Key Points
Standardized flow cytometry allows highly sensitive MRD measurements in virtually all BCP-ALL patients.
If sufficient cells are measured (>4 million), flow cytometric MRD analysis is at least as sensitive as current PCR-based MRD methods.
Abstract
A fully-standardized EuroFlow 8–color antibody panel and laboratory procedure was stepwise designed to measure minimal residual disease (MRD) in B-cell precursor (BCP) acute lymphoblastic leukemia (ALL) patients with a sensitivity of ≤10−5, comparable to real-time quantitative polymerase chain reaction (RQ-PCR)–based MRD detection via antigen-receptor rearrangements. Leukocyte markers and the corresponding antibodies and fluorochromes were selected based on their contribution in separating BCP-ALL cells from normal/regenerating BCP cells in multidimensional principal component analyses. After 5 multicenter design-test-evaluate-redesign phases with a total of 319 BCP-ALL patients at diagnosis, two 8-color antibody tubes were selected, which allowed separation between normal and malignant BCP cells in 99% of studied patients. These 2 tubes were tested with a new erythrocyte bulk-lysis protocol allowing acquisition of high cell numbers in 377 bone marrow follow-up samples of 178 BCP-ALL patients. Comparison with RQ-PCR–based MRD data showed a clear positive relation between the percentage concordant cases and the number of cells acquired. For those samples with >4 million cells acquired, concordant results were obtained in 93% of samples. Most discordances were clarified upon high-throughput sequencing of antigen-receptor rearrangements and blind multicenter reanalysis of flow cytometric data, resulting in an unprecedented concordance of 98% (97% for samples with MRD < 0.01%). In conclusion, the fully standardized EuroFlow BCP-ALL MRD strategy is applicable in >98% of patients with sensitivities at least similar to RQ-PCR (≤10−5), if sufficient cells (>4 × 106, preferably more) are evaluated.
Introduction
Most current treatment protocols for B-cell precursor (BCP) acute lymphoblastic leukemia (ALL) include minimal residual disease (MRD) measurements, generally based on polymerase chain reaction (PCR) analysis of rearranged antigen receptor genes.1-3 Although flow cytometry (FCM) can be used for MRD detection as well,4-9 studies so far indicate that the specificity and sensitivity of FCM-MRD diagnostics are inferior to PCR-based MRD diagnostics.10-13 Nevertheless, we and others have recently shown that the use of 6- or 7-color immunostainings combined with the introduction of new markers and new marker combinations significantly improved FCM-MRD analysis in BCP-ALL patients.10,12 These improvements were particularly related to specificity, whereas the sensitivity still appeared to be lower than for the PCR-based methods. To further improve FCM-based MRD diagnostics, more objective and efficient discrimination of BCP-ALL cells from normal BCP cells and improved sample preparation procedures for acquisition of larger numbers of cells are a prerequisite.
Eight-color immunostainings may contribute to improve flow cytometric MRD detection in BCP-ALL patients. Recently, an 8-color antibody tube was developed in the ALL-REZ-BFM 2002 trial.14 This tube contained 7 antibodies (CD10, CD19, CD20, CD22, CD34, CD45, CD38) and the nucleic acid dye Syto41 and gave concordant MRD results with PCR-MRD data in 86.5% of samples. A Chinese study reported an 8-color antibody tube (CD10, CD19, CD20, CD34, CD38, CD45, CD58, plus CD66c or CD13/CD33 or NG2/CD15) with a sensitivity of 0.001% in 81.6% of patients.8 Shaver et al elegantly analyzed the relative contribution that each marker and/or pair of markers made to detect MRD15 and concluded that a single 8-color tube consisting of CD9, CD10, CD19, CD20, CD34, CD38, CD45, and CD58 could provide as much diagnostic utility as their existing 3-tube panel with 12 markers.
Within the EuroFlow Consortium (EU-FP6, LSHB-CT-2006-018708), we aimed to design standardized 8-color immunophenotyping protocols for multicenter MRD measurement in BCP-ALL and to improve the sensitivity of the assay to ≤10−5 (at least comparable to PCR). First, in order to select the most informative markers in distinguishing BCP-ALL from normal BCP cells, we applied novel software tools and principal component-based analyses.16,17 In each cycle of design-test-evaluate-redesign, the antibody tubes were tested on BCP-ALL samples and normal and/or regenerating bone marrow (BM), followed by assessment of the contribution of each antibody, until satisfactory results were obtained after 5 testing rounds. Second, a flow cytometric protocol for staining and acquisition of large numbers of cells (>4 million) was developed, allowing theoretical sensitivities of at least 0.001% (≤10−5). Finally, the selected antibody tubes and standardized laboratory procedures were prospectively validated on follow-up samples from BCP-ALL patients, using the EuroMRD PCR-MRD methods in parallel as gold standard.2
Materials and methods
BCP-ALL patients and normal controls
Data were collected in 7 EuroFlow centers. BM samples obtained from healthy donors or patients in whom no hematological malignancy could be detected (eg, BM samples submitted for lymphoma staging, neuroblastoma staging) were used as control BM for normal/reactive BCP cells. BM samples obtained from pediatric ALL patients after induction therapy (day 78 of therapy) or 1 year after stop of therapy, proven to be MRD-negative by real-time quantitative polymerase chain reaction (RQ-PCR) analysis, were used as a source of regenerating BCP. In the first part of the study (panel design and optimization), samples from 319 BCP-ALL patients, which were consecutively received during 5 design-test-evaluate-redesign phases (initial phase: n = 69; phase 1: n = 61; phase 2: n = 28; phase 3: n = 78; phase 4: n = 83), were included. In the second part of the study (MRD analysis), 377 follow-up samples obtained from 178 BCP-ALL patients (day 15: n = 111; day 33: n = 139; day 78: n = 107; other time points: n = 20) were included. Patient characteristics are summarized in Table 1. The institutional review board of each participating center approved this study, and informed consent for study participation was obtained from each patient and/or his/her legal guardian.
Patient characteristics
| . | Initial phase . | % . | Phase 1 . | % . | Phase 2 . | % . | Phase 3 . | % . | Phase 4 . | % . | MRD phase . | % . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Number | 69 | 61 | 28 | 78 | 83 | 178 | ||||||
| Age, y | ||||||||||||
| Median | 6 | 5 | 5 | 5 | 3 | 5 | ||||||
| Minimum | 0 | 1 | 1 | 0 | 0 | 0 | ||||||
| Maximum | 76 | 75 | 17 | 17 | 55 | 77 | ||||||
| Sex | ||||||||||||
| Male | 35 | 56 | 40 | 66 | 14 | 50 | 46 | 59 | 43 | 52 | 94 | 53 |
| Female | 27 | 44 | 21 | 34 | 14 | 50 | 32 | 41 | 40 | 48 | 84 | 47 |
| White blood cell count (×109/L) | ||||||||||||
| Median | 13.4 | 8.2 | 13.7 | 10.9 | 15.9 | 8.5 | ||||||
| Minimum | 0.4 | 0.8 | 1.2 | 0.7 | 0.5 | 0.5 | ||||||
| Maximum | 818 | 87.6 | 312 | 595 | 1250 | 900 | ||||||
| No data | 10 | 1 | 0 | 2 | 19 | 11 | ||||||
| Immunophenotype | ||||||||||||
| Pro-B-ALL | 8 | 13 | 2 | 3 | 4 | 14 | 5 | 6 | 6 | 7 | 5 | 3 |
| Common-ALL | 44 | 73 | 48 | 79 | 20 | 71 | 59 | 76 | 56 | 67 | 137 | 77 |
| Pre-B-ALL | 8 | 13 | 11 | 18 | 4 | 14 | 13 | 17 | 21 | 25 | 35 | 20 |
| No data | 7 | 0 | 0 | 1 | 0 | 1 | ||||||
| Genetic data* | ||||||||||||
| TEL-AML1 | 6 | 11 | 15 | 27 | 6 | 27 | 11 | 16 | 11 | 18 | 45 | 26 |
| BCR-ABL | 4 | 6 | 1 | 2 | 0 | 0 | 3 | 4 | 1 | 2 | 4 | 2 |
| E2A-PBX1 | 0 | 0 | 3 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 2 |
| MLL-AF4 | 4 | 6 | 1 | 2 | 2 | 7 | 3 | 4 | 4 | 6 | 3 | 2 |
| MLL other | 2 | 3 | 1 | 2 | 1 | 4 | 1 | 1 | 1 | 2 | 2 | 1 |
| Hyperdiplod | 14 | 25 | 27 | 48 | 6 | 26 | 26 | 37 | 22 | 37 | 67 | 43 |
| Hypodiploid | 0 | 0 | 1 | 2 | 1 | 4 | 1 | 1 | 1 | 2 | 2 | 1 |
| . | Initial phase . | % . | Phase 1 . | % . | Phase 2 . | % . | Phase 3 . | % . | Phase 4 . | % . | MRD phase . | % . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Number | 69 | 61 | 28 | 78 | 83 | 178 | ||||||
| Age, y | ||||||||||||
| Median | 6 | 5 | 5 | 5 | 3 | 5 | ||||||
| Minimum | 0 | 1 | 1 | 0 | 0 | 0 | ||||||
| Maximum | 76 | 75 | 17 | 17 | 55 | 77 | ||||||
| Sex | ||||||||||||
| Male | 35 | 56 | 40 | 66 | 14 | 50 | 46 | 59 | 43 | 52 | 94 | 53 |
| Female | 27 | 44 | 21 | 34 | 14 | 50 | 32 | 41 | 40 | 48 | 84 | 47 |
| White blood cell count (×109/L) | ||||||||||||
| Median | 13.4 | 8.2 | 13.7 | 10.9 | 15.9 | 8.5 | ||||||
| Minimum | 0.4 | 0.8 | 1.2 | 0.7 | 0.5 | 0.5 | ||||||
| Maximum | 818 | 87.6 | 312 | 595 | 1250 | 900 | ||||||
| No data | 10 | 1 | 0 | 2 | 19 | 11 | ||||||
| Immunophenotype | ||||||||||||
| Pro-B-ALL | 8 | 13 | 2 | 3 | 4 | 14 | 5 | 6 | 6 | 7 | 5 | 3 |
| Common-ALL | 44 | 73 | 48 | 79 | 20 | 71 | 59 | 76 | 56 | 67 | 137 | 77 |
| Pre-B-ALL | 8 | 13 | 11 | 18 | 4 | 14 | 13 | 17 | 21 | 25 | 35 | 20 |
| No data | 7 | 0 | 0 | 1 | 0 | 1 | ||||||
| Genetic data* | ||||||||||||
| TEL-AML1 | 6 | 11 | 15 | 27 | 6 | 27 | 11 | 16 | 11 | 18 | 45 | 26 |
| BCR-ABL | 4 | 6 | 1 | 2 | 0 | 0 | 3 | 4 | 1 | 2 | 4 | 2 |
| E2A-PBX1 | 0 | 0 | 3 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 2 |
| MLL-AF4 | 4 | 6 | 1 | 2 | 2 | 7 | 3 | 4 | 4 | 6 | 3 | 2 |
| MLL other | 2 | 3 | 1 | 2 | 1 | 4 | 1 | 1 | 1 | 2 | 2 | 1 |
| Hyperdiplod | 14 | 25 | 27 | 48 | 6 | 26 | 26 | 37 | 22 | 37 | 67 | 43 |
| Hypodiploid | 0 | 0 | 1 | 2 | 1 | 4 | 1 | 1 | 1 | 2 | 2 | 1 |
Genetic data were not available for all patients; percentages refer to positive patients per all analyzed patients.
Immunophenotyping MRD panel design
First, BM samples obtained from 69 BCP-ALL patients at diagnosis were stained with the EuroFlow BCP-ALL antibody panel (23 different antibodies in four 8-color tubes).18 The subsequently designed and optimized MRD tubes were tested during phase 1 to 4 on diagnostic BM samples from BCP-ALL patients using the standardized EuroFlow sample preparation and instrument setup protocols.18,19 Data were analyzed using Infinicyt software by comparing BCP-ALL cells with the nearest normal/reactive BCP subsets using automated population separator (APS) plots (see supplemental Methods, available on the Blood Web site) as illustrated in Figure 1 and supplemental Figures 1 and 2. Regenerating BCP cells from 6 T-ALL patients were used as an additional negative control (supplemental Figure 3).
Data analysis strategy used to optimize the antibody panel for distinguishing between BCP-ALL cells and their nearest normal/reactive BCP counterpart. First, multiple normal/reactive BM samples and/or regenerating BM samples were merged (phase 1: n = 7; phase 2: n = 11; phase 3: n = 14; phase 4: n = 10) and CD19-positive B cells were selected. These were subdivided into 4 B-cell subsets, based on the backbone markers (CD19/CD10/CD20/CD34/CD45): CD34+ pre–B-I cells (light green), CD34−/CD10+/CD20− to dim pre–B-II/immature cells (dark blue), CD34−/CD10+/CD20+ immature/transitional B cells (light blue), and CD34−/CD10−/CD20+ mature B cells (dark green). Dot plots of CD34 vs CD10 (A) and CD10 vs CD20 (B) are shown. The 1 standard deviation (SD) (dashed line) and 2 SD lines (solid line) of the 2 most immature BCP subsets (pre–B-I [light green] and pre–B-II/immature [dark blue]) were displayed in an APS view, which was subsequently fixed (supervised; C). Each individual BCP-ALL case was added to the fixed APS plot, and the normal BCP population nearest to each of the BCP-ALL populations was defined (D). The BCP-ALL cells and nearest normal BCP subset were then visualized in a separate (nonfixed and balanced) APS plot, 1 using the backbone markers only (E) and 1 using all 8 markers (F), by plotting the 1 SD curve and 2 SD curves of the 2 populations. To prevent an influence of the number of cells on the principal component analysis (PCA), we opted to use a balanced PCA, implying a fixed ratio between normal and pathological events. Finally, the separation between the 2 populations was scored based on: no overlap between 2 SD curves: 3 points; overlap of the 2 SD curves: 2 points; overlap of the 2 SD and the 1 SD curve: 1 point; overlap of both 1 SD curves: 0 points. An example of this scoring is shown in supplemental Figure 1. It should be noted that the above described strategy was only used for optimizing the antibody panel for the BCP-ALL MRD tubes and not for actual MRD analyses.
Data analysis strategy used to optimize the antibody panel for distinguishing between BCP-ALL cells and their nearest normal/reactive BCP counterpart. First, multiple normal/reactive BM samples and/or regenerating BM samples were merged (phase 1: n = 7; phase 2: n = 11; phase 3: n = 14; phase 4: n = 10) and CD19-positive B cells were selected. These were subdivided into 4 B-cell subsets, based on the backbone markers (CD19/CD10/CD20/CD34/CD45): CD34+ pre–B-I cells (light green), CD34−/CD10+/CD20− to dim pre–B-II/immature cells (dark blue), CD34−/CD10+/CD20+ immature/transitional B cells (light blue), and CD34−/CD10−/CD20+ mature B cells (dark green). Dot plots of CD34 vs CD10 (A) and CD10 vs CD20 (B) are shown. The 1 standard deviation (SD) (dashed line) and 2 SD lines (solid line) of the 2 most immature BCP subsets (pre–B-I [light green] and pre–B-II/immature [dark blue]) were displayed in an APS view, which was subsequently fixed (supervised; C). Each individual BCP-ALL case was added to the fixed APS plot, and the normal BCP population nearest to each of the BCP-ALL populations was defined (D). The BCP-ALL cells and nearest normal BCP subset were then visualized in a separate (nonfixed and balanced) APS plot, 1 using the backbone markers only (E) and 1 using all 8 markers (F), by plotting the 1 SD curve and 2 SD curves of the 2 populations. To prevent an influence of the number of cells on the principal component analysis (PCA), we opted to use a balanced PCA, implying a fixed ratio between normal and pathological events. Finally, the separation between the 2 populations was scored based on: no overlap between 2 SD curves: 3 points; overlap of the 2 SD curves: 2 points; overlap of the 2 SD and the 1 SD curve: 1 point; overlap of both 1 SD curves: 0 points. An example of this scoring is shown in supplemental Figure 1. It should be noted that the above described strategy was only used for optimizing the antibody panel for the BCP-ALL MRD tubes and not for actual MRD analyses.
Immunophenotyping MRD analyses
The finally selected BCP-ALL MRD tubes were evaluated on BM samples obtained during follow-up of BCP-ALL patients, using an optimized bulk-lysis protocol (see “Results”).20 BM samples were processed according to this new EuroFlow bulk-lysis protocol and subsequently stained using the regular EuroFlow protocol.19 MRD analyses and interpretation were performed locally, and data were subsequently sent to the BCP-ALL-MRD coordinator for central evaluation. Initial FCM-MRD data analysis was performed using 2-dimensional dot plots for sequential gating of BCP-ALL cells, comparable to previous studies using 4 to 6 color stainings.10,12 For this study, we provisionally defined a minimum of 10 clustered events to consider a sample as MRD positive (lower limit of detection, LOD) and a minimum of 40 clustered events for accurate quantitation of the MRD level (lower limit of quantitation, LLOQ).21 Interlaboratory variability in data analysis was evaluated as described in the supplemental Methods and supplemental Figure 4.
RQ-PCR–based MRD analyses
MRD levels were routinely determined by RQ-PCR analysis of rearranged immunoglobulin and/or T-cell receptor (TR) gene rearrangements in laboratories participating in the quality control rounds of the EuroMRD network (see www.EuroMRD.org).3,22-26 RQ-PCR data, performed in triplicate, were analyzed according to the EuroMRD guidelines, using the criteria to prevent false-negative MRD results.2 Because application of these criteria might result in some false-positive RQ-PCR results,2 we performed next-generation sequencing (NGS) to confirm or exclude the presence of MRD in discordant samples considered positive by RQ-PCR but negative by FCM.
NGS-based MRD analyses
NGS was generally performed as described previously.27 Briefly, depending on the IGH, TRG, and/or TRD rearrangements applied as MRD targets in the RQ-PCR analysis, we performed a targeted approach: the follow-up samples were amplified using the multiplex primer set(s) of the relevant immunoglobulin/TR locus only, and data analysis was focused on the specific junctional region sequence (ie, the 1 used for RQ-PCR analysis). The primers for TCRG were newly designed (supplemental Table 1), and individual primer combinations from multiplex PCR were tested for sensitivity using NGS for diluted diagnostic ALL samples from patients with respective V and Jgamma segment combinations, all reaching the sensitivity of 10−5. All data were finally scored as either MRD-positive or MRD-negative.
Results
Design and optimization of 8-color MRD labeling for BCP-ALL
In the initial phase, 5 antibodies (CD19, CD45, CD34, CD10, and CD20) were upfront selected as backbone markers because they allow appropriate BCP gating as well as characterization of several BCP subpopulations and are known to allow discrimination between normal BCP and BCP-ALL cells.10,28-30 To evaluate which other markers could contribute to optimal separation of BCP-ALL cells from normal/reactive BCP cells, the EuroFlow BCP-ALL diagnosis panel18 was applied to 69 BCP-ALL patients as well as to normal/reactive BM samples. Based on principal component analysis (visualized through APS plots)16,17,19 of the BCP-ALL cells vs normal/reactive BCP cells (analyzed per tube), CD9, CD123, CD66c, CD81, CD24, and CD10 appeared to be markers that were most frequently differentially expressed (see supplemental Figure 5). These markers were combined with the 5 backbone markers listed above and complemented with terminal deoxynucleotidyltransferase (TdT) and CD58, both previously reported to be of relevance for BCP-ALL MRD analyses.10,28,29,31 The remaining open position was filled in with surface membrane IgKappa/IgLambda (SmIgK/L), as a potential exclusion maker for more mature BCPs. Fluorochrome positions were primarily determined based on the position of the involved markers in the EuroFlow BCP-ALL panel.18
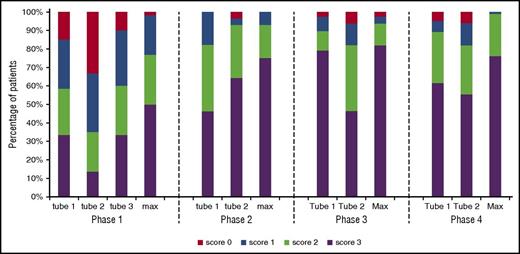
The resulting 3 phase 1 MRD tubes (Table 2) were subsequently tested on 61 consecutive BCP-ALL patients at diagnosis, and the discriminatory power was evaluated by comparing the leukemic BCP with the nearest normal BCP subset in APS plots. Whereas both tube 1 and tube 3 gave good/fair separation in ∼60% of cases, tube 2 was clearly less informative (fair/good separation in <35% of cases) (Figure 2). When the tube providing the best separation for each patient was selected, good/fair separation was observed in 77% of cases. Considering only tube 1 and 3, good/fair separation was still observed in 71% of cases. These data indicate that tube 1 and 3 had complementary value and confirm the limited value of tube 2.
Development and design of the EuroFlow BCP-ALL MRD panelTF2-1
| Phase . | Violet laser . | Blue laser . | Red laser . | |||||
|---|---|---|---|---|---|---|---|---|
| Initial* . | PB . | PO . | FITC . | PE . | PerCP Cy5.5 . | PECy7 . | APC . | APC H7 . |
| CD20 | CD45 | CD58 | CD66c | CD34 | CD19 | CD10 | CD38 | |
| CD9 | CD45 | TdT | CD13 | CD34 | CD19 | CD22 | CD24 | |
| CD21 | CD45 | CD15/CD65 | NG2 | CD34 | CD19 | CD123 | CD81 | |
| 1 | PB | PO | FITC | PE | PerCP Cy5.5 | PECy7 | APC | APC H7 |
| CD20 | CD45 | CD58 | CD66c | CD34 | CD19 | CD10 | CD38 | |
| CD20 | CD45 | TdT | SmIgK/L | CD34 | CD19 | CD10 | CD24 | |
| CD20 | CD45 | CD9 | CD123 | CD34 | CD19 | CD10 | CD81 | |
| 2 | PB | PO | FITC | PE | PerCP Cy5.5 | PE Cy7 | APC | APC H7 |
| CD20 | CD45 | CD9 | CD22 | CD34 | CD19 | CD10 | CD38 | |
| CD20 | CD45 | CD123 | CD66c | CD34 | CD19 | CD10 | CD24 | |
| 3 | PB | PO | FITC | PE | PerCP Cy5.5 | PE Cy7 | APC | APC C750 |
| CD20 | CD45 | CD9 (ML13) | CD66c/CD123 | CD34 | CD19 | CD10 | CD38 | |
| CD20 | CD45 | CD58 | CD22 | CD34 | CD19 | CD10 | CD81 | |
| 4 | PB | PO | FITC | PE | PerCP Cy5.5 | PE Cy7 | APC | APC C750 |
| CD20 | CD45 | CD81 | CD66c/CD123 | CD34 | CD19 | CD10 | CD38 | |
| CD20 | CD45 | CD81 | CD73/CD304 | CD34 | CD19 | CD10 | CD38 | |
| Phase . | Violet laser . | Blue laser . | Red laser . | |||||
|---|---|---|---|---|---|---|---|---|
| Initial* . | PB . | PO . | FITC . | PE . | PerCP Cy5.5 . | PECy7 . | APC . | APC H7 . |
| CD20 | CD45 | CD58 | CD66c | CD34 | CD19 | CD10 | CD38 | |
| CD9 | CD45 | TdT | CD13 | CD34 | CD19 | CD22 | CD24 | |
| CD21 | CD45 | CD15/CD65 | NG2 | CD34 | CD19 | CD123 | CD81 | |
| 1 | PB | PO | FITC | PE | PerCP Cy5.5 | PECy7 | APC | APC H7 |
| CD20 | CD45 | CD58 | CD66c | CD34 | CD19 | CD10 | CD38 | |
| CD20 | CD45 | TdT | SmIgK/L | CD34 | CD19 | CD10 | CD24 | |
| CD20 | CD45 | CD9 | CD123 | CD34 | CD19 | CD10 | CD81 | |
| 2 | PB | PO | FITC | PE | PerCP Cy5.5 | PE Cy7 | APC | APC H7 |
| CD20 | CD45 | CD9 | CD22 | CD34 | CD19 | CD10 | CD38 | |
| CD20 | CD45 | CD123 | CD66c | CD34 | CD19 | CD10 | CD24 | |
| 3 | PB | PO | FITC | PE | PerCP Cy5.5 | PE Cy7 | APC | APC C750 |
| CD20 | CD45 | CD9 (ML13) | CD66c/CD123 | CD34 | CD19 | CD10 | CD38 | |
| CD20 | CD45 | CD58 | CD22 | CD34 | CD19 | CD10 | CD81 | |
| 4 | PB | PO | FITC | PE | PerCP Cy5.5 | PE Cy7 | APC | APC C750 |
| CD20 | CD45 | CD81 | CD66c/CD123 | CD34 | CD19 | CD10 | CD38 | |
| CD20 | CD45 | CD81 | CD73/CD304 | CD34 | CD19 | CD10 | CD38 | |
Markers that were changed as compared with the previous panel are marked in boldface.
Only tubes 1, 3, and 4 of the EuroFlow BCP-ALL diagnosis panel were evaluated, because most markers in tube 2 (Cyµ, SmIgK, SmIgL, SmIgM) were not expected to contribute to separation of normal and malignant BCP cells.
Power to distinguish BCP-ALL cells from their nearest normal BCP counterpart using the EuroFlow BCP-ALL MRD tubes. Data reflect the percentage of patients that reached the specified score, obtained as described in supplemental Figure 1. Briefly, for each patient, BCP-ALL cells and their nearest normal BCP subset were visualized in a (nonfixed and balanced) APS plot showing the median, 1 SD curves, and 2 SD curves for both populations. Each patient was subsequently scored as follows: no overlap between 2 SD curves: 3 points; overlap of the 2 SD curves: 2 points; overlap of the 2 SD and the 1 SD curve: 1 point; overlap of both 1 SD curves: 0 points. Max, the maximal score of the individual tubes. Phase 1: 7 normal/reactive BM samples and 61 BCP-ALL patients; phase 2: 7 normal BM samples, 4 regenerating BM samples, and 28 BCP-ALL patients; phase 3: 5 normal/reactive BM samples, 9 regenerating BM samples, and 78 BCP-ALL patients; phase 4: 10 normal/reactive BM samples and 83 BCP-ALL patients.
Power to distinguish BCP-ALL cells from their nearest normal BCP counterpart using the EuroFlow BCP-ALL MRD tubes. Data reflect the percentage of patients that reached the specified score, obtained as described in supplemental Figure 1. Briefly, for each patient, BCP-ALL cells and their nearest normal BCP subset were visualized in a (nonfixed and balanced) APS plot showing the median, 1 SD curves, and 2 SD curves for both populations. Each patient was subsequently scored as follows: no overlap between 2 SD curves: 3 points; overlap of the 2 SD curves: 2 points; overlap of the 2 SD and the 1 SD curve: 1 point; overlap of both 1 SD curves: 0 points. Max, the maximal score of the individual tubes. Phase 1: 7 normal/reactive BM samples and 61 BCP-ALL patients; phase 2: 7 normal BM samples, 4 regenerating BM samples, and 28 BCP-ALL patients; phase 3: 5 normal/reactive BM samples, 9 regenerating BM samples, and 78 BCP-ALL patients; phase 4: 10 normal/reactive BM samples and 83 BCP-ALL patients.
To evaluate the relevance of each individual marker in discriminating BCP-ALL cells from normal/reactive BCP cells, those markers that received a weight over 10% in the first or second principal component in an APS view of the nearest normal BCP cells and the BCP-ALL cells were selected. CD66c (80% of cases), CD9 (63%), and CD123 (55%) contributed most frequently.
Based on these phase 1 BCP-ALL results, the panel was redesigned: CD58, TdT, SmIgK/L, and CD81 were (at least provisionally) excluded, whereas CD22, which might be important for gating of B-cells in case of CD19-targeting therapies, was included (Table 2).
The phase 2 BCP-ALL MRD tubes were evaluated on diagnostic samples from 28 consecutive BCP-ALL patients. Good/fair separation between BCP-ALL cells and their nearest normal/reactive BCP counterpart was possible in ∼75% of cases in both tubes (Figure 2), showing significant improvement over the phase 1 tubes. If the best score of both tubes was used for each case, over 85% of BCP-ALL cases showed good/fair separation from the corresponding normal BCP subset, and in only 3 cases (12%) separation was poor.
During phase 2, additional studies were performed: (1) Because of nonoptimal (relatively weak) CD9 (MEM61) staining, another CD9 clone (ML13) was evaluated with much stronger results; (2) Two newly available fluorochromes (APC-C750 and APC-A750) showed lower background than APC-H7 (less binding to apoptotic cells; no binding to monocytes); (3) More detailed evaluation of the usefulness of CD81 vs CD24 (reevaluation of phase 1 data) showed that CD81 was more frequently differentially expressed between normal/reactive BCP cells and BCP-ALL cells and showed that CD81 in combination with only the backbone markers resulted in a higher percentage of cases with good separation than CD24 did (31% vs 20%); (4) Because CD66c and CD123 are both virtually negative on normal/reactive BCP cells, we tested whether these markers could be combined in the phycoerythrin (PE) channel and concluded that background levels were not affected by combining these 2 markers (data not shown). Based on these data, the new CD9 clone was included in the MRD panel, APC-H7 was replaced by APC-C750/A750, CD24 was replaced by CD81, and CD66c and CD123 were combined into 1 fluorescence channel. The open fluorescein isothiocyanate (FITC) position was used for further evaluation of CD58. The combined data provided the phase-3 BCP-ALL MRD tubes (Table 2).
The 2 phase 3 BCP-ALL MRD tubes were evaluated on 78 BCP-ALL patients. Overall, tube 1 resulted in good/fair separation in 90% of cases, whereas this was achieved in 82% of cases for tube 2 (Figure 2). In the 3 cases for which tube 1 did not result in good separation, normal/reactive BCP and BCP-ALL cells could be separated in tube 2, mainly due to differential expression of CD81. Further evaluation showed that CD38 (∼35% of cases), CD66c/CD123 (∼30%), and CD81 (∼19%) improved the separation between normal/reactive and malignant BCP cells as compared with the 5 backbone markers only, whereas CD9, CD58, and CD22 had no or limited additional value. CD9 in tube 1 was therefore replaced by CD81-FITC (which demonstrated equally good staining patterns as CD81-APC-C750) and tube 2 was discarded.
Because in a few cases the evaluated MRD tubes still did not yet result in sufficient separation between normal/reactive and malignant BCP cells, we evaluated several other markers reported to be of potential interest for MRD analysis (eg, CD44-FITC, CD27-PE, CD164-FITC, CD73-PE, CD49f-FITC, CD200-PE, CD86-FITC, and Drebrin-PE).32-36 Based on initial testing on diagnostic BCP-ALL samples, CD73 and CD304 appeared to be most promising based on the level and frequency of overexpression (∼20% for CD73 and ∼40% for CD304) and their stability during follow-up (data not shown). Because it appeared not to be possible to combine these 2 markers with CD66c and CD123 in a single fluorescence channel (due to too high background levels), a second tube was designed; this tube was identical to tube 1 but with CD73/CD304 instead of CD66c/CD123 in the PE channel (Table 2).
The 2 phase 4 BCP-ALL MRD tubes were run on 83 consecutive diagnostic BCP-ALL samples. Overexpression of CD66c/CD123 or CD73/CD304 was observed in 45% and 46% of cases, respectively; 31% of cases did not show overexpression of either CD66c/CD123 or CD73/CD304. Tube 1 resulted in good/fair separation in 89% of cases, whereas this was attained in 82% of cases for tube 2 (Figure 2). If the best score of both tubes was used, 99% of cases showed good/fair separation between the BCP-ALL cells and the nearest normal/reactive BCP subset. Therefore, tube 1 and tube 2 were complementary to each other, and one might either decide at diagnosis which tube is best for monitoring the particular patient or use both tubes to have an extra internal control and more precise measurements. These 2 optimized tubes were considered to be final and ready for further evaluation in follow-up samples of BCP-ALL patients.
Optimization of the flow cytometric MRD sample preparation protocol
We aimed for a sensitivity of ≤10−5, at least comparable to the sensitivity reached in RQ-PCR–based MRD analysis. If a cluster of 10 to 40 BCP-ALL cells should be present to consider a sample as positive, one should acquire at least 4 million cells in order to reach the required sensitivity. Because the cellularity of BM samples obtained during the early phases of treatment is frequently low,37 staining whole BM samples using the regular EuroFlow protocols would not allow acquisition of millions of cells. We therefore designed and tested a new EuroFlow erythrocyte bulk-lysis procedure: sufficiently large volumes of BM, ie, containing >10 million cells, are lysed, and the leukocytes are subsequently resuspended in a small volume of washing buffer. This new protocol allowed staining of 10 million cells in 100 µL of cell suspension per tube (supplemental Table 2). Evaluation of this new protocol showed that the percentage of doublets did not increase, that the number of evaluable leukocytes increased significantly, and that there were no major differences in cellular composition as compared with the regular EuroFlow staining protocol (Figure 3). Given the large increase in the number of cells stained with this new approach, all antibody titers were reevaluated; modifications appeared not to be necessary.
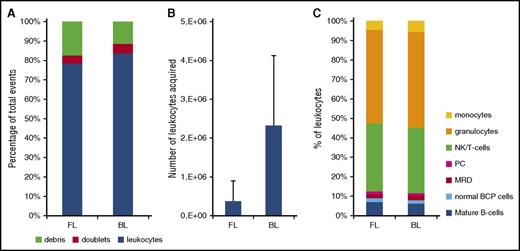
Evaluation of the EuroFlow bulk-lysis protocol. For reasons of comparison, each of the BM samples (day 15: n = 15; day 33: n = 15; day 78: n = 12) was processed according to the standard EuroFlow protocol (FL) and in parallel according to the EuroFlow bulk-lysis protocol (BL). (A) Number of leukocytes, debris, and doublets, calculated as percentage of acquired events. Using the bulk-lysis method, significantly less debris (P = .032 by paired Student t test) and significantly more leukocytes (P = .03 by paired Student t test) were measured. There were no significant differences between the 2 methods for the percentage of doublets. (B) Absolute number of leukocytes acquired. Using BL, on average 12-fold more leukocytes could be acquired (P < .0001). Please note that we included relatively many day 15 samples in order to be able to evaluate the impact of the 2 methods on the MRD levels as well. However, these day 15 samples generally have a very low white blood cell count; consequently, the number of leukocytes acquired after BL is still relatively low in a subset of samples. (C) Distribution of leukocyte subpopulations, defined as percentage of leukocytes. By paired Student t test (2-sided), small but statistically significant differences were observed for T/NK cells (mean: 24% vs 26%, P = .0047), granulocytes (mean: 33% vs 38%, P < .001), and monocytes (mean: 3.2% vs 4.5%, P < .001), whereas no significant differences were observed for the remaining populations. Of note, in 2 samples, MRD was only detected using the bulk-lysis method (0.013% and 0.018%) but not using the whole BM method. In the 11 samples MRD positive by both methods, MRD levels were not significantly different from each other (paired Student t test: P = .30), with mean values of 6.3% and 6.7% by whole BM and bulk-lysed BM method, respectively. Correlation analysis showed a Spearman r of 0.964 (95% confidence interval: 0.857-0.991; P < .0001). PC, plasma cells.
Evaluation of the EuroFlow bulk-lysis protocol. For reasons of comparison, each of the BM samples (day 15: n = 15; day 33: n = 15; day 78: n = 12) was processed according to the standard EuroFlow protocol (FL) and in parallel according to the EuroFlow bulk-lysis protocol (BL). (A) Number of leukocytes, debris, and doublets, calculated as percentage of acquired events. Using the bulk-lysis method, significantly less debris (P = .032 by paired Student t test) and significantly more leukocytes (P = .03 by paired Student t test) were measured. There were no significant differences between the 2 methods for the percentage of doublets. (B) Absolute number of leukocytes acquired. Using BL, on average 12-fold more leukocytes could be acquired (P < .0001). Please note that we included relatively many day 15 samples in order to be able to evaluate the impact of the 2 methods on the MRD levels as well. However, these day 15 samples generally have a very low white blood cell count; consequently, the number of leukocytes acquired after BL is still relatively low in a subset of samples. (C) Distribution of leukocyte subpopulations, defined as percentage of leukocytes. By paired Student t test (2-sided), small but statistically significant differences were observed for T/NK cells (mean: 24% vs 26%, P = .0047), granulocytes (mean: 33% vs 38%, P < .001), and monocytes (mean: 3.2% vs 4.5%, P < .001), whereas no significant differences were observed for the remaining populations. Of note, in 2 samples, MRD was only detected using the bulk-lysis method (0.013% and 0.018%) but not using the whole BM method. In the 11 samples MRD positive by both methods, MRD levels were not significantly different from each other (paired Student t test: P = .30), with mean values of 6.3% and 6.7% by whole BM and bulk-lysed BM method, respectively. Correlation analysis showed a Spearman r of 0.964 (95% confidence interval: 0.857-0.991; P < .0001). PC, plasma cells.
Evaluation of the EuroFlow BCP-ALL MRD tube
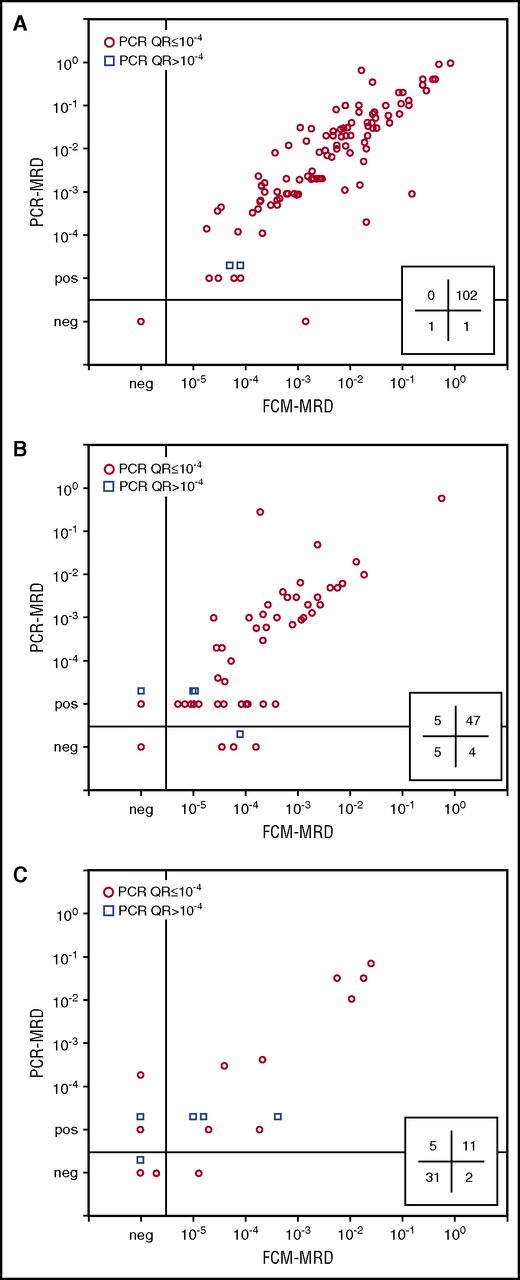
To evaluate whether the newly designed high-throughput EuroFlow BCP-ALL MRD strategy performed well, we tested the final MRD tubes on follow-up samples of BCP-ALL patients. Based on the immunophenotype of the BCP-ALL cells at diagnosis, 1 MRD tube was selected and subsequently used for MRD evaluation. First, flow cytometric MRD data obtained in 178 BCP-ALL patients were compared with routinely obtained PCR-MRD data. As shown in Figure 4A, the concordance between the FCM-MRD data and PCR-based MRD data was highly dependent on the number of cells acquired by FCM. In addition, the sensitivity of FCM-MRD (percentage samples positive by both FCM and PCR relative to samples positive by PCR) significantly increased when higher cell numbers were acquired (Figure 4B). Therefore, only samples in which MRD could clearly be detected by FCM-MRD or samples that had sufficient cells acquired for reaching a sensitivity of ≤10−5 were included in the subsequent analyses. Based on an LLOQ of 40 events, at least 4 × 106 cells should be acquired, which was possible in 227 out of 377 samples (60%). FCM-MRD data obtained in these patients were comparable to PCR-based MRD results in 93% of samples (Figure 5; Table 3). All but 1 of the 17 discordant samples (7 FCM+/PCR− and 10 FCM−/PCR+) had MRD levels <10−4 (supplemental Table 2). Bland-Altman analysis showed higher PCR-based MRD values with a mean difference of 0.34 log or a factor of 2.2 (supplemental Figure 6).
Performance of FCM-MRD vs PCR-based MRD is dependent on the number of acquired cells. (A) The percentage of discordant cases by FCM-MRD and PCR-MRD is shown for individual time points (day 15, day 33, and day 78) as well as for all samples together. Data are presented for variable numbers of acquired cells: all samples (independent of cell number) and samples with at least 1, 2, 3, 4, or 5 million cells acquired. (B) The sensitivity of FCM-MRD relative to PCR-MRD is shown for individual time points (day 15, day 33, and day 78) as well as for all samples together. Data are presented for variable numbers of acquired cells: all samples (independent of cell number; n = 377) and samples with at least 1 (n = 330), 2 (n = 287), 3 (n = 255), 4 (n = 227), or 5 million cells (n = 191) acquired. Sensitivity is calculated as the number of samples positive by both FCM and PCR divided by the total number of samples positive by PCR (ie, the reference method).
Performance of FCM-MRD vs PCR-based MRD is dependent on the number of acquired cells. (A) The percentage of discordant cases by FCM-MRD and PCR-MRD is shown for individual time points (day 15, day 33, and day 78) as well as for all samples together. Data are presented for variable numbers of acquired cells: all samples (independent of cell number) and samples with at least 1, 2, 3, 4, or 5 million cells acquired. (B) The sensitivity of FCM-MRD relative to PCR-MRD is shown for individual time points (day 15, day 33, and day 78) as well as for all samples together. Data are presented for variable numbers of acquired cells: all samples (independent of cell number; n = 377) and samples with at least 1 (n = 330), 2 (n = 287), 3 (n = 255), 4 (n = 227), or 5 million cells (n = 191) acquired. Sensitivity is calculated as the number of samples positive by both FCM and PCR divided by the total number of samples positive by PCR (ie, the reference method).
Comparison of MRD data obtained by 8-color EuroFlow FCM and routinely obtained molecular MRD data. Flow cytometric MRD data were compared with molecular MRD data and are shown for samples obtained at day 15 (A), day 33 (B), or day 78 (C). In the lower right part of each panel, the number of FCM−/PCR+, FCM+/PCR+, FCM+/PCR−, and FCM−/PCR− is indicated. Only samples in which MRD could clearly be detected by FCM-MRD or samples which had sufficient cells acquired for reaching a sensitivity of ≤10−5 (ie, ≥ 4 × 106 cells acquired) were included in the analyses. Based on a LOD of 10 events, the sensitivity of FCM-MRD was 2.5 × 10−6; the quantitative range (based on a LLOQ of 40 events) is 10−5. Consequently, FCM-MRD data between 2.5 × 10−6 and 10−5 should be considered positive, but are below the limit of quantitation.
Comparison of MRD data obtained by 8-color EuroFlow FCM and routinely obtained molecular MRD data. Flow cytometric MRD data were compared with molecular MRD data and are shown for samples obtained at day 15 (A), day 33 (B), or day 78 (C). In the lower right part of each panel, the number of FCM−/PCR+, FCM+/PCR+, FCM+/PCR−, and FCM−/PCR− is indicated. Only samples in which MRD could clearly be detected by FCM-MRD or samples which had sufficient cells acquired for reaching a sensitivity of ≤10−5 (ie, ≥ 4 × 106 cells acquired) were included in the analyses. Based on a LOD of 10 events, the sensitivity of FCM-MRD was 2.5 × 10−6; the quantitative range (based on a LLOQ of 40 events) is 10−5. Consequently, FCM-MRD data between 2.5 × 10−6 and 10−5 should be considered positive, but are below the limit of quantitation.
Concordance between flow cytometric and molecular MRD dataTF3-1
| . | Day 15 . | Day 33 . | Day 78 . | Other TP . | Total . | % (Total %) . | |
|---|---|---|---|---|---|---|---|
| Concordant | |||||||
| FCM+/PCR+ | 102 | 47 | 11 | 9 | 169 | 74 | (93) |
| FCM−/PCR− | 1 | 5 | 31 | 4 | 41 | 18 | |
| Discordant | |||||||
| FCM−/PCR+ | 0 | 5 | 5 | 0 | 10 | 4 | (7) |
| FCM+/PCR− | 1 | 4 | 2 | 0 | 7 | 3 | |
| Total | 104 | 61 | 49 | 13 | 227 | 100 | |
| . | Day 15 . | Day 33 . | Day 78 . | Other TP . | Total . | % (Total %) . | |
|---|---|---|---|---|---|---|---|
| Concordant | |||||||
| FCM+/PCR+ | 102 | 47 | 11 | 9 | 169 | 74 | (93) |
| FCM−/PCR− | 1 | 5 | 31 | 4 | 41 | 18 | |
| Discordant | |||||||
| FCM−/PCR+ | 0 | 5 | 5 | 0 | 10 | 4 | (7) |
| FCM+/PCR− | 1 | 4 | 2 | 0 | 7 | 3 | |
| Total | 104 | 61 | 49 | 13 | 227 | 100 | |
Only samples in which MRD could clearly be detected by FCM-MRD or samples which had at least 4 × 106 cells acquired were included in the analyses. Based on an LLOQ of 40 events, this allowed a sensitivity of at least 10−5.
TP, time points.
Detailed evaluation of discordant cases
To evaluate the discordant cases, several additional analyses were performed. First, FCM fetal calf serum data files were blindly distributed to 4 laboratories for reanalysis of the FCM-MRD data. Out of the 7 cases initially scored positive by FCM-MRD and negative by PCR-MRD, 6 were interpreted as negative by all 4 centers upon FCM-MRD reanalysis, whereas 1 sample (day 15) was consistently scored positive by all 4 centers. Second, RQ-PCR–MRD data were checked for cases negative by FCM-MRD and positive by PCR-MRD. In 8 of 10 cases (all confirmed to be FCM-MRD–negative by reanalysis in different centers), PCR-MRD data were considered positive based on a single well in a single target. To further evaluate whether this low-level positivity was potentially caused by nonspecific amplification, NGS was used to confirm the possible presence of the leukemia-specific antigen receptor rearrangement. In 7 out of 9 available samples, NGS-MRD was negative, whereas MRD positivity could be confirmed in the remaining 2 patients. Thus, 6 of 17 (35%) discordant cases were due to initial misinterpretation of the FCM-MRD data and at least 7 of 17 (41%) were due to overinterpretation of PCR-MRD data; the remaining 4 cases appeared to be truly discordant cases (supplemental Table 3). After these additional evaluations, the actual concordance increased to 98%. If only samples with MRD levels <0.01% were included, 97% gave concordant results.
Discussion
After 5 phases of optimization, we finally selected two 8-color antibody tubes that only differed for the markers present in the PE channel (CD66c/CD123 vs CD73/CD304). These 2 tubes are comparable with 8-color BCP-ALL MRD tubes recently used in other studies, because all proposed panels include CD19, CD10, CD20, CD34, and CD45. In our study, these markers were considered backbone markers from start onwards, based on our previous experience.10,12,18,28-30 Also, CD38 is present in all proposed panels and was proven to be relevant in our present study as well as in the studies of Karawajew et al14 and Shaver et al.15 The remaining 2 positions were completed with different markers: CD9, CD13/CD33, CD15/NG2, CD58 (2 studies), CD66c (2 studies), CD73, CD81, CD123, CD304, and a nucleic acid dye. In our analysis, CD9, CD58, and CD22 appeared be of limited value and therefore were discarded, while Shaver and colleagues, who also applied mathematical modeling systems, identified these as important MRD markers.15 However, they did not test CD66c, CD73, CD123, or CD304, and the difference between the contribution of CD9 and CD81 in their study was limited.15 CD15/NG2 might be relevant in ALL with MLL gene rearrangements, mainly occurring in infants.38 These cases are rare and frequently present with a pro-B-ALL immunophenotype that can relatively easily be distinguished from normal BCP cells and plasma cells.39 In our study, we tested and finally selected 4 markers that are frequently abnormally expressed on BCP-ALL cells: CD66c (associated with BCR-ABL and hyperdiploidy),40,41 CD123 (associated with hyperdiploidy42 ), CD73,36 and CD304 (possibly associated with TEL-AML1).33 By combining 2 of these markers in a single fluorescence channel, abnormal expression could be identified in ∼70% of BCP-ALL patients. Furthermore, in combination with the backbone markers and CD81, BCP-ALL cells could clearly be distinguished from normal BCP cells in virtually all patients. Thus, after multiple phases of multicenter testing of a wide range of leukocyte markers, antibody clones, and fluorochrome-conjugated reagents using objective novel software tools, we were able to select 2 highly effective BCP-ALL MRD tubes.
It remains to be evaluated whether the designed BCP-ALL MRD tubes can also be used during antibody-based therapies. Especially Blinatumomab and chimeric antigen receptor–T-cells (targeting CD19) may hamper the gating of BCP based on CD19. Although alternative gating strategies can be applied (eg, based on CD10, CD34, and/or CD45), one could also decide to add CD24 and/or CD22 to the current tubes (transforming it into a 10-color tube) in MRD-based trials involving Blinatumomab.43 Addition of CD24 and CD22 will also have the advantage that the earliest BCP cells, expressing CD24 and/or CD22 but not yet CD19,44 can be identified; this may be of relevance for the identification of all BCP cells in regenerating BM samples.
In order to obtain MRD data with good sensitivity, acquisition of large numbers of cells appears to be a prerequisite. There is no consensus yet about the number of cells needed for a population. Most studies in BCP-ALL indicate a minimum number between 10 and 50 events, whereas a recent consensus report on MRD detection in multiple myeloma patients defined 20 and 50 cells as the LOD and LLOQ, respectively.14,21,45 Consequently, a sensitivity of 10−5 (generally reached in PCR-MRD and NGS-MRD analysis) requires acquisition of ≥106 cells, preferably ≥5 × 106 cells. We therefore developed the new Euroflow bulk lysis protocol, allowing acquisition of such high cell numbers. Although the bulk lysis protocol contains several washing steps, which will likely result in some cell loss, there is no evidence for selective loss of BCP-ALL cells, given the high concordance between the final FCM-MRD results and the PCR-MRD results. To our best knowledge, other FCM-MRD studies so far have not acquired ≥4 × 106 events and therefore could not have reached the same sensitivity as shown here, although the study by the ALL-REZ-BFM 2002 trial group comes close.14 Our data clearly show that acquisition of large numbers of cells (≥4 × 106) is a prerequisite for obtaining good sensitivities and data that are truly comparable to PCR-MRD data.
We finally tested the newly designed and optimized BCP-ALL MRD tubes in combination with the bulk lysis protocol on follow-up samples from BCP-ALL patients and compared the FCM-MRD data with PCR-MRD data. The concordance between both methods (in the absence of any cutoff) was extremely high (93%) and was significantly better than in previous studies (82.3%10 and 86.5%14 ). As mentioned above, this increased concordance is likely due to the higher sensitivity of the current study, resulting from the higher number of cells analyzed. Detailed evaluation of the discordant samples using NGS-based approaches showed that several discrepant cases were due to overinterpretation of the PCR-MRD data, as the involved leukemia-specific immunoglobulin/TR rearrangements could not be detected by (qualitative) NGS analysis. Consequently, it is most likely that in these discordant cases the positive RQ-PCR MRD data, interpreted according to the EuroMRD criteria for prevention of false-negative MRD results, are due to nonspecific amplification,2,10,46,47 and that these samples actually were MRD-negative.
Next to these “false-positive PCR-MRD” samples, part of the initially discordant cases could be explained by “false-positive FCM-MRD” results: samples were initially scored MRD-positive (generally at very low levels of <0.01%), but these samples were consistently scored MRD-negative upon blind reanalysis at 4 different centers. Interpretation of FCM-MRD data, especially at MRD levels <0.01%, is still expert-based and depends on the number of events in the suspected population, their distance from normal, and the homogeneity of the suspected population (clustering of suspected cells). Although the number of events may easily be defined, distance from normal and homogeneity of the population are more complex to be objectively defined. Novel software tools, including automated gating approaches and maturation pathway analysis (Supplemental Figure 7) are currently being developed within the EuroFlow Consortium and will facilitate more standardized and objective FCM-MRD measurements in the near future.37 Preliminary maturation-based FCM-MRD data showed very good concordance with PCR-MRD data, although further improvements are needed for detection of low levels of MRD (<0.01%).
The NGS-MRD analyses and reanalyses of FCM-MRD data increased the concordance to an unprecedented rate of 98%; the remaining discordant cases are likely due to statistical variation around the detection limits of both assays.14 Therefore, the here presented EuroFlow FCM-MRD strategy proved to be highly sensitive (at least comparable to PCR-MRD) and fast and allows standardized quantification of MRD in virtually all BCP-ALL patients. By increasing the number of acquired cells to 107 (eg, by running both tubes with 5 × 106 cells), the sensitivity and robustness likely can even be further increased.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This research was performed within the EuroFlow Consortium, which started with an EU-FP6 grant (LSHB-CT-2006-018708) and obtained sustainability by protecting and licensing intellectual property, thereby obtaining royalties, which are exclusively being used for supporting the EuroFlow research program (chairmen: J.J.M.v.D. and A.O.). The authors gratefully acknowledge Tomas Kalina and the technicians of the Laboratory for Medical Immunology, Erasmus MC, for technical assistance and thank Christa Homburg and colleagues for performing molecular MRD analyses of part of the Dutch patients and Marieke Comans-Bitter for organizational support.
E.F. was supported by the Grant Agency of the Czech Republic (project of Centre of Excellence No. P302/12/G101). L.S., T.S., and P.T. were supported by European Research Area Network (ERA-NET) PrioMedChild, grant 40-41800-98-027. E.M. was supported by Ministry of Health of the Czech Republic, grant 15-28525A. M.K. was supported by the University Hospital Motol, Prague, Czech Republic (00064203). E.S.d.C., Q.L., and A.O. acknowledge the Bilateral Cooperation Program between Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (Brasília, Brazil) and Dirección General de Políticas Universitárias–Ministério de Educación, Cultura y Deportes (Madrid, Spain) (311/15). E.S.d.C. acknowledges Research Foundation of the State of Rio de Janeiro, Rio de Janeiro, Brazil (E26/110.105/2014, E26/102.191/2013) and Conselho Nacional de Desenvolvimento Científico e Tecnológico–CNPQ of Brazil (400194/2014-7). G.G., C.B., and P.B. were supported by Fondazione Tettamanti. The authors gratefully acknowledge the contribution of the EuroClonality/EuroMRD NGS network (chair: A. W. Langerak) for support with the NGS data. The research for this manuscript was in part performed within the framework of the Erasmus Postgraduate School Molecular Medicine.
Authorship
Contribution: V.H.J.v.d.V., E.M., J.J.M.v.D., and A.O. designed the research; P.T., E.M., Q.L., G.E.G., A.O., J.J.M.v.D., and V.H.J.v.d.V. developed the methodology and the research strategies; P.T. and V.H.J.v.d.V. interpreted the data; P.T., E.M., L.S., A.J.v.d.S.-G., C.B., E.O., G.G., M. Bartels, E.S.d.C., M.N., P.B., T.S., L.L., G.E.G., A.O., and V.H.J.v.d.V. analyzed the FCM data; P.T., E.M., L.S., A.J.v.d.S.-G., M. Bartels, M.N., E.S., O.H., E.S.d.C., J.G.t.M., C.B., E.O., P.B., and L.L. performed FCM experiments; M.K., E.F., J.T., and M. Brüggemann designed and performed the NGS experiments and analyzed NGS data; and V.H.J.v.d.V. wrote the paper; all authors reviewed and approved the manuscript.
Conflict-of-interest disclosure: J.J.M.v.D. has performed contract research for Roche, Amgen, and BD Biosciences; V.H.J.v.d.V. has performed contract research for Roche, Amgen, Pfizer, Janssen, and BD Biosciences and received consultancy fees from Celgene. G.E.G. is an employee of Cytognos SL, Salamanca, Spain. The remaining authors declare no competing financial interests.
A complete list of the members of the EuroFlow Consortium appears in “Appendix.”
Correspondence: Jacques J. M. van Dongen, Department of Immunology, Erasmus MC, University Medical Center Rotterdam, Wytemaweg 80, 3015 CN Rotterdam, The Netherlands; e-mail: j.j.m.van_dongen@lumc.nl.
Appendix: study group members
The members of the EuroFlow Consortium are: Coordination: J. J. M. van Dongen, W. M. Bitter. Participants: V. H. J. van der Velden, A. W. Langerak, P. Theunissen, J. te Marvelde, J. Schilperoord-Vermeulen, A. E. Bras, M. van der Burg, M. Wentink, S. Posthumus (Erasmus MC, Rotterdam, The Netherlands), A. Orfao, J. Almeida, Q. Lecrevisse, J. Flores-Montero, S. Matarraz, María-Belén Vidriales, J. J. Pérez-Morán, N. Puig, M. Pérez-Andrés, L. Martín, E. Blanco (USAL, Salamanca, Spain), M. Gomes da Silva, A. Medina Almeida, J. Caetano, T. Faria (IPO, Lisbon, Portugal), M. Kneba, M. Brüggemann, M. Ritgen, S. Böttcher , M. Szczepanowski, E. Harbst, M. Ipsen (UNIKIEL, Kiel, Germany), E. Macintyre, V. Asnafi, A. Trinquand, L. Lhermitte (AP-HP, Paris, France), J. Trka, O. Hrusak, T. Kalina, E. Mejstrikova, D. Thurner, M. Novakova (DPH/O, Prague, Czech Republic), T. Szczepanski, L. Sędek, J. Bulsa, A. Sonsala (SUM, Zabrze, Poland), E. Sobral da Costa, C. E. Pedreira, F. Pinto Mariz, T. Sigaud S. Palmereira (UFRJ, Rio de Janeiro, Brazil), M. Lima, A. H. Santos (CHP, Porto, Portugal), E. Sonneveld, A. J. van der Sluijs-Gelling (DCOG, The Hague, The Netherlands). Affiliated participants: G. Gaipa, C. Buracchi, C. Bugarin (HSGerardo, Monza, Italy), M. Roussel (CHUP, Rennes, France), L. Campos-Guyotat, C. Aanei (CHU, Saint-Etienne, France), N. Villamor (UB, Barcelona, Spain), J. Philippé, C. Bonroy, B. Denys, A. Willems, S. de Sy (UZG, Ghent, Belgium), P. Fernandez (KSA, Aarau, Switzerland), M. Vlkova, J. Nechvatalova (FNUSA, Brno, Czech Republic), A. E. Sousa, A. L. Serra-Caetano (IMM, Lisboa, Portugal), O. Speer, J. Trück (KISPI, Zürich, Switzerland). Associated SME: CYTOGNOS, Salamanca, Spain (M. Martín-Ayuso, J. Bensadon, F. Martin de Lara, G. Grigore, P. Peñalosa, J. Verde, R. Fluxa, P. Manzano).
References
Author notes
P.T. and E.M. contributed equally to this study.

![Figure 1. Data analysis strategy used to optimize the antibody panel for distinguishing between BCP-ALL cells and their nearest normal/reactive BCP counterpart. First, multiple normal/reactive BM samples and/or regenerating BM samples were merged (phase 1: n = 7; phase 2: n = 11; phase 3: n = 14; phase 4: n = 10) and CD19-positive B cells were selected. These were subdivided into 4 B-cell subsets, based on the backbone markers (CD19/CD10/CD20/CD34/CD45): CD34+ pre–B-I cells (light green), CD34−/CD10+/CD20− to dim pre–B-II/immature cells (dark blue), CD34−/CD10+/CD20+ immature/transitional B cells (light blue), and CD34−/CD10−/CD20+ mature B cells (dark green). Dot plots of CD34 vs CD10 (A) and CD10 vs CD20 (B) are shown. The 1 standard deviation (SD) (dashed line) and 2 SD lines (solid line) of the 2 most immature BCP subsets (pre–B-I [light green] and pre–B-II/immature [dark blue]) were displayed in an APS view, which was subsequently fixed (supervised; C). Each individual BCP-ALL case was added to the fixed APS plot, and the normal BCP population nearest to each of the BCP-ALL populations was defined (D). The BCP-ALL cells and nearest normal BCP subset were then visualized in a separate (nonfixed and balanced) APS plot, 1 using the backbone markers only (E) and 1 using all 8 markers (F), by plotting the 1 SD curve and 2 SD curves of the 2 populations. To prevent an influence of the number of cells on the principal component analysis (PCA), we opted to use a balanced PCA, implying a fixed ratio between normal and pathological events. Finally, the separation between the 2 populations was scored based on: no overlap between 2 SD curves: 3 points; overlap of the 2 SD curves: 2 points; overlap of the 2 SD and the 1 SD curve: 1 point; overlap of both 1 SD curves: 0 points. An example of this scoring is shown in supplemental Figure 1. It should be noted that the above described strategy was only used for optimizing the antibody panel for the BCP-ALL MRD tubes and not for actual MRD analyses.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/129/3/10.1182_blood-2016-07-726307/4/m_blood726307f1.jpeg?Expires=1769086603&Signature=Qgx5-UOJYqh-oJywaApzRK-v3qe-yqbnpAZwiImWQOQ-oJ2w4eeJosv5FjsXh9HeqbedwSVhz1Tzwg3aiffNc0CVHZRfYPwLy-ErqY4aqxG30me4sLB3YPYmIVg-JMpEaEk7p8ox8LP6eAFG1NINBVP7OVEqk1bELoaa-kNZz~7CN17YLiF~o9n31YWDr8y~5hPTW0IP8IzZhuwKaYIhAWYR1Tv0HGReyi3pIyFUD4El6INkF9CVq2KOrChXeLQ7U0Z8zSQwXcIcd1HHbLpV0LqS5-vakmM-UYC6Y6m-rUd4FvaGXYfx~27zRi6V9CobrTrmqpZtEQsWiNLD3aCQrA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)