In this issue of Blood, Sebastian et al provide evidence that the immunomodulatory drugs (IMiDs) inhibit intracellular decomposition of H2O2 resulting in oxidative stress and that the individual cellular antioxidative capacity of multiple myeloma (MM) cells predicts response to this widely used class of drugs.1
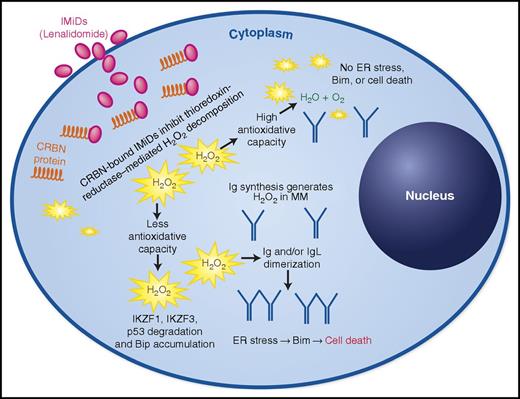
Cereblon-bound IMiDs cause oxidative stress by inhibition of key enzymes of intracellular H2O2 decomposition, leading to immunoglobulin dimerization, thereby resulting in proapoptotic endoplasmic reticulum (ER) stress. Measuring the antioxidative capacity can predict IMiD sensitivity in MM cells. Ig, immunoglobulin. See Figure 7J in the article by Sebastian et al that begins on page 991.
Cereblon-bound IMiDs cause oxidative stress by inhibition of key enzymes of intracellular H2O2 decomposition, leading to immunoglobulin dimerization, thereby resulting in proapoptotic endoplasmic reticulum (ER) stress. Measuring the antioxidative capacity can predict IMiD sensitivity in MM cells. Ig, immunoglobulin. See Figure 7J in the article by Sebastian et al that begins on page 991.
Alongside proteasome inhibitors (PIs), the thalidomide analogs, lenalidomide and pomalidomide, are key agents in the current treatment of MM and have an increasing role as powerful combination partners in the dawning era of immunotherapy. However, mechanisms of action and resistance are still not fully understood. It has recently been shown that the antitumor effect of all IMiDs requires binding to the ubiquitously expressed E3 ligase, cereblon (CRBN), leading to degradation of the zinc finger proteins IZKF1/Ikaros and IZKF3/Aiolos, mediating abrogation of IRF4 and Myc expression.2,3 Additional downstream sequelae of this IMiD/CRBN interaction include the destabilization of the CD147-MCT1 complex.4 As a result, either the absence of CRBN or the mutations within, for example, the IMiD-binding domain, lead to IMiD resistance.5,6 However, these studies cannot explain how MM cells become resistant to IMiDs despite the continued expression of wild-type CRBN.
Sebastian et al here turn to the long-known observation that thalidomide triggers oxidative stress.7 They exposed MM cell lines with different degrees of IMiD sensitivity to H2O2 and assessed their capacity to decompose this oxidative reagent by simply monitoring oxygen bubble formation. The inverse correlation observed between IMiD sensitivity and oxygen production led them to a more sophisticated analysis using flow cytometry, taking advantage of the autofluorescent properties of flavin adenine dinucleotide (FAD) and NADPH (reduced nicotinamide adenine dinucleotide phosphate) to assess cellular antioxidative capacity in a more standardized, quantitative assay. They demonstrated a striking inverse correlation between IMiD sensitivity and antioxidative capacity in both a variety of MM cell lines and primary patient samples (see figure).
Although this potential predictive biomarker will need to be prospectively validated in clinical trials, the authors went on to show that IMiDs increase peroxides in a CRBN-dependent manner by inhibiting the enzyme, thioredoxin reductase. Although the direct mechanism by which this enzyme is inhibited by IMiDs remains elusive, their further studies using direct inhibitors of thioredoxin reductase strongly suggest that this enzyme is a promising therapeutic target in MM, even more so as the efficacy of these direct inhibitors appeared to be CRBN independent.
Furthermore, the authors found that oxidative stress triggered by IMiDs increased the formation of dimers between immunoglobulin light chains. This excess of misfolded protein overrides the cellular chaperone capacity, thereby inducing ER stress, which finally results in the proapoptotic activation of the BH3-only protein, BIM (see figure). The recognition of this cascade is another important finding of this study as it finally identifies a mechanistic link between exposure to IMiDs and the induction of proteotoxic stress, a well-known “Achilles’ heel” of immunoglobulin-producing MM cells, and might help explain why IMiDs are more effective in plasma cell malignancies than in most other B-cell disorders.8
Thinking 1 step ahead, the other major therapeutic strategy in MM, the inhibition of proteasomes, is also well known to strongly increase proteotoxic stress by inhibiting the proteasomal degradation of misfolded proteins. It is therefore no surprise that not only were the authors not able to confirm the synergistic efficacy of combining IMiDs and PIs, as known from clinical trials, but also they found an augmented production of ER stress and proapoptotic sequelae. This observation is of great interest as there has been an ongoing debate regarding the mechanisms underlying this clinically observed synergy even though the main mechanism of action of IMiDs on MM cells, as previously understood, required the ubiquitination and proteasomal degradation of IZKF1/3, which would be abrogated by adding PIs.
As with any research that introduces new concepts, this work by Sebastian et al will stimulate several lines of investigation, which are likely to include elucidation of the mechanism by which thioredoxin reductase is inhibited by IMiD-bound CRBN, validation of antioxidative capacity as a robust predictive biomarker within clinical trials, the identification of clinical grade agents that directly target reactive oxygen decomposition to overcome or prevent IMiD resistance, and, finally, further investigation of this proposed cascade of mechanisms within cells of the (immune-) microenvironment.
Most importantly, it will be essential to clarify the clinical relevance of this proposed mechanism of resistance to IMiDs in the context of those of which we are currently aware, including loss of CRBN expression, the occurrence of CRBN-IZKF1/3 mutations, and differences in IMiD-induced immune cell activation.5,6,9
Conflict-of-interest disclosure: M.S.R. has received research funding and consultancy fees from Celgene, Novartis, Amgen, Morphosys, Takeda, and Janssen.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal