Key Points
MYC and BCL2 genetic alterations are associated with COO subtype-specific clinical effect in R-CHOP-treated DLBCL.
Abstract
The clinical significance of MYC and BCL2 genetic alterations in diffuse large B-cell lymphoma (DLBCL), apart from translocations, has not been comprehensively investigated using high-resolution genetic assays. In this study, we profiled MYC and BCL2 genetic alterations using next-generation sequencing and high-resolution SNP array in 347 de novo DLBCL cases treated with R-CHOP (rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone) at the British Columbia Cancer Agency. Cell-of-origin (COO) subtype was determined by Lymph2Cx digital gene expression profiling. We showed that the incidence of MYC/BCL2 genetic alterations and their clinical significance were largely dependent on COO subtypes. It is noteworthy that the presence of BCL2 gain/amplification is significantly associated with poor outcome in activated B-cell-like and BCL2 translocation with poor outcome in germinal center B-cell subtypes, respectively. Both have prognostic significance independent of MYC/BCL2 dual expression and the International Prognostic Index (IPI). Furthermore, the combination of BCL2 genetic alterations with IPI identifies markedly worse prognostic groups within individual COO subtypes. Thus, high-resolution genomic assays identify extremely poor prognostic groups within each COO subtype on the basis of BCL2 genetic status in this large, uniformly R-CHOP-treated population-based cohort of DLBCL. These results suggest COO subtype-specific biomarkers based on BCL2 genetic alterations can be used to risk-stratify patients with DLBCL treated with immunochemotherapy.
Introduction
MYC and BCL2 are critical driver genes for non-Hodgkin lymphoma, including diffuse large B-cell lymphoma (DLBCL). These 2 genes play important roles in normal B-cell differentiation and tumorigenesis by affecting diverse cellular processes such as apoptosis, proliferation, growth, cell cycle control, cell migration, and metabolism.1-4 The main molecular mechanisms underlying their deregulation include translocations, gains or amplifications, and perhaps mutations; however, comprehensive genetic profiling of MYC and BCL2 and their effect on survival have not yet been fully investigated.
The clinical significance of MYC and BCL2 translocations (MYCTR and BCL2TR) in DLBCL have been extensively studied, and although the prognostic effect of BCL2TR is controversial,5-7 MYCTR predicts an inferior outcome in most studies.8-10 There is also general consensus that patients with DLBCL with concurrent MYCTR and BCL2TR, so-called double-hit lymphoma (DHIT), have an extremely aggressive clinical course.11-14 In the new 2016 World Health Organization classification, these cases are separated from DLBCL, not otherwise specified.15 In contrast, the prognostic significance of other genetic alterations is less clear. For increased copy number (CN) of MYC and BCL2, highly discordant results regarding their incidence and prognostic effects have been reported, which may be attributable to the small and heterogeneous treatment cohorts evaluated, as well as inconsistent definitions of increased CN based on cytogenetic approaches.16-21 There is also little information on the spectrum and clinical effect of MYC and BCL2 mutations, especially their prognostic significance in the era of immunochemotherapy.22-25 Importantly, no study has evaluated all 3 genetic aberrations including mutation, CN alteration, and translocation in the same cohort where the treatment is held constant. Finally, whereas the cooperating effects of MYC and BCL2 in lymphomagenesis have been shown,11,26-28 the interactions of the various MYC and BCL2 genetic alterations have not been comprehensively investigated.
Two major subtypes of DLBCL based on cell of origin (COO) are recognized, referred to as the activated B-cell-like (ABC) subtype and the germinal center B-cell-like (GCB) subtype, which have distinct underlying biology and clinical behavior.29-31 Although specific MYC/BCL2 genetic alterations occur at different frequencies between COO subtypes, limited information on the prognostic effect in the individual subtypes is available.7,9,17,18 More recent studies have shown that MYC and BCL2 dual-protein expression (DPE) may identify an inferior survival group in DLBCL and is more common in ABC-DLBCL.26-28,32 The characterization of these factors may allow risk-adapted treatments to be considered, but the full contribution of MYC/BCL2 genetic alterations to pathogenesis, including DPE, have not been elucidated.
Recent developments in high-throughput genetic technologies have the potential to provide an accurate assessment of genetic alterations of DLBCL. Here, we conducted a comprehensive genetic analysis of MYC and BCL2, using next-generation sequencing and high-resolution SNP arrays on a population-based cohort of 347 patients with de novo DLBCL uniformly treated with R-CHOP, allowing us to describe the clinically relevant MYC/BCL2 genetic alterations specific to the COO subtypes.
Patients and method
Patient cohort
The BC Cancer Agency (BCCA) Lymphoid Cancer database was searched to identify all patients with de novo DLBCL who were diagnosed between 1985 and 2011. From 4063 DLBCL cases, 347 patients with de novo DLBCL were included in a final cohort after meeting the following criteria: 16 years of age or older; treated with curative intent at the BCCA, using R-CHOP (rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone) for 3 to 8 cycles, depending on stage, with or without radiotherapy; available complete clinical and laboratory data; and a fresh frozen diagnostic biopsy sample. DLBCL was defined using the 2008 World Health Organization classification,33 as determined by standardized review by expert hematopathologists (A.M., P.F., and R.D.G.). Patients were excluded if they had primary mediastinal large B-cell lymphoma, primary central nervous system lymphoma, or central nervous system involvement at the time of diagnosis, a history of an indolent lymphoma, or positive HIV serology. Clinical information including International Prognostic Index (IPI) factors were also collected and categorized into 2 IPI risk groups based on its scores: low/low-intermediate group (IPI score, 0-2) or high/high intermediate group (IPI score, 3-5). This study was reviewed and approved by the University of British Columbia-BCCA Research Ethics Board in accordance with the Declaration of Helsinki.
Mutation analysis
Mutation status was determined by deep targeted resequencing. Three hundred forty-seven fresh frozen tumor and 67 matched normal samples were sequenced using next-generation sequencing. Library construction was performed using DNA extracted from fresh frozen samples with Truseq Custom Amplicon kits (Illumina, San Diego, CA), as well as the Fluidigm Access Array system (Fluidigm, San Francisco, CA), according to manufacturer’s instructions. The pooled samples were sequenced on an Illumina MiSeq instrument with paired-end 2 × 150-bp reads. Sequencing reads were processed via an in-house pipeline using Mutascope (v1.02). Further methodologic details are provided in the supplemental Methods, available on the Blood Web site.
Copy number analysis
Affymetrix Human SNP6.0 Arrays (Affymetrix, Santa Clara, CA) were used to determine CN status in fresh frozen biopsies from 341 cases. Six cases were excluded because of the low quality of SNP array data. Gene-specific CN changes of MYC and BCL2 were predicted using OncoSNP (v1.3), as previously described.34,35 Patients with CN gain and/or amplification of each gene being studied were combined into a single group designated MYCGA or BCL2GA, respectively. The genomic regions with significant focal CN gain and loss (q < 0.25) were determined using GISTIC 2.0.36 Further methodologic details are provided in the supplemental text.
IHC and FISH analyses on tissue microarrays
The analyses of immunohistochemistry (IHC), fluorescent in situ hybridization (FISH), and Lymph2Cx assay were performed on formalin-fixed paraffin-embedded tissue (FFPET) biopsies of 341 cases. Six cases were not evaluated, as sufficient FFPET materials were not available. IHC staining on the 4-μm slides of tissue microarray was performed for MYC, BCL2, CD10, BCL6, and IRF4 (antibodies and cutoffs are listed in supplemental Table 2) on the Benchmark XT platform (Ventana, AZ) and scored independently by 3 expert hematopathologists (A.M., P.F., and R.D.G.). FISH analysis was performed using commercially available dual-color break-apart probes (probes are listed in supplemental Table 3). Two people (S.B.-N. and D.E.) independently scored at least 100 nuclei per case. The cases displaying break-apart signals in ≥5% of cells were considered to have a translocation. Discrepant cases were further examined by A.M. and D.W.S. to reach consensus.
Determination of cell of origin classification
Digital gene expression profiling (GEP) was performed to assign COO, using the Lymph2Cx 20-gene GEP assay on the NanoString platform (NanoString Technologies) for the cases in which tumor content was ≥10%, as previously described.37,38 Two hundred nanograms of RNA extracted from FFPET samples were hybridized overnight at 65°C with probes used to quantitate the 20 genes that contribute to the Lymph2Cx assay. The COO score was calculated based on the model previously described37 and assigned to ABC, GCB, and unclassified categories. According to these procedures, COO was successfully assigned in 323 cases. One hundred eighty-three cases were assigned to the GCB subtype, 104 cases were assigned to ABC, and 36 were unclassified.
Statistical analysis
χ2 test and t-tests were used to assess the differences in patient clinical and phenotypic characteristics, according to MYC/BCL2 genetic alterations. Correlations among MYC/BCL2 genetic alterations were assessed using Fisher’s exact test with multiple testing corrections by Benjamini-Hochberg procedures. The Kaplan-Meier method was used to estimate the time to progression (TTP; progression/relapse or death from lymphoma or acute treatment toxicity), progression-free survival (PFS; progression/relapse or death from any cause), disease-specific survival (DSS; death from lymphoma or acute treatment toxicity), and overall survival (OS; death from any cause), with all endpoints measured from the time of pathologic diagnosis and log-rank test performed to compare survival curves. Univariate and multivariate Cox proportional hazard regression models were used to evaluate proposed prognostic factors. All reported P values are 2-sided, and those less than 0.05 were considered statistically significant. All the analyses were performed using R software v3.2.3.
Results
Genetic analysis of MYC and BCL2
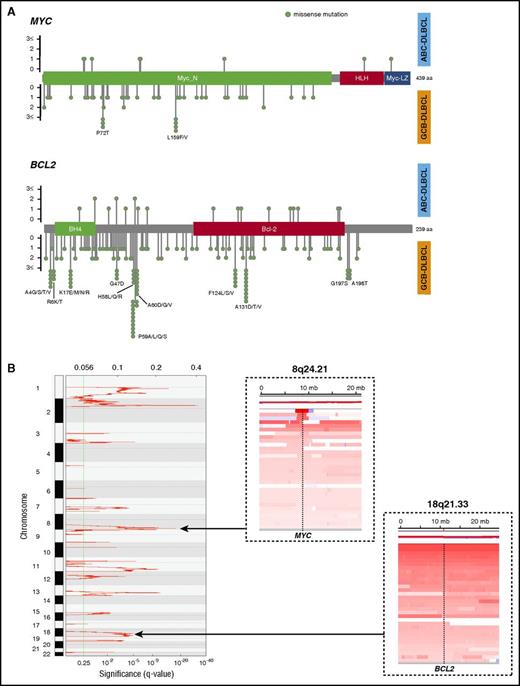
MYC and BCL2 were sequenced at an average depth of 663-fold in 347 DLBCL cases. Overall, 62 single nucleotide variants (SNVs) were detected in MYC (hereafter referred to as MYCMUT) in 29 cases (8%; 5 in ABC [5%], 19 in GCB [10%], and 2 in unclassifiable cases [6%]; P= .09 [ABC vs GCB]) and 190 SNVs in BCL2 (hereafter referred to as BCL2MUT) in 87 cases (25%; 10 in ABC [10%], 66 in GCB [36%], and 7 in unclassifiable cases [19%]; P< .001 [ABC vs GCB]). All SNVs were missense mutations, and 10 cases with MYCMUT and 41 with BCL2MUT contained multiple mutations. In GCB-DLBCL, we observed hotspot mutations (more than 2 SNVs) with MYC (P72 and L159) and BCL2 (A4, R6, K17, G47, H58, P59, A60, F124, A131, G197, and A198), whereas no hotspots were identified in ABC-DLBCL (Figure 1A). Of note, all C to G transition mutations in MYC and 56/68 (82%) in BCL2 targeted WRCY motifs, suggesting that these mutations likely occurred as a consequence of somatic hypermutation (SHM).39
Feature of MYC and BCL2 mutations and CN alterations. (A) Lollipop plots showing the type and location of MYC and BCL2 mutations. Top plot represents mutations observed in ABC-DLBCL cases, and bottom plot represents those observed in GCB-DLBCL cases. (B) GISTIC gain score plot (left). Copy number heat map visualized by Integrative Genomics Viewer (IGV), illustrating gain affecting the MYC locus (dashed line; top right) on chromosome 8 and BCL2 locus (dashed line; bottom right) on chromosome 18 in representative DLBCL samples.
Feature of MYC and BCL2 mutations and CN alterations. (A) Lollipop plots showing the type and location of MYC and BCL2 mutations. Top plot represents mutations observed in ABC-DLBCL cases, and bottom plot represents those observed in GCB-DLBCL cases. (B) GISTIC gain score plot (left). Copy number heat map visualized by Integrative Genomics Viewer (IGV), illustrating gain affecting the MYC locus (dashed line; top right) on chromosome 8 and BCL2 locus (dashed line; bottom right) on chromosome 18 in representative DLBCL samples.
The frequency of the cases with MYCGA and BCL2GA were 20% (69/341) and 25% (85/341), respectively. In particular, MYC gains were situated within a focal amplified peak at chromosome 8q24.21 that was detected by GISTIC analysis (q = 1.28E−12). In contrast, BCL2 gains typically occurred as part of broad region of CN aberration involving chromosome 18q21.33 (Figure 1B).
MYCTR and BCL2TR were observed in 15% (50/325) and 30% (90/300) of evaluable cases, respectively. Twenty-five cases (8%) harbored concurrent translocations of MYC and BCL2, and thus were determined to be DHIT. MYCTR occurred in 19% of GCB-DLBCL cases, in contrast to 11% of the ABC-DLBCL cases. BCL2TR were also seen more frequently in the GCB-DLBCL cases (46% in GCB-DLBCL vs 5% in ABC-DLBCL).
Clinical and phenotypic characteristics according to MYC and BCL2 genetic alterations
The patients with BCL2MUT, BCL2GA, BCL2TR, and MYCTR were more likely to be female (P = .02) and older (>60 years old; P= .008) and have high Eastern Cooperate Oncology Group performance scores (>1; P = .03) and elevated serum lactate dehydrogenase levels (P= .04), respectively. In contrast, the presence of MYC genetic alterations except MYCTR and DHIT were not associated with any individual IPI factors (Table 1). There were no significant differences between gain and amplification of either MYC or BCL2 in their clinical and phenotypic characteristics or their protein expression levels (supplemental Table 4). In agreement with previous studies,16-18 MYCTR, MYCGA, BCL2MUT, and BCL2TR were significantly more frequently encountered in GCB-DLBCL (P = .047, P = .001, P < .001, and P < .001, respectively), whereas BCL2GA was seen more frequently in ABC-DLBCL (P < .001). Importantly, all 25 DHIT cases were found in GCB-DLBCL (P < .001)
Clinical and phenotypic characteristics according to MYC and BCL2 genetic alterations
| Characteristic . | MYC mutation . | p . | MYC Gain/AMP . | P . | MYC translocation . | P . | BCL2 mutation . | P . | BCL2 Gain/AMP . | P . | BCL2 translocation . | P . | MYC/BCL2 DHIT . | P . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Yes (n = 29) n (%) . | No (n = 318) n (%) . | Yes (n = 69) n (%) . | No (n = 269) n (%) . | Yes (n = 50) n (%) . | No (n = 275) n (%) . | Yes (n = 87) n (%) . | No (n = 260) n (%) . | Yes (n = 85) n (%) . | No (n = 253) n (%) . | Yes (n = 90) n (%) . | No (n = 210) n (%) . | Yes (n = 25) n (%) . | No (n = 272) n (%) . | ||||||||
| Age | |||||||||||||||||||||
| >60 y | 18 (62) | 187 (59) | .73 | 40 (58) | 161 (60) | .77 | 35 (70) | 159 (58) | .11 | 55 (62) | 150 (58) | .45 | 61 (72) | 140 (55) | .008 | 59 (66) | 119 (57) | .15 | 16 (61) | 161 (59) | .82 |
| Sex | |||||||||||||||||||||
| Male | 16 (55) | 202 (63) | .37 | 48 (70) | 163 (61) | .17 | 33 (66) | 169 (61) | .54 | 46 (52) | 172 (66) | .018 | 56 (66) | 155 (61) | .45 | 51 (57) | 136 (65) | .19 | 16 (61) | 169 (62) | .95 |
| Stage | |||||||||||||||||||||
| III, IV | 17 (59) | 167 (53) | .59 | 41 (60) | 139 (53) | .25 | 25 (53) | 145 (53) | .95 | 46 (52) | 138 (54) | .74 | 50 (61) | 130 (52) | .15 | 52 (58) | 110 (53) | .38 | 14 (56) | 147 (54) | .88 |
| LDH | |||||||||||||||||||||
| >ULN | 17 (61) | 148 (51) | .35 | 35 (60) | 126 (50) | .17 | 28 (67) | 127 (49) | .038 | 48 (61) | 117 (49) | .079 | 39 (51) | 122 (53) | .85 | 47 (57) | 96 (49) | .23 | 13 (59) | 129 (51) | .47 |
| ECOG PS | |||||||||||||||||||||
| 2 or more | 12 (41) | 102 (32) | .33 | 22 (32) | 89 (33) | .86 | 17 (36) | 86 (31) | .51 | 35 (40) | 79 (31) | .13 | 27 (33) | 84 (33) | .88 | 36 (40) | 58 (28) | .033 | 9 (36) | 84 (31) | .62 |
| Extranodal sites | |||||||||||||||||||||
| 2 or more | 6 (21) | 50 (16) | .51 | 10 (15) | 42 (16) | .82 | 5 (11) | 45 (16) | .32 | 13 (15) | 43 (17) | .64 | 11 (13) | 41 (16) | .53 | 9 (10) | 37 (18) | .094 | 1 (4) | 45 (17) | .095 |
| IPI score | .22 | .60 | .34 | .40 | .33 | .071 | .86 | ||||||||||||||
| Low (0-1) | 10 (35) | 102 (34) | 19 (29) | 89 (35) | 13 (30) | 95 (36) | 24 (28) | 88 (36) | 24 (30) | 84 (35) | 21 (24) | 77 (38) | 7 (30) | 91 (35) | |||||||
| Intermediate (2-3) | 10 (35) | 143 (48) | 34 (52) | 117 (45) | 19 (43) | 124 (47) | 44 (52) | 109 (45) | 35 (44) | 116 (48) | 47 (54) | 89 (44) | 12 (52) | 122 (46) | |||||||
| High (4-5) | 9 (30) | 56 (19) | 12 (19) | 51 (20) | 12 (27) | 48 (18) | 17 (22) | 48 (19) | 20 (26) | 43 (17) | 19 (22) | 36 (18) | 4 (17) | 51 (19) | |||||||
| Cell of origin* | .088 | .001 | .047 | <.001 | <.001 | <.001 | <.001 | ||||||||||||||
| ABC | 5 (19) | 99 (34) | 11 (17) | 87 (35) | 11 (22) | 92 (35) | 10 (12) | 94 (39) | 46 (58) | 52 (22) | 6 (7) | 89 (43) | 0 | 93 (35) | |||||||
| GCB | 19 (73) | 164 (55) | 48 (74) | 132 (53) | 35 (71) | 142 (53) | 66 (80) | 117 (49) | 24 (30) | 156 (66) | 78 (88) | 98 (47) | 25 (100) | 140 (53) | |||||||
| Unclassified | 2 (8) | 34 (11) | 6 (9) | 30 (12) | 3 (6) | 32 (12) | 7 (8) | 29 (12) | 9 (11) | 27 (12) | 5 (5) | 21 (10) | 0 | 31 (12) | |||||||
| MYC IHC | |||||||||||||||||||||
| Positive | 17 (63) | 117 (39) | .015 | 33 (51) | 98 (39) | .079 | 36 (73) | 96 (36) | <.001 | 30 (37) | 104 (42) | .35 | 44 (54) | 87 (37) | .005 | 38 (44) | 88 (42) | .82 | 20 (80) | 105 (39) | <.001 |
| BCL2 IHC | |||||||||||||||||||||
| Positive | 18 (67) | 199 (67) | .99 | 45 (69) | 164 (65) | .53 | 38 (78) | 172 (64) | .064 | 65 (80) | 152 (62) | .003 | 69 (86) | 140 (59) | <.001 | 80 (91) | 115 (56) | <.001 | 23 (92) | 170 (64) | .004 |
| MYC/BCL2 IHC | |||||||||||||||||||||
| DPE | 15 (56) | 85 (28) | .004 | 23 (35) | 74 (30) | .36 | 30 (61) | 68 (25) | <.001 | 26 (32) | 74 (30) | .70 | 41 (51) | 56 (24) | <.001 | 33 (38) | 61 (30) | .16 | 18 (72) | 75 (28) | <.001 |
| Characteristic . | MYC mutation . | p . | MYC Gain/AMP . | P . | MYC translocation . | P . | BCL2 mutation . | P . | BCL2 Gain/AMP . | P . | BCL2 translocation . | P . | MYC/BCL2 DHIT . | P . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Yes (n = 29) n (%) . | No (n = 318) n (%) . | Yes (n = 69) n (%) . | No (n = 269) n (%) . | Yes (n = 50) n (%) . | No (n = 275) n (%) . | Yes (n = 87) n (%) . | No (n = 260) n (%) . | Yes (n = 85) n (%) . | No (n = 253) n (%) . | Yes (n = 90) n (%) . | No (n = 210) n (%) . | Yes (n = 25) n (%) . | No (n = 272) n (%) . | ||||||||
| Age | |||||||||||||||||||||
| >60 y | 18 (62) | 187 (59) | .73 | 40 (58) | 161 (60) | .77 | 35 (70) | 159 (58) | .11 | 55 (62) | 150 (58) | .45 | 61 (72) | 140 (55) | .008 | 59 (66) | 119 (57) | .15 | 16 (61) | 161 (59) | .82 |
| Sex | |||||||||||||||||||||
| Male | 16 (55) | 202 (63) | .37 | 48 (70) | 163 (61) | .17 | 33 (66) | 169 (61) | .54 | 46 (52) | 172 (66) | .018 | 56 (66) | 155 (61) | .45 | 51 (57) | 136 (65) | .19 | 16 (61) | 169 (62) | .95 |
| Stage | |||||||||||||||||||||
| III, IV | 17 (59) | 167 (53) | .59 | 41 (60) | 139 (53) | .25 | 25 (53) | 145 (53) | .95 | 46 (52) | 138 (54) | .74 | 50 (61) | 130 (52) | .15 | 52 (58) | 110 (53) | .38 | 14 (56) | 147 (54) | .88 |
| LDH | |||||||||||||||||||||
| >ULN | 17 (61) | 148 (51) | .35 | 35 (60) | 126 (50) | .17 | 28 (67) | 127 (49) | .038 | 48 (61) | 117 (49) | .079 | 39 (51) | 122 (53) | .85 | 47 (57) | 96 (49) | .23 | 13 (59) | 129 (51) | .47 |
| ECOG PS | |||||||||||||||||||||
| 2 or more | 12 (41) | 102 (32) | .33 | 22 (32) | 89 (33) | .86 | 17 (36) | 86 (31) | .51 | 35 (40) | 79 (31) | .13 | 27 (33) | 84 (33) | .88 | 36 (40) | 58 (28) | .033 | 9 (36) | 84 (31) | .62 |
| Extranodal sites | |||||||||||||||||||||
| 2 or more | 6 (21) | 50 (16) | .51 | 10 (15) | 42 (16) | .82 | 5 (11) | 45 (16) | .32 | 13 (15) | 43 (17) | .64 | 11 (13) | 41 (16) | .53 | 9 (10) | 37 (18) | .094 | 1 (4) | 45 (17) | .095 |
| IPI score | .22 | .60 | .34 | .40 | .33 | .071 | .86 | ||||||||||||||
| Low (0-1) | 10 (35) | 102 (34) | 19 (29) | 89 (35) | 13 (30) | 95 (36) | 24 (28) | 88 (36) | 24 (30) | 84 (35) | 21 (24) | 77 (38) | 7 (30) | 91 (35) | |||||||
| Intermediate (2-3) | 10 (35) | 143 (48) | 34 (52) | 117 (45) | 19 (43) | 124 (47) | 44 (52) | 109 (45) | 35 (44) | 116 (48) | 47 (54) | 89 (44) | 12 (52) | 122 (46) | |||||||
| High (4-5) | 9 (30) | 56 (19) | 12 (19) | 51 (20) | 12 (27) | 48 (18) | 17 (22) | 48 (19) | 20 (26) | 43 (17) | 19 (22) | 36 (18) | 4 (17) | 51 (19) | |||||||
| Cell of origin* | .088 | .001 | .047 | <.001 | <.001 | <.001 | <.001 | ||||||||||||||
| ABC | 5 (19) | 99 (34) | 11 (17) | 87 (35) | 11 (22) | 92 (35) | 10 (12) | 94 (39) | 46 (58) | 52 (22) | 6 (7) | 89 (43) | 0 | 93 (35) | |||||||
| GCB | 19 (73) | 164 (55) | 48 (74) | 132 (53) | 35 (71) | 142 (53) | 66 (80) | 117 (49) | 24 (30) | 156 (66) | 78 (88) | 98 (47) | 25 (100) | 140 (53) | |||||||
| Unclassified | 2 (8) | 34 (11) | 6 (9) | 30 (12) | 3 (6) | 32 (12) | 7 (8) | 29 (12) | 9 (11) | 27 (12) | 5 (5) | 21 (10) | 0 | 31 (12) | |||||||
| MYC IHC | |||||||||||||||||||||
| Positive | 17 (63) | 117 (39) | .015 | 33 (51) | 98 (39) | .079 | 36 (73) | 96 (36) | <.001 | 30 (37) | 104 (42) | .35 | 44 (54) | 87 (37) | .005 | 38 (44) | 88 (42) | .82 | 20 (80) | 105 (39) | <.001 |
| BCL2 IHC | |||||||||||||||||||||
| Positive | 18 (67) | 199 (67) | .99 | 45 (69) | 164 (65) | .53 | 38 (78) | 172 (64) | .064 | 65 (80) | 152 (62) | .003 | 69 (86) | 140 (59) | <.001 | 80 (91) | 115 (56) | <.001 | 23 (92) | 170 (64) | .004 |
| MYC/BCL2 IHC | |||||||||||||||||||||
| DPE | 15 (56) | 85 (28) | .004 | 23 (35) | 74 (30) | .36 | 30 (61) | 68 (25) | <.001 | 26 (32) | 74 (30) | .70 | 41 (51) | 56 (24) | <.001 | 33 (38) | 61 (30) | .16 | 18 (72) | 75 (28) | <.001 |
Bold indicates significance.
ECOG PS, Eastern Cooperate Oncology Group performance status; Gain/AMP: gain and/or amplification; LDH, lactate dehydrogenase; ULN, upper level of normal.
P value is based on the comparison between the ABC and GCB subtypes.
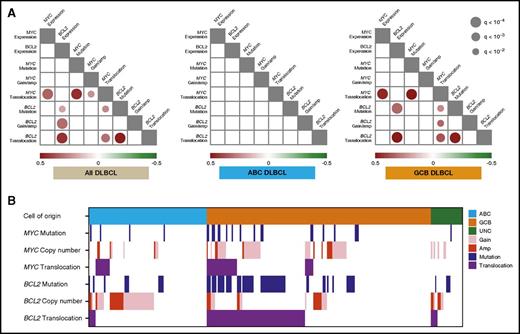
There were highly significant correlations of all 3 BCL2 genetic alterations with BCL2 protein expression across the entire DLBCL cohort. When analyzed within COO subtypes, BCL2 protein expression was significantly associated with BCL2MUT and BCL2TR in GCB-DLBCL. The positive correlation of BCL2MUT with BCL2 protein expression was probably explained by its frequent co-occurrence with BCL2TR. In contrast, only MYCTR was strongly correlated with MYC protein expression in the entire DLBCL cohort and GCB-DLBCL (Figure 2A). When excluding the cases with MYC gain, the cases with MYC amplification trend to having higher MYC protein expression compared with CN normal samples, but this did not reach statistical significance (P = .06, Fisher’s exact test).
Spectrum and correlation of MYC and BCL2 genetic alterations in DLBCL. (A) Statistically significant (q < 0.01) positive (red) and negative (green) correlations among MYC/BCL2 genetic alterations and MYC/BCL2 protein expression in the entire DLBCL cohort (left), ABC-DLBCL (center), and GCB-DLBCL (right), detected by pairwise calculations of Fisher’s exact test. The size and color for each circle indicates the level of significance, as expressed by the q-value and correlation, respectively. (B) The distribution and interaction of MYC/BCL2 genetic alterations, according to COO subtypes.
Spectrum and correlation of MYC and BCL2 genetic alterations in DLBCL. (A) Statistically significant (q < 0.01) positive (red) and negative (green) correlations among MYC/BCL2 genetic alterations and MYC/BCL2 protein expression in the entire DLBCL cohort (left), ABC-DLBCL (center), and GCB-DLBCL (right), detected by pairwise calculations of Fisher’s exact test. The size and color for each circle indicates the level of significance, as expressed by the q-value and correlation, respectively. (B) The distribution and interaction of MYC/BCL2 genetic alterations, according to COO subtypes.
Correlation between MYC and BCL2 genetic alterations
Figure 2 also shows the co-occurrence pattern of MYC/BCL2 genetic alterations. In GCB-DLBCL, 25 (71%) of 35 cases with MYCTR had concurrent BCL2TR. Forty-eight (73%) of 66 cases with BCL2MUT co-occurred with BCL2TR in GCB-DLBCL (Figure 2B). In the correlation analysis, strong positive correlations between mutations and translocations of both MYC and BCL2 were observed in all cases with DLBCL, as well as within GCB-DLBCL, suggesting that SHM occurs frequently in the context of these translocations. As expected, MYC and BCL2 translocations significantly co-occurred in GCB-DLBCL (P < .0001), and of interest, BCL2 mutation was significantly correlated to all MYC genetic alterations in all DLBCL. This was a result of significant correlations within GCB-DLBCL, with no strong correlations observed in ABC-DLBCL (Figure 2A). We also observed negative correlations between BCL2GA and BCL2MUT (P = .03, Fisher’s exact test) in all DLBCL, whereas this was not significant after adjustment for multiple-tests.
Prognostic significance of MYC and BCL2 genetic alterations
There were no significant differences in baseline characteristics between the current study group and the overall BC Cancer Registry-based population of patients with de novo DLBCL treated with R-CHOP at the BCCA within the same calendar period (n = 1177), with the exception that there was a significantly lower proportion of patients with 2 or more extranodal sites in the study cohort (supplemental Table 5). The outcomes in the study cohort were also not significantly different from the registry-based population (supplemental Figure 1), showing that the nature of the study cohort appears representative of the DLBCL population in BC as a whole.
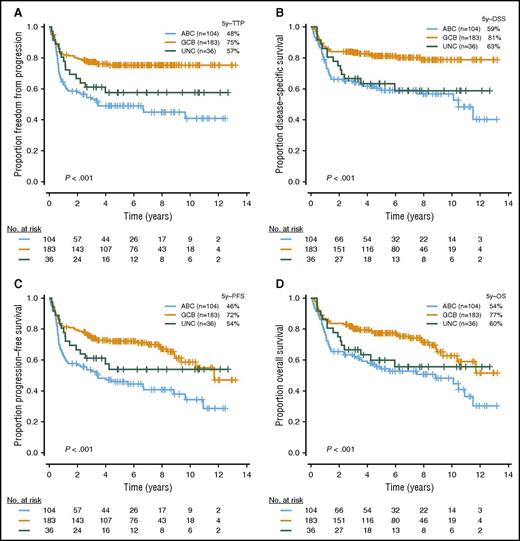
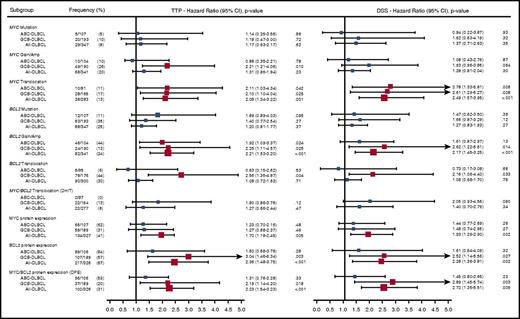
With a median follow-up in living patients of 6.2 years and a minimal follow-up of 6 months for living patients, ABC-DLBCL had significantly inferior outcomes compared with GCB-DLBCL (Figure 3; log-rank P < .001 for TTP, PFS, DSS, and OS). High or high intermediate-IPI was also associated with shorter survival (supplemental Figure 2; log-rank P < .001 for TTP, PFS, DSS, and OS). Given that most of the individual MYC/BCL2 genetic alterations were highly correlated with 1 or the other COO subtype, which, in turn, were strongly associated with survival outcomes, we investigated the prognostic value of the MYC/BCL2 genetic alterations in the entire cohort, as well as individual COO subtypes. In univariate analysis, the presence of MYCTR or BCL2GA was significantly associated with inferior TTP (P = .04 and .02, respectively) within ABC-DLBCL. In GCB-DLBCL, the patients with MYCGA, MYCTR, BCL2GA, or BCL2TR experienced significantly worse outcomes (P = .01, P = .03, P = .03, and P = .004 for TTP, respectively), and DHIT had a trend to lower TTP and DSS (P = .12 and P = .08, respectively) (Figure 4). For MYCMUT and BCL2MUT, there were no significant differences in clinical outcome between the cases with clonal and subclonal mutations (supplemental Figure 3).
Outcomes in patients with DLBCL treated with R-CHOP, according to COO subtypes. Kaplan Meier curves represent (A) TTP, (B) DSS, (C) PFS, and (D) OS, according to COO subtypes.
Outcomes in patients with DLBCL treated with R-CHOP, according to COO subtypes. Kaplan Meier curves represent (A) TTP, (B) DSS, (C) PFS, and (D) OS, according to COO subtypes.
Univariate analysis of MYC/BCL2 genetic alterations. Forest plot summarizes the results of univariate analyses (TTP and DSS). Red squares represent significant hazard ratios. The size for each square indicates the level of hazard ratios.
Univariate analysis of MYC/BCL2 genetic alterations. Forest plot summarizes the results of univariate analyses (TTP and DSS). Red squares represent significant hazard ratios. The size for each square indicates the level of hazard ratios.
Next, we analyzed whether the negative prognostic effect of MYC/BCL2 genetic alterations was dependent on DPE and IPI, which were both strongly associated with outcome in the entire DLBCL cohort. When multivariate analyses incorporating IPI and DPE were individually applied to BCL2GA and MYCTR in ABC-DLBCL, BCL2GA remained significantly associated with inferior TTP, PFS, and OS (P = .03, P = .03, and P= .04, respectively), and a trend to lower DSS was observed (P = .08), whereas MYCTR was not significantly associated with outcomes independent of these factors. In GCB-DLBCL, the independent prognostic effect of BCL2TR was seen in TTP, PFS, and OS (P = .02, P = .007, and P = .03, respectively), while not observed for MYCTR, MYCGA, or BCL2GA (Table 2; supplemental Table 6). Of interest, pairwise analysis showed that DPE did not have prognostic significance independent of BCL2GA in ABC-DLBCL or BCL2TR in GCB-DLBCL (supplemental Table 7). Furthermore, BCL2GA further defined poor prognostic cases within DPE in ABC-DLBCL (P = .014 for TTP and P = .012 for DSS), whereas BCL2TR was not associated with prognosis in DPE GCB-DLBCL, as most of DPE cases were BCL2TR-positive.
Multivariate analysis of TTP, including prognostic genetic alterations, IPI, and DPE
| COO subtype, model, and variables . | HR (95% CI) . | P value . |
|---|---|---|
| ABC-DLBCL | ||
| Model 1 | ||
| BCL2 Gain/AMP (+ vs −) | 1.9 (1.1-3.4) | .03 |
| IPI (high, 3-5 vs low, 0-2) | 2.0 (1.1-3.6) | .02 |
| DPE (+ vs −) | 1.1 (0.6-1.9) | .81 |
| Model 2 | ||
| MYC TR (+ vs −) | 1.6 (0.7-3.5) | .23 |
| IPI (high, 3-5 vs low, 0-2) | 2.0 (1.2-3.6) | .01 |
| DPE (+ vs −) | 1.2 (0.7-2.2) | .51 |
| GCB-DLBCL | ||
| Model 1 | ||
| BCL2 TR (+ vs −) | 2.4 (1.2-4.3) | .02 |
| IPI (high, 3-5 vs low, 0-2) | 3.5 (1.8-6.6) | <.001 |
| DPE (+ vs −) | 1.5 (0.7-3.2) | .27 |
| Model 2 | ||
| MYC TR (+ vs −) | 1.9 (0.9-4.0) | .10 |
| IPI (high, 3-5 vs low, 0-2) | 4.4 (2.3-8.2) | <.001 |
| DPE (+ vs −) | 1.8 (0.9-3.8) | .12 |
| Model 3 | ||
| BCL2 Gain/AMP (+ vs −) | 2.1 (0.97-4.4) | .06 |
| IPI (high, 3-5 vs low, 0-2) | 4.3 (2.3-8.1) | <.001 |
| DPE (+ vs −) | 2.2 (1.1-4.5) | .02 |
| Model 4 | ||
| MYC Gain/AMP (+ vs −) | 1.8 (0.9-3.4) | .08 |
| IPI (high, 3-5 vs low, 0-2) | 4.2 (2.2-7.7) | <.001 |
| DPE (+ vs −) | 2.3 (1.1-4.5) | .02 |
| COO subtype, model, and variables . | HR (95% CI) . | P value . |
|---|---|---|
| ABC-DLBCL | ||
| Model 1 | ||
| BCL2 Gain/AMP (+ vs −) | 1.9 (1.1-3.4) | .03 |
| IPI (high, 3-5 vs low, 0-2) | 2.0 (1.1-3.6) | .02 |
| DPE (+ vs −) | 1.1 (0.6-1.9) | .81 |
| Model 2 | ||
| MYC TR (+ vs −) | 1.6 (0.7-3.5) | .23 |
| IPI (high, 3-5 vs low, 0-2) | 2.0 (1.2-3.6) | .01 |
| DPE (+ vs −) | 1.2 (0.7-2.2) | .51 |
| GCB-DLBCL | ||
| Model 1 | ||
| BCL2 TR (+ vs −) | 2.4 (1.2-4.3) | .02 |
| IPI (high, 3-5 vs low, 0-2) | 3.5 (1.8-6.6) | <.001 |
| DPE (+ vs −) | 1.5 (0.7-3.2) | .27 |
| Model 2 | ||
| MYC TR (+ vs −) | 1.9 (0.9-4.0) | .10 |
| IPI (high, 3-5 vs low, 0-2) | 4.4 (2.3-8.2) | <.001 |
| DPE (+ vs −) | 1.8 (0.9-3.8) | .12 |
| Model 3 | ||
| BCL2 Gain/AMP (+ vs −) | 2.1 (0.97-4.4) | .06 |
| IPI (high, 3-5 vs low, 0-2) | 4.3 (2.3-8.1) | <.001 |
| DPE (+ vs −) | 2.2 (1.1-4.5) | .02 |
| Model 4 | ||
| MYC Gain/AMP (+ vs −) | 1.8 (0.9-3.4) | .08 |
| IPI (high, 3-5 vs low, 0-2) | 4.2 (2.2-7.7) | <.001 |
| DPE (+ vs −) | 2.3 (1.1-4.5) | .02 |
Bold indicates significance.
CI, confidence interval; Gain/AMP: gain and/or amplification; HR, hazard ratio; TR, translocation.
We also analyzed the prognostic effect of composite genetic alterations. The ABC-DLBCL cases with either MYCTR or BCL2GA had significantly inferior outcomes (5-year TTP, 34%; P = .003; supplemental Figure 4A) compared with those without these genetic alterations. In GCB-DLBCL, the presence of any genetic alteration (MYCGA, MYCTR, BCL2GA, and/or BCL2TR) was significantly associated with worse outcome (5-year TTP, 65%; P = .0007; supplemental Figure 4B). Of note, the group of GCB-DLBCL cases without any these genetic alterations had a TTP and DSS of more than 90%. Pairwise analysis also revealed that these COO-specific composite genetic alterations were associated with outcomes independent of DPE, and the prognostic effect of DPE was not retained (supplemental Table 8).
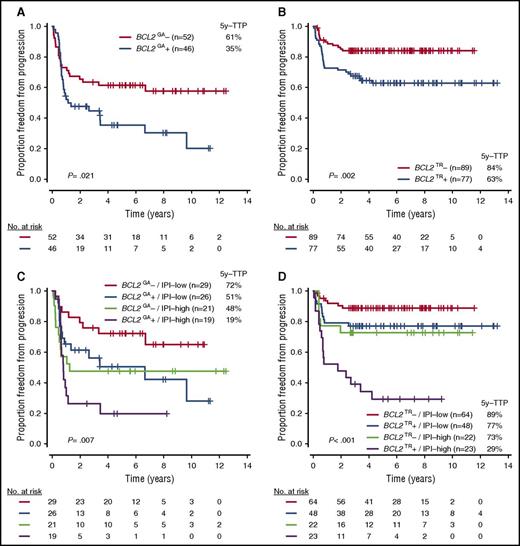
Because BCL2GA and BCL2TR were independently associated with outcome, as well as frequently altered in ABC-DLBCL and GCB-DLBCL, respectively (Figures 5A-B; supplemental Figure 5), we investigated whether the incorporation of these biomarkers and IPI further stratified the cases within the COO subtypes. Notably, the prognostic power of BCL2TR in GCB-DLBCL was particularly evident when examining patients with a high IPI score (scores, 3-5). Indeed, the 5-year TTP of this risk group was 29%, which was similar to the group in patients with ABC-DLBCL with BCL2GA and high IPI (5-year TTP, 19%; Figures 5C-D; supplemental Figure 6). Importantly, BCL2TR had prognostic significance independent of DHIT in GCB-DLBCL (supplemental Table 9), and when excluding the cases with DHIT, BCL2TR and high IPI still had a strong effect on the prognosis of GCB-DLBCL (5-year TTP, 27%; supplemental Figure 7).
TTP according to BCL2 genetic alterations with or without IPI risk groups in each COO subtype. (A-D) TTP according to presence of BCL2GA in ABC-DLBCL (A), BCL2TR in GCB-DLBCL (B), BCL2GA with IPI in ABC-DLBCL (C), and BCL2TR with IPI in GCB-DLBCL (D).
TTP according to BCL2 genetic alterations with or without IPI risk groups in each COO subtype. (A-D) TTP according to presence of BCL2GA in ABC-DLBCL (A), BCL2TR in GCB-DLBCL (B), BCL2GA with IPI in ABC-DLBCL (C), and BCL2TR with IPI in GCB-DLBCL (D).
Discussion
In this large, population registry-based cohort of patients with de novo DLBCL uniformly treated with R-CHOP, we used high-resolution genetic analyses to provide comprehensive genomic profiling of MYC and BCL2 and determine their prognostic significance across well-defined COO subtypes. Our findings highlight that, in ABC and GCB subtypes, the presence of BCL2GA and BCL2TR, respectively, portend poor outcomes independent of IPI and dual protein expression status, and can be used to define markedly poor outcome groups in models incorporating IPI risk scores. Furthermore, the incidence and interactions of MYC/BCL2 genetic alterations and their effects on protein expression were largely dependent on COO subtypes, indicating that their clinical effect should be separately assessed within COO subtypes, as the cases with ABC-DLBCL are associated with inferior outcome compared with those of GCB-DLBCL.31,38 Indeed, the size of our study cohort and GEP-based COO assignment allowed us to identify poor prognostic subgroups of MYCGA and BCL2TR in GCB-DLBCL, which were not found when looking at the DLBCL cohort as a whole. To date, only a few studies have evaluated the prognostic effect of MYC/BCL2 genetic alterations related to COO subtype defined by GEP.7,16,18 With the emergence of COO subtype-specific therapies,40,41 such biomarkers could help to further define the patients suitable for alternative treatment approaches and/or enrollment on clinical trials testing novel agents alone or in combination.
The prognostic importance of dual MYC and BCL2 protein expression has been reported in a number of studies,26-28 but the biological and clinical contribution of MYC/BCL2 genetic alterations to this phenotype remain unknown. Interestingly, in the present study, survival of the patients with DPE was not significantly worse when adjusted for prognostic MYC/BCL2 genetic alterations. In particular, BCL2GA was significantly associated with inferior outcome within DPE of ABC-DLBCL. These data suggest that MYC/BCL2 genetic alterations are more relevant to outcome than protein expression. Thus, further systematic genetic analysis will be needed to clarify the biological basis of aggressive clinical behavior within the DPE cases.
We also demonstrated that incorporation of BCL2 genetic alterations with the IPI further stratified outcomes within each COO subtype. This is particularly relevant in the GCB-DLBCL, where BCL2TR with high IPI defined a subset of cases with an extremely inferior prognosis compared with the remaining cases. Importantly, this approach can identify the patients with significantly inferior prognosis who have less than 30% 5-year TTP and includes 25% of ABC-DLBCL and 14% of GCB-DLBCL. This contrasts with DHIT, accounting for only 10% of cases, exclusively within GCB-DLBCL. These data support the notion that integration of genetic data with clinical risk factors improves risk stratification, capable of identifying groups at high risk for treatment failure.
Interestingly, in the present study, the outcomes of DHIT cases were not as poor as those described in most previous studies.11-14 This may be explained by our population registry-based strategy, in which assays for MYC/BCL2 translocations were performed in all cases.32 In some of the previous retrospective studies, it is likely that FISH was performed selectively, enriching for cases with unusual morphology, high proliferation, or high-risk clinical features. In addition, some of these previous studies included B-cell lymphoma unclassifiable and transformed DLBCL, which have different clinical courses from de novo DLBCL and were excluded from the present study. These explanations are supported by the similar prognosis of DHIT cases (2-year OS; 60%-70%) reported from 2 prospective clinical trials with de novo DLBCL treated with immunochemotherapy, where FISH was performed in all available samples.9,42 Larger prospective studies are warranted to assess the clinical effect of DHIT cases and resolve this growing controversy.
We observed a slightly higher mutation rate of MYC and BCL2 compared with previous studies.43-46 This is partially explained by the inclusion of possible false discovery as a result of the lack of paired normal samples in a proportion of the cases. Another limitation of this study is a lack of the evaluation of prognostic effect of MYC/BCL2 genetic alterations in an independent cohort. In particular, the prognostic significance of the combination of BCL2 genetic alterations and IPI are novel observations, which need validation in other cohort and clinical trials before proposing them as clinically useful biomarkers.
To date, there have been discrepant results of cytogenetic studies evaluating the incidence and prognostic effect of increased CN of MYC and BCL2 in DLBCL.16-21 These differences may be attributed to the technical challenges of performing and interpreting FISH on whole sections or tissue microarray slides. In order to overcome these limitations, we used a genetic approach based on the analysis of high-resolution SNP arrays and CN assigned using OncoSNP, which has been well validated for the copy number calls.34,35 Using OncoSNP allowed us to detect low-level CN increases, integrated with tumor ploidy status and purity,35 which are not usually taken into account using FISH-based cytogenetic approaches. Recently, next-generation sequencing-based assays to assess CNAs in FFPET have been used to explore the clinical utility and effect on management of solid cancers, as well as hematologic malignancies.47,48 Future studies are therefore needed to develop the optimal assay for CN detection, ideally using a platform broadly applicable to routine clinical practice.
In conclusion, this study describes the first comprehensive genomic profile of MYC/BCL2 genetic alterations using next-generation sequencing and high-resolution SNP arrays. Notably, the combination of BCL2 genetic alteration and IPI separated patients into groups with significantly different prognoses in individual COO subtypes. Once our findings have been externally validated, the path forward for introducing robust COO-specific biomarkers into routine clinical practice will be realized. Our data also have therapeutic implications regarding the potential utility of targeting these genes. In a recently published study,49 venetoclax, a BCL2 antagonist, was shown to have substantial antitumor activity in patients with B-cell lymphomas. Development of agents targeting MYC is also an area of active research.50 Thus, our results, when combined with the emergence of novel agents, further strengthens the need for comprehensive molecular profiling of MYC and BCL2 within COO subtypes in DLBCL cases at diagnosis to identify those patients more likely to benefit from alternative treatment approaches.
Presented as an oral presentation at the 57th annual meeting of the American Society of Hematology, Orlando, FL, 5 December 2015.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This study was supported by a Program Project Grant from the Terry Fox Research Institute (Grant No. 1023 to R.D.G.). D.E. is supported by fellowships from the Michael Smith Foundation for Health Research, Canadian Institutes of Health Research and Japanese Society for the Promotion of Science. D.W.S. is supported by the British Columbia Cancer Foundation.
Authorship
Contribution: D.E., R.D.G., and D.W.S. designed and performed the research, analyzed and interpreted data, and wrote the paper; A.M. and P.F. performed pathological review of cases; A.M., S.B.-N., and D.E. performed FISH; D.E. performed library construction and the sequencing run; B.M. and M.B. performed RNA and DNA extractions; C.H. analyzed next-generation sequencing data; H.P.S., D.L., and S.S. analyzed SNP array data; F.C.C., A.B., S.P.S., and R.D.M. provided bioinformatics assistance; R.K., K.J.S., L.H.S., and J.M.C. supervised assembly of clinical data, reviewed the manuscript, and provided editorial input; and M.A.M. and C.S. participated in the original design of the project and provided editorial input
Conflict-of-interest disclosure: J.M.C., R.D.G., and D.W.S. are inventors on a patent held by the National Cancer Institute that has been licensed by NanoString Technologies. S.P.S is a founder and shareholder of Contextual Genomics Inc. The remaining authors declare no competing financial interests.
Correspondence: David W. Scott, British Columbia Cancer Research Centre, 675 West 10th Ave, Room 12-114, Vancouver, BC V5Z 1L3, Canada; e-mail: dscott8@bccancer.bc.ca.
References
Author notes
R.D.G. and D.W.S. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal