Key Points
NOD1 ligand administration restores hematopoietic precursor pools in germ-free mice to the levels seen in specific pathogen-free animals.
NOD1 ligand–NOD1 signaling promotes steady-state hematopoiesis indirectly through the induction of cytokines by MSCs.
Abstract
The microbiota is known to influence the generation of hematopoietic progenitors, although the pathways underlying this process are still poorly understood. NOD1 and NOD2 are intracellular sensors for both Gram-positive and Gram-negative bacteria, but their role in steady-state hematopoiesis has never been characterized. We observed that stimulation with NOD1 or NOD2 ligand had no effect on the survival/proliferation of hematopoietic precursors. Nonetheless, NOD1, but not NOD2, ligand induced expression of multiple hematopoietic cytokines (interleukin-7 [IL-7], Flt3L, stem cell factor [SCF], ThPO, and IL-6) from bone marrow mesenchymal stromal cells (MSCs) in vitro. Moreover, in vivo administration of NOD1 ligand to germ-free mice restored the numbers of hematopoietic stem cells and precursors in bone marrow as well as serum concentrations of IL-7, Flt3L, SCF, and ThPO to the levels displayed by specific pathogen-free control animals. Based on these findings, we propose that NOD1 signaling in MSCs serves as an important pathway underlying the requirement for microbiota in the maintenance of steady-state hematopoiesis. This function is distinct from that triggered by lipopolysaccharide in both its broad effects on multiple progenitors and specific targeting of MSCs as cytokine producing intermediates.
Introduction
Although experiments in germ-free (GF) mice have revealed that the sensing of secreted products or metabolites from commensal bacteria contributes to peripheral immune responses,1-4 relatively little is known about how the microbiota regulates hematopoiesis in the bone marrow (BM) compartment.5 Two previous studies indicated that basal stimulation by gut microbiota regulates granulopoiesis in GF mice to promote generation of a “pool-in-reserve” of myeloid cells within the BM.6,7 Although the microbiota ligand(s) involved were not identified, the work of Balmer et al implicated the NF-κB signaling pathway downstream of MyD88 in this process.6 Indeed, different Toll-like receptor (TLR) ligands induce hematopoietic stem cell (HSC) cycling, expansion of HSC and progenitor populations, as well as promote a shift toward myeloid differentiation.8,9 Nevertheless, naïve Myd88−/−, TRIF−/−, or Myd88−/−TRIF−/− mice, which are unable to respond to bacterial components via TLR, do not display significant changes in hematopoietic cell composition,10 suggesting that other bacterial sensors may be more important in or contribute to the influence of the microbiota on steady-state hematopoiesis. Here, we focused on the possible role of the NOD family of pathogen recognition receptors. NOD1 and NOD2 are involved in intracellular sensing of pathogenic and commensal bacteria through recognition of 2 distinct peptidoglycan components (meso-diaminopimelic acid or muramyl dipeptide), which in contrast to lipopolysaccharide (LPS), are derived from cell walls of both Gram-positive and Gram-negative bacteria.11 However, although NOD2 expression is limited primarily to myeloid cells, NOD1 is widely expressed in multiple cell lineages, making its possible contribution to hematopoiesis more likely.11
Study design
Animals
NOD1−/− mice12 were kindly provided by Dr. Mazier Divangahi (McGill University). NOD2−/−,13 specific pathogen–free (SPF), and GF C57BL/6 mice were obtained from Taconic Farms. All mice were housed at American Association for the Accreditation of Laboratory Animal Care–accredited SPF facilities at the National Institute of Allergy and Infectious Diseases (NIAID) in accordance with animal study proposals approved by the Institute’s Animal Care and Use Committee. GF C57BL/6 mice were bred and maintained at the NIAID gnotobiotic animal facility.
FACS analysis and cell sorting
Generation of mesenchymal stromal cells
Mesenchymal stromal cells (MSCs) were prepared by culturing whole BM cells using a MesenCult proliferation kit with Mesenpure (Stem Cell Technologies). When confluent, cells were harvested and separated into CD45+ macrophages (BMMϕ) and CD45neg mesenchymal stromal cells (MSC) by AutoMACS (Milteny Biotec).
Quantitative polymerase chain reaction analysis
Total RNA was isolated using RNeasy (QIAGEN). Reverse transcription was carried out with Superscript II RT (Invitrogen) and gene expression analysis performed using SYBER Green-base real-time quantitative polymerase chain reaction, with the primers indicated (supplemental Table 2).
Detection of NOD1 and NOD2 ligand in serum
These molecules were quantitated with a bioassay using human embryonic kidney cells stably transfected with an NF-κB–inducible reporter construct plus mouse NOD1 or NOD2 (InvivoGen) and standard curves constructed with different concentrations of C12-iE-DAP or MDP (InvivoGen).
Data analysis
The statistical significance of differences between paired groups was analyzed by Student t test or, in the case of multiple groups, by one-way analysis of variance with Dunnett’s multiple comparison tests.
Results and discussion
To evaluate the effects of the microbiota on the major hematopoietic precursors in BM, we compared the absolute number and phenotype of hematopoietic stem and precursor cells (HSPCs) in BM from GF and conventional SPF mice. GF animals displayed a reduced number of HSCs, multipotential progenitors (MPPs), and common lymphoid progenitors (CLPs) (Figure 1A), demonstrating that the microbiota regulates not only the numbers of granulocyte precursors but also the size of the entire pool of HSPCs in BM.6 As expected, sera from GF mice displayed significantly lower levels of NOD1 and NOD2 ligands than did sera from SPF mice (Figure 1B).
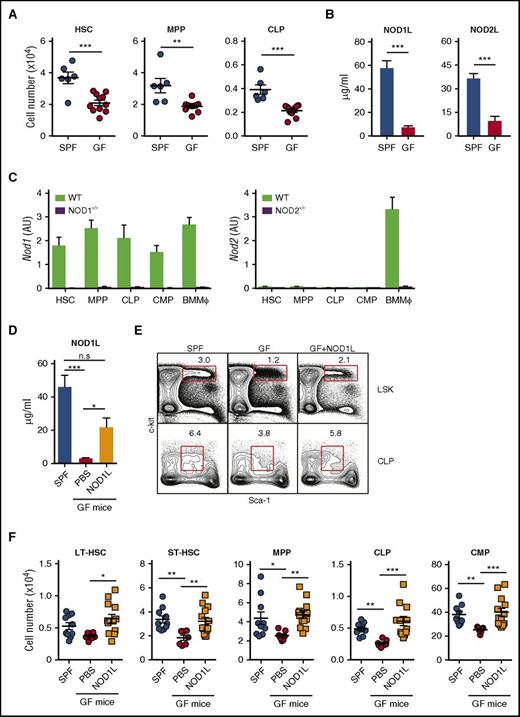
NOD1L administration restores numbers of HSPCs in GF mice to the levels observed in SPF mice. (A) Levels of HSC, MPP, and CLP in SPF vs GF mice. Each symbol indicates the absolute number of LSK or CLP in BM from individual SPF (n = 6) and GF mice (n = 11). The horizontal bars represent the mean values for each group. (B) Serum levels of NOD1 or NOD2 ligand in naïve SPF and GF mice. Bars represent the mean ± standard error of the mean (SEM) of the values obtained in the reporter gene assay used (n = 8 per group). (C) Levels of Nod1 and Nod2 mRNA expression in indicated in BM HSPC populations isolated from WT, NOD1−/−, or NOD2−/− mice (n = 10). BMMϕ from WT animals were included as positive controls. Bars represent the mean ± standard deviation (SD) of triplicate values of Nod1 or Nod2 expression levels relative to the Rplp2 housekeeping gene. (D) NOD1 ligand levels in GF mice (n = 5-9) given either synthetic NOD1 ligand (C12-iE-DAP, 100 μg) or phosphate-buffered saline by gavage every 2 or 3 days for 2 weeks. (E-F) HSPC populations in GF mice after gavage with NOD1 ligand (C12-iE-DAP, 100 μg) as noted before. (E) The representative dot plots shown were gated on lineageneg and IL-7Rαneg (upper panels) or IL-7Rα+ (lower panels) cells, respectively. (F) Absolute numbers of LT-HSC, ST-HSC, MPP, CLP, and CMP in SPF mice, GF mice, and GF mice treated with NOD1 ligand. The symbols represent the cell numbers for the individual animals in each group (n = 7-12). The bars represent the means ± SEM of these values. *P < .05; **P < .01; ***P < .001 denote the statistical significance of the differences in group means. The data shown in (A), (B), (D), and (F) are pooled from 2 to 3 independently performed experiments, whereas the values shown in (C) are from an experiment representative of 2 performed.
NOD1L administration restores numbers of HSPCs in GF mice to the levels observed in SPF mice. (A) Levels of HSC, MPP, and CLP in SPF vs GF mice. Each symbol indicates the absolute number of LSK or CLP in BM from individual SPF (n = 6) and GF mice (n = 11). The horizontal bars represent the mean values for each group. (B) Serum levels of NOD1 or NOD2 ligand in naïve SPF and GF mice. Bars represent the mean ± standard error of the mean (SEM) of the values obtained in the reporter gene assay used (n = 8 per group). (C) Levels of Nod1 and Nod2 mRNA expression in indicated in BM HSPC populations isolated from WT, NOD1−/−, or NOD2−/− mice (n = 10). BMMϕ from WT animals were included as positive controls. Bars represent the mean ± standard deviation (SD) of triplicate values of Nod1 or Nod2 expression levels relative to the Rplp2 housekeeping gene. (D) NOD1 ligand levels in GF mice (n = 5-9) given either synthetic NOD1 ligand (C12-iE-DAP, 100 μg) or phosphate-buffered saline by gavage every 2 or 3 days for 2 weeks. (E-F) HSPC populations in GF mice after gavage with NOD1 ligand (C12-iE-DAP, 100 μg) as noted before. (E) The representative dot plots shown were gated on lineageneg and IL-7Rαneg (upper panels) or IL-7Rα+ (lower panels) cells, respectively. (F) Absolute numbers of LT-HSC, ST-HSC, MPP, CLP, and CMP in SPF mice, GF mice, and GF mice treated with NOD1 ligand. The symbols represent the cell numbers for the individual animals in each group (n = 7-12). The bars represent the means ± SEM of these values. *P < .05; **P < .01; ***P < .001 denote the statistical significance of the differences in group means. The data shown in (A), (B), (D), and (F) are pooled from 2 to 3 independently performed experiments, whereas the values shown in (C) are from an experiment representative of 2 performed.
When measured by quantitative polymerase chain reaction, HSCs, MPPs, CLPs, and common myeloid progenitors (CMPs) displayed significant levels of NOD1 but not NOD2 mRNA (Figure 1C). Therefore, we chose to treat GF mice with NOD1 ligand by peroral gavage, resulting in a systemic elevation in its concentration (Figure 1D).15 Importantly, this treatment augmented the number of long-term (LT)-HSCs, short-term (ST)-HSCs, MPP, GMPs, and CLPs in the BM of NOD1 ligand–treated GF mice to levels indistinguishable from those observed in BM from SPF animals (Figure 1E-F) without increasing serum levels of proinflammatory cytokines (data not shown). These results demonstrated that NOD1 ligand positively regulates the absolute number of HSPCs in BM. Moreover, NOD1−/− but not NOD2−/− mice displayed reduced numbers of HSPCs supporting the involvement of NOD1 signaling in steady-state hematopoiesis (supplemental Figure 2).
The influence of the microbiota on hematopoiesis in BM may result from a direct effect on HSPCs or occur indirectly through their detection by niche supporting cells.16 To test whether NOD1 ligand stimulation directly affects expansion of HSPCs, we sort-purified HSCs, MPPs, and CLPs from the BM of wild-type (WT) mice and cultured them in the presence of NOD1 ligand. We found that NOD1 ligand alone failed to stimulate significant proliferation of HSCs, MPPs, or CLPs, or to enhance cytokine-induced expansion of the same cells (Figure 2A). We further tested the capacity of NOD1 ligand to promote differentiation of myeloid cells from either Lineageneg Sca-1+c-kit+ (LSK) cells, CMPs, or CLPs. In contrast to LPS, NOD1 ligand stimulation failed to promote the generation of either myeloid cell type (supplemental Figure 3A-C).
NOD1 ligand stimulation of MSCs induces secretion of cytokines that promote proliferation of HSPCs. (A) FACS-purified HSC, MPP, and CLP were cultured for 48 hours in the presence of NOD1 (10 μg/mL) or NOD2 (10 μg/mL) ligand alone or in combination with SCF (10 ng/mL), Flt3L (10 ng/mL), or ThPO (10 ng/mL) in the case of HSC and MPP, and with IL-7 (10 ng/mL) and Flt3L in the case of CLP. [3H]-thymidine was added during the last 16 hours of culture. Bars represent mean ± SD of thymidine incorporation for triplicate cultures. N.D., not determined. (B) Expression of Nod1 and Nod2 mRNA in cultured CD45neg MSC and CD45+ Mϕ from WT, NOD1−/−, or NOD2−/− mice. Bars represent mean ± SD of triplicate values of Nod1 and Nod2 expression relative to RPLP2 expression. (C) MSCs generated from WT or NOD1−/− mice were stimulated with NOD1 ligand (C12-iE-DAP, 10 μg/mL) or LPS (10 μg/mL) and 72 hours later harvested for RNA extraction. Bars represent mean ± SD of triplicate measurements of Nod1 relative to Rplp2 expression for each stimulus. (D) Expression of mRNA for IL-3, IL-6, IL-7, SCF, Thpo, and Flt3L in WT MSCs treated with NOD1 ligand (10 μg/mL) or LPS (10 μg/mL). Bars represent the means ± SD of triplicate values of mRNA expression relative to Hprt for each cytokine. (E-F) Sorted LSK (2 × 103/well), HSC (2 × 103/well), CLP (2 × 103/well), or CMP (4 × 103/well) from WT mice were stimulated for 48 hours with the culture supernatants harvested from WT (WT CS) or NOD1−/− (KO CS) MSC cultures treated with NOD1 ligand (10 μg/mL) for 72 hours (E). [3H]-thymidine (0.5 μCi/well, New England Nuclear Corp., sp act: 2 Ci/mmol) was added at 48 hours and the incorporated radioactivity measured 16 hours later. Bars represent the mean ± SD of the values from triplicate cultures. The data shown in (A-E) are from 1 representative out of 3 experiments performed. (F) Serum levels of IL-7, SCF, ThPO, and Flt3L in GF mice after gavage with NOD1 ligand. The concentrations of IL-7, Flt3L, ThPO, and SCF in culture supernatants or serum were measured by Quantikine ELISA kit (R&D systems). Bars represent the mean ± SEM of the ELISA values obtained from the individual SPF, GF, and NOD1 ligand–treated GF mice shown in Figure 1F and pooled from 2 independently performed replicate experiments. *P < .05; **P < .01; ***P < .001.
NOD1 ligand stimulation of MSCs induces secretion of cytokines that promote proliferation of HSPCs. (A) FACS-purified HSC, MPP, and CLP were cultured for 48 hours in the presence of NOD1 (10 μg/mL) or NOD2 (10 μg/mL) ligand alone or in combination with SCF (10 ng/mL), Flt3L (10 ng/mL), or ThPO (10 ng/mL) in the case of HSC and MPP, and with IL-7 (10 ng/mL) and Flt3L in the case of CLP. [3H]-thymidine was added during the last 16 hours of culture. Bars represent mean ± SD of thymidine incorporation for triplicate cultures. N.D., not determined. (B) Expression of Nod1 and Nod2 mRNA in cultured CD45neg MSC and CD45+ Mϕ from WT, NOD1−/−, or NOD2−/− mice. Bars represent mean ± SD of triplicate values of Nod1 and Nod2 expression relative to RPLP2 expression. (C) MSCs generated from WT or NOD1−/− mice were stimulated with NOD1 ligand (C12-iE-DAP, 10 μg/mL) or LPS (10 μg/mL) and 72 hours later harvested for RNA extraction. Bars represent mean ± SD of triplicate measurements of Nod1 relative to Rplp2 expression for each stimulus. (D) Expression of mRNA for IL-3, IL-6, IL-7, SCF, Thpo, and Flt3L in WT MSCs treated with NOD1 ligand (10 μg/mL) or LPS (10 μg/mL). Bars represent the means ± SD of triplicate values of mRNA expression relative to Hprt for each cytokine. (E-F) Sorted LSK (2 × 103/well), HSC (2 × 103/well), CLP (2 × 103/well), or CMP (4 × 103/well) from WT mice were stimulated for 48 hours with the culture supernatants harvested from WT (WT CS) or NOD1−/− (KO CS) MSC cultures treated with NOD1 ligand (10 μg/mL) for 72 hours (E). [3H]-thymidine (0.5 μCi/well, New England Nuclear Corp., sp act: 2 Ci/mmol) was added at 48 hours and the incorporated radioactivity measured 16 hours later. Bars represent the mean ± SD of the values from triplicate cultures. The data shown in (A-E) are from 1 representative out of 3 experiments performed. (F) Serum levels of IL-7, SCF, ThPO, and Flt3L in GF mice after gavage with NOD1 ligand. The concentrations of IL-7, Flt3L, ThPO, and SCF in culture supernatants or serum were measured by Quantikine ELISA kit (R&D systems). Bars represent the mean ± SEM of the ELISA values obtained from the individual SPF, GF, and NOD1 ligand–treated GF mice shown in Figure 1F and pooled from 2 independently performed replicate experiments. *P < .05; **P < .01; ***P < .001.
In addition to hematopoietic cells, BM consists of various nonhematopoietic cell types that form the BM niche and not only provide a structural scaffold but also play an important role in regulating the proliferation and differentiation rates of HSC required for optimal hematopoiesis.17 The latter functions are performed primarily by MSCs that produce a wide variety of cytokines, many of which also have a role in steady-state homeostasis.18,19 To examine whether NOD1 stimulation affects the function of MSCs in HSC regulation, we generated MSCs from BM cells of WT or NOD1−/− mice (supplemental Figure 4A).20
We found that both in vitro cultured and ex vivo isolated MSCs selectively express only NOD1, whereas cultured CD45+Mϕ displayed both NOD1 and NOD2 mRNAs (Figure 2B and supplemental Figure 4B). We next stimulated culture-derived MSCs with NOD1 ligand or LPS. Interestingly, NOD1 ligand, but not LPS, stimulation increased NOD1 mRNA expression (Figure 2C). In the same experiment, mRNA expression for various cytokines critical for maintenance of HSPCs was also evaluated (Figure 2D). As previously reported,21,22 LPS stimulation induced IL-6 and tumor necrosis factor (TNF)-α mRNA expression, but little or none of the other cytokine mRNAs assayed. In contrast, NOD1 stimulation under the same conditions induced IL-3, IL-7, Flt3L, SCF, and ThPO in addition to IL-6 and TNF-α mRNA expression. As expected, none of the cytokine mRNAs was induced by NOD1 ligand in NOD1−/− MSCs or WT CD45+Mϕ (supplemental Figure 5A,C). Stimulation with NOD2 ligand failed to induce cytokine responses in MSCs, while triggering IL-6 secretion by CD45+Mϕ (supplemental Figure 5B,D). These results revealed a unique property of NOD1 ligand stimulation in triggering production of multiple hematopoietic cytokines in MSC. NOD1 ligand stimulation enhanced IL-6 production by LPS-stimulated MSCs (supplemental Figure 6), supporting the concept that TLR4 and NOD1 signaling pathways act independently and mediate distinct outcomes.
To test the hypothesis that NOD1-stimulated MSCs produce cytokines, which in turn are responsible for inducing HSPC proliferation, we set up a bioassay with FACS-purified LSK, CLP, and CMP cultured in the presence of supernatants obtained from either WT or NOD1−/− MSCs stimulated with NOD1 ligand. The culture supernatants from WT, but not NOD1−/−, MSCs induced significant proliferation of all 3 HSPC populations tested (Figure 2E), demonstrating that NOD1 ligand stimulation induces the production of cytokines from MSCs needed for supporting the maintenance of HSPCs. Antibody neutralization experiments implicated IL-6, SCF, Flt3L, and IL-7 in the proliferation of MPPs, CMPs, and CLPs induced by supernatants from NOD1 ligand–stimulated MSCs with different cytokines or combinations of cytokines playing distinct roles depending on the target cell population (supplemental Figure 7).
To confirm that NOD1 ligand from microbiota affects production of hematopoietic cytokines in vivo, we measured the amount of these cytokines in the sera of SPF mice, GF mice, and GF animals treated with NOD1 ligand. Although serum levels of IL-3 and IL-6 were undetectable in all 3 groups, the amounts of IL-7, Flt3L, SCF, and ThPO were significantly lower in sera from GF mice compared with SPF animals (Figure 2F). Importantly, oral treatment of GF mice with NOD1 ligand restored serum levels of IL-7, Flt3L, SCF, and ThPO to the same levels observed in SPF mice. These findings demonstrated that NOD1 ligand administration to GF mice induces systemic increase in hematopoietic cytokines, which in turn may contribute to the expansion of HSPC pools and the maintenance of steady-state hematopoiesis.
Because of the wide tissue distribution of NOD1, its stimulation may have many different outcomes depending on the cell type triggered. In contrast, TLR4 signaling is most often associated with production of a common set of pro-inflammatory cytokines such as IL-6 or TNF-α. Based on the results presented here, LPS or NOD1 ligand–stimulated BM MSCs are both likely to promote hematopoiesis (supplemental Figure 8). LPS as a potent inducer of IL-6 favors myelopoiesis, whereas NOD1 ligand augments the numbers of all HSPCs. Nevertheless, the partial effects of either MyD8810 or NOD1 deficiency (supplemental Figure 2) on hematopoiesis in steady state suggest that in the presence of the microbiota, these pathways can have redundant functions. Future studies of the relative importance and interaction of these 2 innate recognition pathways could lead to novel therapeutic approaches for the treatment of hematopoietic disorders.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Lara Mittereder, Sara Hieny, and Sandy Oland for their excellent technical assistance; Calvin Eigsti for performing FACS sorting; the NIAID Gnotobiotic Animal Facility staff, in particular Dana Trageser-Cesler, Cesar Acevedo, and Jess LeGrand, for invaluable help in maintaining GF animals; and Mazier Divangahi (McGill University) for providing the NOD1−/− mice used in this study.
This work was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health, and by a fellowship from the Uehara Memorial Foundation (C.I.).
Authorship
Contribution: C.I., D.J., and A.S. designed the study; C.I., D.J., and N.B. performed experiments; N.B. and Y.B. provided GF mice and advice on their use; C.I. and D.J. analyzed the data and performed the statistics; and C.I., D.J., and A.S. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dragana Jankovic, BG33 RM 1W10A3, 33 North Dr, Bethesda, MD 20814; e-mail: djankovic@niaid.nih.gov.
References
Author notes
A.S. and D.J. are considered co-senior authors.

![Figure 2. NOD1 ligand stimulation of MSCs induces secretion of cytokines that promote proliferation of HSPCs. (A) FACS-purified HSC, MPP, and CLP were cultured for 48 hours in the presence of NOD1 (10 μg/mL) or NOD2 (10 μg/mL) ligand alone or in combination with SCF (10 ng/mL), Flt3L (10 ng/mL), or ThPO (10 ng/mL) in the case of HSC and MPP, and with IL-7 (10 ng/mL) and Flt3L in the case of CLP. [3H]-thymidine was added during the last 16 hours of culture. Bars represent mean ± SD of thymidine incorporation for triplicate cultures. N.D., not determined. (B) Expression of Nod1 and Nod2 mRNA in cultured CD45neg MSC and CD45+ Mϕ from WT, NOD1−/−, or NOD2−/− mice. Bars represent mean ± SD of triplicate values of Nod1 and Nod2 expression relative to RPLP2 expression. (C) MSCs generated from WT or NOD1−/− mice were stimulated with NOD1 ligand (C12-iE-DAP, 10 μg/mL) or LPS (10 μg/mL) and 72 hours later harvested for RNA extraction. Bars represent mean ± SD of triplicate measurements of Nod1 relative to Rplp2 expression for each stimulus. (D) Expression of mRNA for IL-3, IL-6, IL-7, SCF, Thpo, and Flt3L in WT MSCs treated with NOD1 ligand (10 μg/mL) or LPS (10 μg/mL). Bars represent the means ± SD of triplicate values of mRNA expression relative to Hprt for each cytokine. (E-F) Sorted LSK (2 × 103/well), HSC (2 × 103/well), CLP (2 × 103/well), or CMP (4 × 103/well) from WT mice were stimulated for 48 hours with the culture supernatants harvested from WT (WT CS) or NOD1−/− (KO CS) MSC cultures treated with NOD1 ligand (10 μg/mL) for 72 hours (E). [3H]-thymidine (0.5 μCi/well, New England Nuclear Corp., sp act: 2 Ci/mmol) was added at 48 hours and the incorporated radioactivity measured 16 hours later. Bars represent the mean ± SD of the values from triplicate cultures. The data shown in (A-E) are from 1 representative out of 3 experiments performed. (F) Serum levels of IL-7, SCF, ThPO, and Flt3L in GF mice after gavage with NOD1 ligand. The concentrations of IL-7, Flt3L, ThPO, and SCF in culture supernatants or serum were measured by Quantikine ELISA kit (R&D systems). Bars represent the mean ± SEM of the ELISA values obtained from the individual SPF, GF, and NOD1 ligand–treated GF mice shown in Figure 1F and pooled from 2 independently performed replicate experiments. *P < .05; **P < .01; ***P < .001.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/129/2/10.1182_blood-2016-06-723742/4/m_blood723742f2.jpeg?Expires=1767763946&Signature=r8pIGpQy617RYjKOHNfxymgdks1UkrQolTgp7Onc58uXe-i3Kaq1AxL6xqOb27PQ4EGeeEyX3SnU-2NzloI0tmeTLV-58VtdcFWMY9i69ZhbVYyUKpk2zRli9qhtYaj2hyadh1cZ5fLXGGOFrq9fgixULvNTGo4H4fWkuN7VHza8QwTAsvPsNbTpv6-bUa83t7mIgjGvqoB7-4MNezpN4FqCPnkNhnhNV0zI8iN4kZQBD913Ivaj7aJVXuBkHMzmdD8GkBAm4wz1vSQS9fCJmgA4rgBSxz4pLjk6OH3V9R2Ru3uTikLn3skczjA8Wprr7iBobHc4JjNNG~qkXEKnow__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal