressed as a percentage of control (mean ± standard deviation, n = 3). (C) OPM2-NT and OPM2-CRBN knockdown (shCRBN) cells were treated with lenalidomide for 3 days. Cell lysates were prepared, separated by electrophoresis, and immunoblotted as indicated. (D) Lenalidomide induced XBP-1 mRNA splicing in CRBN-positive cells but showed minimal splicing in shCRBN cells. OPM2-NT and shCRBN cells were treated with lenalidomide (10 μM) for 3 days. Reverse transcription polymerase chain reaction was performed to evaluate XBP-1 mRNA splicing, a marker of ER stress. (E) OPM2-NT and shCRBN cells were treated for 6 days with lenalidomide (10 μM). Cell lysates were prepared, separated by electrophoresis, and immunoblotted as indicated. Lenalidomide-induced, ER stress–mediated PARP degradation was more prominent at day 6 in OPM2-NT cells compared with shCRBN cells. (F) OCIMY5 cells overexpressing CRBN progressively accumulated IgL dimers after lenalidomide treatment and induced ER stress. OCIMY5-vector and CRBN cells were treated with lenalidomide (10 μM) for different periods (24-96 hours). Cell lysates were prepared, separated by electrophoresis, and immunoblotted as indicated. (G) Lenalidomide-sensitive (MM.1S) and lenalidomide-resistant (MM.1S.res) cell lines were treated with lenalidomide (10 μM) for 3 days. Cell lysates were prepared and immunoblotted as indicated. (H) CRBN-expressing but with high antioxidative capacity exhibiting MM cell line, RPMI-8226, treated with lenalidomide (10 μM) for 3 days. Cell lysates were prepared and immunoblotted as indicated. Blots are representative of 3 independent experiments. All data are shown as mean ± SEM for a minimum of 3 independent experiments. **P < .01 compared with control; ***P < .001 compared with control.
Lenalidomide-mediated intracellular H2O2 elevation induces ER stress. (A) OPM2-NT and shCRBN cells were treated for 72 hours with lenalidomide (10 μM) or DMSO (control). Cell lysates were prepared, separated by electrophoresis, and immunoblotted with the indicated antibodies. (B) OPM2-NT and OPM2-shCRBN knockdown cells were incubated with lenalidomide (10 μM) for 48 hours. Secreted and intracellular IgL-λ levels were determined via enzyme-linked immunosorbent assay. Data are expressed as a percentage of control (mean ± standard deviation, n = 3). (C) OPM2-NT and OPM2-CRBN knockdown (shCRBN) cells were treated with lenalidomide for 3 days. Cell lysates were prepared, separated by electrophoresis, and immunoblotted as indicated. (D) Lenalidomide induced XBP-1 mRNA splicing in CRBN-positive cells but showed minimal splicing in shCRBN cells. OPM2-NT and shCRBN cells were treated with lenalidomide (10 μM) for 3 days. Reverse transcription polymerase chain reaction was performed to evaluate XBP-1 mRNA splicing, a marker of ER stress. (E) OPM2-NT and shCRBN cells were treated for 6 days with lenalidomide (10 μM). Cell lysates were prepared, separated by electrophoresis, and immunoblotted as indicated. Lenalidomide-induced, ER stress–mediated PARP degradation was more prominent at day 6 in OPM2-NT cells compared with shCRBN cells. (F) OCIMY5 cells overexpressing CRBN progressively accumulated IgL dimers after lenalidomide treatment and induced ER stress. OCIMY5-vector and CRBN cells were treated with lenalidomide (10 μM) for different periods (24-96 hours). Cell lysates were prepared, separated by electrophoresis, and immunoblotted as indicated. (G) Lenalidomide-sensitive (MM.1S) and lenalidomide-resistant (MM.1S.res) cell lines were treated with lenalidomide (10 μM) for 3 days. Cell lysates were prepared and immunoblotted as indicated. (H) CRBN-expressing but with high antioxidative capacity exhibiting MM cell line, RPMI-8226, treated with lenalidomide (10 μM) for 3 days. Cell lysates were prepared and immunoblotted as indicated. Blots are representative of 3 independent experiments. All data are shown as mean ± SEM for a minimum of 3 independent experiments. **P < .01 compared with control; ***P < .001 compared with control.
We postulated that the inhibition of IgG-λ protein secretion (due to H2O2-induced high dimerization) led to an ER stress response in CRBN-positive cells. After 3 days of treatment with lenalidomide, an ER stress response occurred in OPM2-NT but not in CRBN-knockdown cells (Figure 4C-D). Immunoblotting showed decreased XBP-1 unspliced and increased XBP-1 spliced proteins in CRBN-positive (nontarget) but not in CRBN-knockdown cells (Figure 4C). Another ER stress marker, GRP78/BiP, accumulated after lenalidomide treatment in CRBN-positive cells (Figure 4C). To further confirm that lenalidomide induced ER stress, we analyzed mRNA splicing of XBP-1 after lenalidomide treatment and saw that splicing was more evident in OPM2-NT than CRBN-knockdown cells (Figure 4D) after 3 days of lenalidomide treatment. The inositol-requiring enzyme 1 protein level markedly decreased in CRBN-positive cells after lenalidomide treatment (Figure 4C), indicating elevated inositol-requiring enzyme 1 endoribonuclease activity during ER stress.29
We detected minimal apoptosis in OPM2-NT cells after 3 days of drug treatment, shown by low levels of poly(adenosine 5′-diphosphate-ribose) polymerase (PARP) cleavage (Figure 4C). Therefore, we treated OPM2-NT and OPM2-shCRBN cells with lenalidomide for 6 days and analyzed the PARP levels. Immunoblots showed nearly complete degradation of PARP proteins (cleaved PARP could not be detected because it was completely degraded by day 6) in CRBN-positive, lenalidomide-sensitive cells (Figure 4E). Moreover, to better understand lenalidomide-mediated IgL dimerization and accumulation as a mediator of time-dependent progressive ER stress, we treated CRBN-overexpressing OCIMY5 cells with lenalidomide and showed progressive IgL-λ dimer accumulation, ER stress, and PARP degradation (a sign of later-stage apoptosis; Figure 4F), whereas the OCIMY5 parent cell line was not responsive to lenalidomide (Figure 2G,I). We analyzed other isogenic HMCLs, MM.1S (lenalidomide sensitive) and MM.1Sres (lenalidomide resistant), and found lenalidomide-induced, ER stress–mediated accumulation of Bip protein in CRBN-positive cells but not in CRBN-negative cells (Figure 4G). We further showed that wild-type CRBN-expressing RPMI-8226 with high antioxidative capacity (Figure 1A-F) was resistant to lenalidomide because of no elevated intracellular H2O2-mediated IgL dimerization and ER stress (Bip accumulation) even though IKZF1 and IKZF3 were degraded at day 3 of treatment (Figure 4H).
Moreover, the limited IgL knockdown in OPM2 and H929 cells reduced lenalidomide sensitivity (supplemental Figure 4C,E,G) and decreased IgL-λ dimerization after 3 days of lenalidomide treatment, compared with control cells (supplemental Figure 4D,F,H). Next, we asked whether NAC treatment could reduce lenalidomide-induced IgL dimerization and cytotoxicity, even though NAC did not rescue lenalidomide-induced IKZF1 degradation (supplemental Figure 5A). MM.1S cells treated with NAC (20 mM) exhibited reduced IgL dimerization and also rescued from lenalidomide-induced growth inhibition (supplemental Figure 5B-C). We further noted that increasing concentrations of NAC shown some degree of cell proliferation inhibition on its own in the MM.1S cell line (supplemental Figure 5D).
Lenalidomide-induced ER stress triggers cytotoxicity by activating BH3 protein Bim in MM
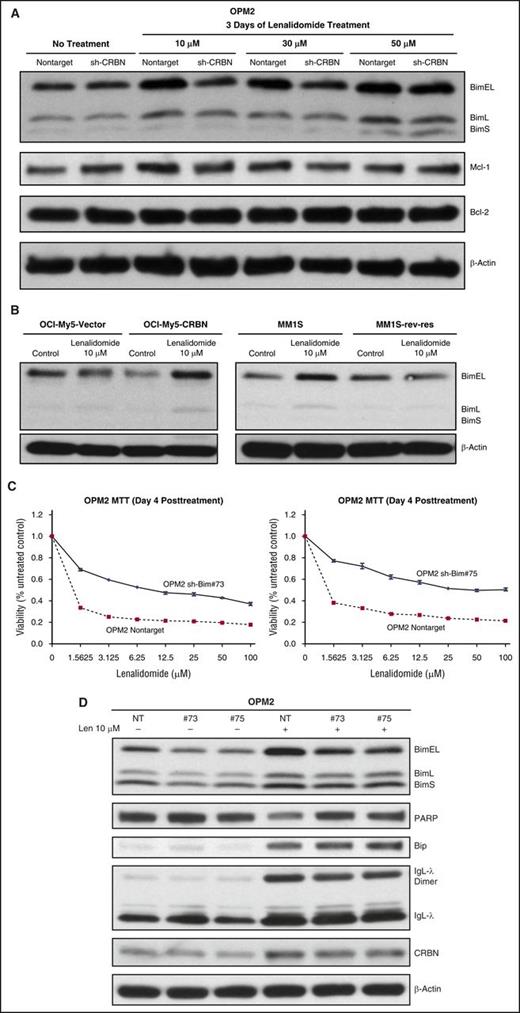
Bim activation induced apoptosis after lenalidomide treatment in CRBN-positive MM cells. CRBN-expressing and CRBN-knockdown OPM2 cells were treated with lenalidomide for 3 days, and cell lysates were immunoblotted and probed for various pro- and antiapoptotic proteins. BH3-only protein Bim was activated after lenalidomide-induced ER stress (Figure 5A). Bim has 3 isoforms, BimS, BimL, and BimEL; their differing proapoptotic potencies are partly due to differences in their interactions with the dynein motor complex.30 We observed accumulation of Bim, especially BimEL, after lenalidomide treatment in CRBN-positive, lenalidomide-sensitive cells. Mcl1 and Bcl2 antiapoptotic proteins did not change markedly after lenalidomide treatment (Figure 5A). We analyzed other CRBN-negative and CRBN-positive isogenic cell lines and confirmed Bim activation (Figure 5B). To confirm Bim involvement in lenalidomide-induced apoptosis, we used stable shRNA expression to knockdown Bim in OPM2 cells. We established 2 different OPM2 clones (numbers 73 and 75) with downregulated Bim and treated them with lenalidomide. Because lenalidomide induced late apoptosis in OPM2 cells, we performed a day-4 viability assay. Both Bim knockdown clones were less sensitive to lenalidomide than control cells (Figure 5C). Moreover, Bim knockdown cells underwent less PARP degradation compared with control cells (Figure 5D). Bim knockdown did not inhibit IgL-λ dimerization and accumulation or subsequent ER stress after lenalidomide treatment. Bim knockdown cells also showed elevated Bip protein accumulation (a marker of ER stress), like nontarget (control) cells after lenalidomide treatment, but ER stress–mediated apoptosis was diminished in Bim knockdown clones (Figure 5D). Thus, BimEL appeared to be a downstream effector of lenalidomide-induced, ER stress–mediated apoptosis in MM.
Lenalidomide-induced ER stress triggers cytotoxicity by activating BH3 protein Bim in MM. (A) OPM2-NT and OPM2-shCRBN cells were treated with increasing concentrations of lenalidomide (10, 30, and 50 μM) for 3 days. Cell lysates were prepared, separated by electrophoresis, and immunoblotted as indicated. The proapoptotic BH3-only protein Bim isoforms predominantly accumulated after lenalidomide treatment in CRBN-positive cells. (B) Additional CRBN-positive and CRBN-negative isogenic cell lines (OCIMY5 CRBN overexpressing, MM.1S sensitive and resistant to lenalidomide) were treated with lenalidomide for 3 days. Cell lysates were prepared and immunoblotted with the indicated antibodies. Bim activation was predominantly mediated by lenalidomide in CRBN-positive cells but not in CRBN-negative or CRBN knockdown cells. (C) Knockdown of Bim-mediated resistance to lenalidomide. Bim knockdown OPM2 clone numbers 73 and 75, and NT control cells were treated with different concentrations of lenalidomide for 4 days, and cell viability was determined with the MTT assay. Each experimental condition was performed in triplicate and repeated at least twice. (D) OPM2-NT and OPM2-shBim clone numbers 73 and 75 were treated with or without lenalidomide (10 μM) for 72 hours. Cell lysates were prepared, separated by electrophoresis, and immunoblotted as indicated. Blots are representative of 3 independent experiments.
Lenalidomide-induced ER stress triggers cytotoxicity by activating BH3 protein Bim in MM. (A) OPM2-NT and OPM2-shCRBN cells were treated with increasing concentrations of lenalidomide (10, 30, and 50 μM) for 3 days. Cell lysates were prepared, separated by electrophoresis, and immunoblotted as indicated. The proapoptotic BH3-only protein Bim isoforms predominantly accumulated after lenalidomide treatment in CRBN-positive cells. (B) Additional CRBN-positive and CRBN-negative isogenic cell lines (OCIMY5 CRBN overexpressing, MM.1S sensitive and resistant to lenalidomide) were treated with lenalidomide for 3 days. Cell lysates were prepared and immunoblotted with the indicated antibodies. Bim activation was predominantly mediated by lenalidomide in CRBN-positive cells but not in CRBN-negative or CRBN knockdown cells. (C) Knockdown of Bim-mediated resistance to lenalidomide. Bim knockdown OPM2 clone numbers 73 and 75, and NT control cells were treated with different concentrations of lenalidomide for 4 days, and cell viability was determined with the MTT assay. Each experimental condition was performed in triplicate and repeated at least twice. (D) OPM2-NT and OPM2-shBim clone numbers 73 and 75 were treated with or without lenalidomide (10 μM) for 72 hours. Cell lysates were prepared, separated by electrophoresis, and immunoblotted as indicated. Blots are representative of 3 independent experiments.
Lenalidomide and bortezomib-mediated oxidative stress in MM cell death
Proteasome inhibitors, including bortezomib, are also reported to induce oxidative stress and inhibition of oxidized protein degradation in MM and related cancers.31-35 Recently, proteasome inhibitors are reported to directly induce oxidative and ER stresses in MM cells via transcriptional repression of a gene encoding mitochondrial thioredoxin reductase (TXNRD2).36 Therefore, we postulated that dual treatment with lenalidomide and bortezomib is more clinically effective due to mutually enhanced oxidative stress.
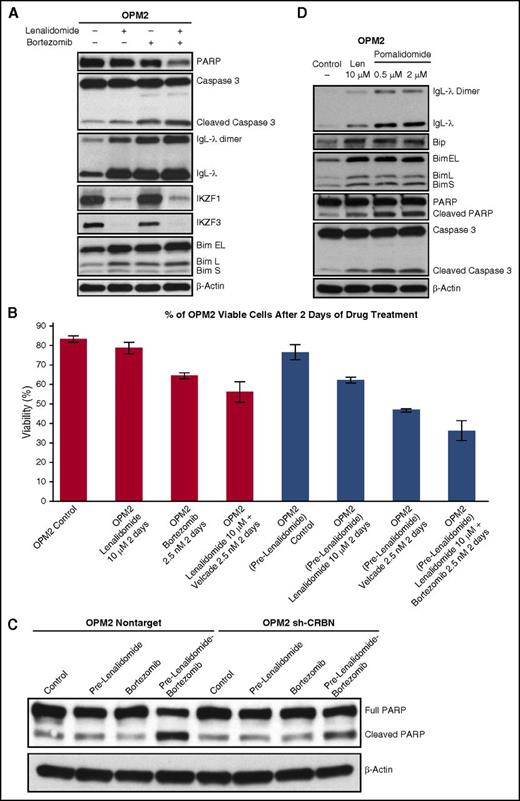
We treated OPM2 with lenalidomide, bortezomib, and lenalidomide plus bortezomib for 3 days to identify better described downstream signaling. We found that concurrent administration of lenalidomide and bortezomib augmented the accumulation of IgL dimers and Bim (Figure 6A). Interestingly, bortezomib treatment alone also increased IgL-λ monomers and dimers, further indicating induction of oxidative stress and inhibition of degradation of oxidized immunoglobulins. Because lenalidomide-mediated MM cytotoxicity is a slow process (Figure 4F) compared with bortezomib, we propose that lenalidomide should precede bortezomib in the clinic; we pretreated MM cells with lenalidomide and then treated them with bortezomib, showing increased sensitivity (Figure 6B). Next, we analyzed whether CRBN-positive cells were more prone to apoptosis after lenalidomide pretreatment followed by bortezomib treatment. OPM2-NT and OPM2-shCRBN cells were pretreated with lenalidomide for 2 days, and cells were washed and then treated with bortezomib for an additional 2 days. Lenalidomide pretreatment enhanced bortezomib-induced PARP cleavage in OPM2 CRBN-positive cells, but cleavage was reduced in CRBN knockdown cells (Figure 6C), suggesting that when cells are resistant to lenalidomide, the synergistic effects with bortezomib also cease.
Lenalidomide- and bortezomib-mediated oxidative stress in MM cell death. (A) OPM2 cells treated with lenalidomide (2 μM), bortezomib (2.5 nM), or lenalidomide plus bortezomib for 3 days. Cell lysates were prepared, separated by electrophoresis, and immunoblotted as indicated. (B) OPM2 cells, with or without 2 days of pretreatment with lenalidomide (10 μM), were treated with bortezomib (2.5 nM), lenalidomide (10 μM), and bortezomib plus lenalidomide for an additional 2 days. Samples were collected and stained with Annexin V-FITC and propidium iodide. Fluorescence-activated cell sorting (FACS) analysis for apoptosis showed the lowest viability with lenalidomide pretreatment followed by bortezomib plus lenalidomide. (C) Pretreatment with lenalidomide enhances bortezomib-mediated apoptosis in CRBN-positive MM cells. OPM2-NT and OPM2-shCRBN cells, with or without pretreatment with lenalidomide for 2 days, were treated with bortezomib (2.5 nM) for an additional 2 days. Cell lysates were prepared, separated by electrophoresis, and immunoblotted as indicated. Lenalidomide pretreatment enhanced bortezomib-induced apoptosis of CRBN-positive cells but had a less pronounced effect in CRBN-knockdown cells. (D) OPM2 cells were treated with lenalidomide (10 µM) or with increasing concentrations of pomalidomide (0.5 and 2 μM) for 3 days. Cell lysates were prepared, separated by electrophoresis, and immunoblotted as indicated. Pre, pretreatment with.
Lenalidomide- and bortezomib-mediated oxidative stress in MM cell death. (A) OPM2 cells treated with lenalidomide (2 μM), bortezomib (2.5 nM), or lenalidomide plus bortezomib for 3 days. Cell lysates were prepared, separated by electrophoresis, and immunoblotted as indicated. (B) OPM2 cells, with or without 2 days of pretreatment with lenalidomide (10 μM), were treated with bortezomib (2.5 nM), lenalidomide (10 μM), and bortezomib plus lenalidomide for an additional 2 days. Samples were collected and stained with Annexin V-FITC and propidium iodide. Fluorescence-activated cell sorting (FACS) analysis for apoptosis showed the lowest viability with lenalidomide pretreatment followed by bortezomib plus lenalidomide. (C) Pretreatment with lenalidomide enhances bortezomib-mediated apoptosis in CRBN-positive MM cells. OPM2-NT and OPM2-shCRBN cells, with or without pretreatment with lenalidomide for 2 days, were treated with bortezomib (2.5 nM) for an additional 2 days. Cell lysates were prepared, separated by electrophoresis, and immunoblotted as indicated. Lenalidomide pretreatment enhanced bortezomib-induced apoptosis of CRBN-positive cells but had a less pronounced effect in CRBN-knockdown cells. (D) OPM2 cells were treated with lenalidomide (10 µM) or with increasing concentrations of pomalidomide (0.5 and 2 μM) for 3 days. Cell lysates were prepared, separated by electrophoresis, and immunoblotted as indicated. Pre, pretreatment with.
We wanted to extend our observations to pomalidomide, a third in class IMiD. We treated OPM2 cells with lenalidomide and also with 2 low concentrations of pomalidomide and found that pomalidomide was even more potent than lenalidomide in its induction of immunoglobulin light chain dimerization as an indicator of increased intracellular H2O2 elevation, induction of ER stress (Bip accumulation), and Bim-mediated apoptosis (Figure 6D).
Direct TrxR/Trx inhibitors bypass CRBN requirement unlike lenalidomide for MM therapy
We hypothesized that if CRBN is required for lenalidomide to inhibit TrxR, using direct TrxR inhibitor or targeting Trx, the downstream effector could potentially obviate CRBN and in some cases overcome resistance. TrxR inhibitors and Trx inhibitors have previously been reported by others to have activity in MM cells.37-39
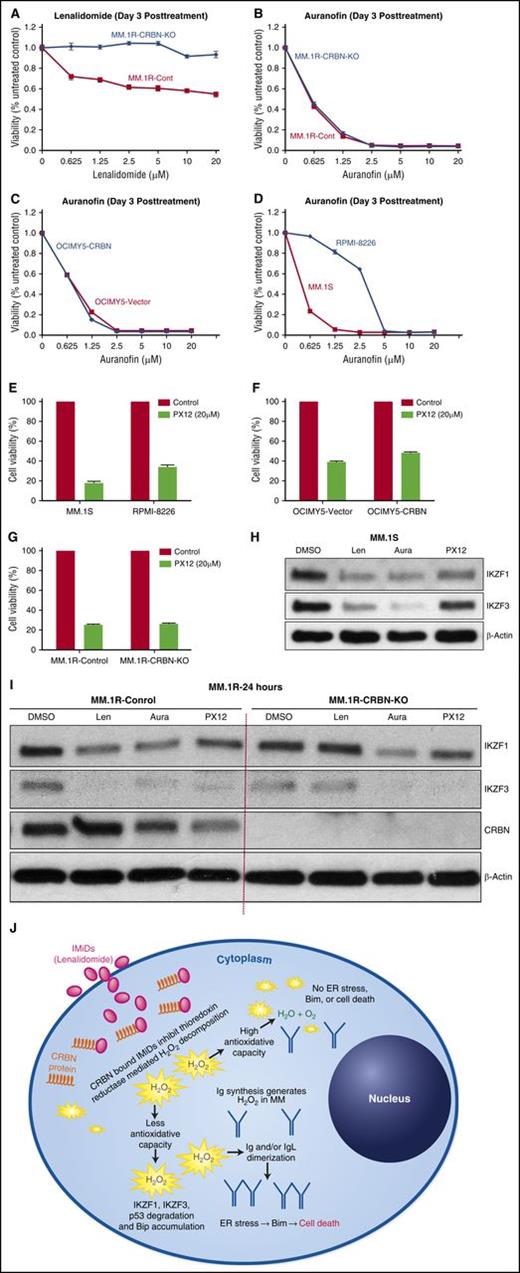
The direct TrxR inhibitor auranofin, an organic gold compound used in clinic as treatment of rheumatoid arthritis,40 was very effective in inducing cell death in MM cell lines, in a CRBN-independent fashion. Using MM.1R isogeneic cells with and without CRBN, and the CRBN-overexpressing OCIMY-5 cell line (transfected with wild-type CRBN), we showed that auranofin treatment caused cytotoxicity irrespective of CRBN expression, while lenalidomide was only effective in CRBN-expressing cells (Figure 7A-C). As with lenalidomide, the sensitivity of CRBN-expressing cell lines to auranofin correlated with their basal antioxidative capacity (MM.1S cells are sensitive as they have lower antioxidative capacity, whereas RPMI-8226 is resistant due to its high antioxidative capacity) (Figure 7D). We also found that targeting Trx with the direct inhibitor PX12 at a clinically achievable concentration of 20 µM41 overcame this cellular antioxidative capacity and was cytotoxic in both RPMI-8226 and MM.1S cell lines (Figure 7E); both express CRBN but have differential sensitivity to lenalidomide (Figure 1A-E). This effect was CRBN independent. Accordingly, isogenic MM cell lines with and without CRBN protein expression showed equal sensitivity to PX12 (Figure 7F-G). Clinically achievable concentrations of auranofin and PX12 induced cell signals similar to those of lenalidomide, including degradation of IKZF1, IKZF3 in MM.1S cell line (Figure 7H) and also in CRBN isogenic MM.1R cells (Figure 7I).
Direct TrxR/Trx inhibitors bypass CRBN requirement unlike lenalidomide for MM therapy. (A) MM.1R-control and MM.1R-CRBN-KO cells were seeded and incubated with lenalidomide at the indicated concentration for 3 days, and MTT assays were performed. (B) MM.1R-control and MM.1R-CRBN-KO cells were seeded and incubated with auranofin at the indicated concentration for 3 days, and MTT assays were performed. (C-D) OCIMY5-vector, OCIMY5-CRBN, MM.1S, and RPMI-8226 cells were seeded and incubated with auranofin at the indicated concentration for 3 days, and MTT assays were performed. (E-G) MM.1S, RPMI-8226, OCIMY5 (vector and CRBN), and MM.1R (control and KO-CRBN) cells were seeded and incubated with 20 µM concentration PX12 for 3 days, and cell viability was measured by MTT assay. Each experimental condition was performed in triplicate and repeated at least twice. (H-I) MM.1S and MM.1R (control and KO-CRBN) cell lines were treated with lenalidomide (10 μM), auranofin (1 μM), or PX12 (20 μM) for 24 hours. Cell lysates were prepared, separated by electrophoresis, and immunoblotted with the indicated antibodies. Blots are representative of 3 independent experiments. (J) Schematic representation of lenalidomide activity in MM. MM cells overproduce immunoglobulins, which generate high quantities of H2O2 through intramolecular and intermolecular disulfide bond formation. In addition, lenalidomide-bound CRBN also causes intracellular elevation of H2O2 by inhibiting intracellular TrxR. Cells with high antioxidative capacity are resistant to apoptosis from H2O2-mediated oxidative stress. For cells with lower antioxidative capacity, this stress leads to the degradation of IKZF1 and IKZF3, immunoglobulin dimerization, and subsequent ER stress–mediated, Bim-dependent apoptosis. Aura, auranofin; Len, lenalidomide.
Direct TrxR/Trx inhibitors bypass CRBN requirement unlike lenalidomide for MM therapy. (A) MM.1R-control and MM.1R-CRBN-KO cells were seeded and incubated with lenalidomide at the indicated concentration for 3 days, and MTT assays were performed. (B) MM.1R-control and MM.1R-CRBN-KO cells were seeded and incubated with auranofin at the indicated concentration for 3 days, and MTT assays were performed. (C-D) OCIMY5-vector, OCIMY5-CRBN, MM.1S, and RPMI-8226 cells were seeded and incubated with auranofin at the indicated concentration for 3 days, and MTT assays were performed. (E-G) MM.1S, RPMI-8226, OCIMY5 (vector and CRBN), and MM.1R (control and KO-CRBN) cells were seeded and incubated with 20 µM concentration PX12 for 3 days, and cell viability was measured by MTT assay. Each experimental condition was performed in triplicate and repeated at least twice. (H-I) MM.1S and MM.1R (control and KO-CRBN) cell lines were treated with lenalidomide (10 μM), auranofin (1 μM), or PX12 (20 μM) for 24 hours. Cell lysates were prepared, separated by electrophoresis, and immunoblotted with the indicated antibodies. Blots are representative of 3 independent experiments. (J) Schematic representation of lenalidomide activity in MM. MM cells overproduce immunoglobulins, which generate high quantities of H2O2 through intramolecular and intermolecular disulfide bond formation. In addition, lenalidomide-bound CRBN also causes intracellular elevation of H2O2 by inhibiting intracellular TrxR. Cells with high antioxidative capacity are resistant to apoptosis from H2O2-mediated oxidative stress. For cells with lower antioxidative capacity, this stress leads to the degradation of IKZF1 and IKZF3, immunoglobulin dimerization, and subsequent ER stress–mediated, Bim-dependent apoptosis. Aura, auranofin; Len, lenalidomide.
Discussion
IMiDs bind to CRBN, and this binding is a prerequisite for teratogenic and anti-MM activity.10,42 Here, we show that the presence of wild-type CRBN does not necessarily indicate sensitivity to IMiDs; its presence is necessary, but not sufficient. Our data showed that cellular antioxidative capacity determines lenalidomide sensitivity in MM cells expressing CRBN (Figure 7J). Accordingly, CRBN-expressing cells that have the highest capacity to decompose H2O2 were resistant to lenalidomide. Moreover, we developed a new method for determining total cellular antioxidative capacity, which can be used to predict which MM patients might benefit from IMiDs-based therapy. Furthermore, we discovered that the Trx/TrxR system-mediated intracellular H2O2 decomposition is inhibited by lenalidomide. Future studies are required to understand how it occurs and whether other peroxidases (eg, Trx-dependent peroxidases) are also inhibited by IMiDs. We are confident that CRBN retains IMiDs in intracellular compartments, induces chemical changes in the cellular redox status, and inhibits intracellular H2O2 decomposition. We and others have found that CRBN levels increase after lenalidomide treatment, so it likely follows that the intracellular concentration of IMiDs will increase with time. Future studies are required to elucidate how CRBN expression is required for IMiDs to optimally inhibit TrxR. These discoveries also support the use of TrxR and/or Trx inhibitors for future MM therapy,38 which can bypass the CRBN requirement. Moreover, how some MM cells with similar CRBN expression have different antioxidative capacities necessitates further studies to better understand MM redox balance. Finally, lenalidomide-mediated elevation of intracellular H2O2 is particularly toxic to MM because of their high production of immunoglobulin proteins. Our findings not only could help identify patients who are most likely to benefit from IMiDs but might also facilitate the design of new drugs that diminish cellular antioxidative capacity and might increase sensitivity to IMiDs.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Carole Viso and Tammy Brehm-Gibson for assistance with fluorescence-activated cell sorting analysis and Jessica Smith for help with experimental laboratory procedures. Bonnie Schimek helped edit figures and designed the graphic illustration. They thank the Center for Individualized Medicine, Mayo Clinic, for their funding contribution (S.S.) for developing biomarkers for predicting IMiDs response in MM. S.S. thanks the Mayo Clinic Specialized Program of Research Excellence Multiple Myeloma for Career Enhancement Award.
This work is supported by National Institutes of Health, National Cancer Institute grant R01 CA83724, Eastern Cooperative Oncology Group grant CA 21115T, and grants from Predolin Foundation, Mayo Clinic Cancer Center, and the Mayo Foundation.
Authorship
Contribution: S.S. designed and performed research, analyzed data, and wrote the manuscript; R.F. was the principal investigator, oversaw the project, and wrote the manuscript with S.S.; Y.X.Z. and C.-X.S. generated initial constructs for CRBN knockdown and overexpression and validated them in vitro; A.K.S., P.L.B., M.C., S.C.P., E.B., S.A.V.W., and G.J.A. provided necessary reagents and experimental advice.
Conflict-of-interest disclosure: S.S. and R.F. have filed a US patent professional application for the method of using cellular antioxidative capacity to predict clinical response to IMiD treatment. R.F. has received a patent for the prognostication of MM based on genetic categorization of the disease. R.F. has received consulting fees from Celgene, Genzyme, BMS, Bayer, Lilly, Onyx, Binding Site, Novartis, Sanofi, Millennium, and AMGEN. He also has sponsored research from Onyx and is a member of the Scientific Advisory Board of Adaptive Biotechnologies. R.F. is a Clinical Investigator of the Damon Runyon Cancer Research Fund.
Correspondence: Rafael Fonseca, Division of Hematology and Medical Oncology, Mayo Clinic, 13400 E Shea Blvd, Scottsdale, AZ 85259; e-mail: fonseca.rafael@mayo.edu.