Key Points
Two circulating exosomal microRNAs, let-7b and miR-18a, improved survival prediction in patients with MM.
Circulating exosomal miRNAs enhanced the stratification of patients with high-risk factors.
Abstract
Exosomes, secreted by several cell types, including cancer cells, can be isolated from the peripheral blood and have been shown to be powerful markers of disease progression in cancer. In this study, we examined the prognostic significance of circulating exosomal microRNAs (miRNAs) in multiple myeloma (MM). A cohort of 156 patients with newly diagnosed MM, uniformly treated and followed, was studied. Circulating exosomal miRNAs were isolated and used to perform a small RNA sequencing analysis on 10 samples and a quantitative reverse transcription polymerase chain reaction (qRT-PCR) array on 156 samples. We studied the relationship between miRNA levels and patient outcomes, including progression-free survival (PFS) and overall survival (OS). We identified miRNAs as the most predominant small RNAs present in exosomes isolated from the serum of patients with MM and healthy controls by small RNA sequencing of circulating exosomes. We then analyzed exosomes isolated from serum samples of 156 patients using a qRT-PCR array for 22 miRNAs. Two of these miRNAs, let-7b and miR-18a, were significantly associated with both PFS and OS in the univariate analysis and were still statistically significant after adjusting for the International Staging System and adverse cytogenetics in the multivariate analysis. Our findings support the use of circulating exosomal miRNAs to improve the identification of patients with newly diagnosed MM with poor outcomes. The results require further validation in other independent prospective MM cohorts.
Introduction
Multiple myeloma (MM) is a hematological malignancy characterized by a clonal proliferation of plasma cells in the bone marrow microenvironment.1 The clinical and biological heterogeneities of this malignancy lead to variable responses to therapy and outcomes.2 With a vast increase in therapeutic choices in MM and improved outcomes, the issue of risk stratification to dissect this heterogeneity is becoming more critical because it may lead to tailored therapies for different groups of patients.
A large number of prognostic biomarkers have been identified, including markers that reflect tumor burden, stage of disease, or tumor biology (such as chromosomal abnormalities or gene expression signatures) and factors present in the host that indicate fitness for therapy.2 The most widely used prognostic factors in MM are currently the International Staging System3 (ISS), based on albumin and β-2 microglobulin (B2M) levels in the peripheral blood at the time of diagnosis, and chromosomal abnormalities such as translocation t(4;14), 17p deletion, and 1q21 amplification.4 A revised ISS has been proposed that includes high-risk cytogenetics for improved characterization of patients with poor survival.5 However, despite these advances, patients within similar prognostic groups display heterogeneous outcomes, indicating that current prognostic factors used in MM are suboptimal in stratifying patients with high-risk features. Combining information about cytogenetic abnormalities and the ISS with other molecular markers may therefore further improve their prognostic value.
Exosomes are 50- to 140-nm nanovesicles that contain proteins and nucleic acids, such as microRNAs (miRNAs).6 They are actively secreted by several cell types, including cancer cells, and can be isolated from the peripheral blood, making them attractive for use as biomarkers of disease progression and risk stratification. Exosomes have been reported to promote tumorigenesis in many cancer types, particularly through transfer of miRNAs.7-9 Remarkably, tumor-derived exosomes contain their own miRNA-associated machinery and display a cell-independent capacity for processing precursor miRNAs into mature miRNAs. This phenomenon mediates an efficient and rapid silencing of messenger RNA (mRNA) in target cells, thus promoting oncogenesis.9
Of note, miRNAs are small noncoding RNAs that are assumed to regulate > 50% of all protein-coding genes.10 They predominantly act as translational repressors by binding to the 3′ untranslated region of their targeted mRNAs, which are deregulated in most cancer types,11 including MM,12,13 and have a role in the development and progression of many cancers.14-18
Circulating miRNAs are generated via 2 main mechanisms: cell death by apoptosis or necrosis, leading to the release of miRNAs bound to AGO proteins, and an active process by secretion of exosomes containing miRNAs.19 Thus, exosomal miRNAs could represent a more specific molecular biomarker than cell-free miRNAs. However, the clinical significance of circulating exosomal miRNAs in MM has not been examined. In this study, we aimed to characterize circulating exosomal miRNAs in MM and determine their impact on patient outcomes.
Materials and methods
Plasma samples from patients with MM
We obtained 156 serum samples from the Intergroupe Francophone du Myélome collected between 14 June 2006 and 16 December 2008 for this study. All patients were newly diagnosed with MM, uniformly followed and treated with a combination of bortezomib and dexamethasone, followed by high-dose melphalan and autologous hematopoietic stem-cell transplantation. None of the patients received therapy before the collection of blood samples. Criteria for diagnosis, clinical staging, and risk stratification were assessed according to the International Myeloma Working Group guidelines.20 Patients provided written informed consent in accordance with the Declaration of Helsinki. In addition, samples from 5 healthy volunteers (for age-matched comparison) were used for the preliminary RNA sequencing study.
Circulating exosome isolation
We isolated circulating exosomes as described previously.21 Serum was extracted from blood drawn on dry tubes. The starting volume of serum was 0.5 mL, which was diluted into 2 mL of phosphate-buffered saline before differential centrifugation and precipitation reagent. Exosomes were isolated from frozen serum samples using a combined centrifugation and exosome isolation reagent method (supplemental Figure 1A, available on the Blood Web site). Serum was isolated by centrifugation at 300 g for 10 minutes, and further spun down at 2000 g for 10 minutes and 10 000 g for 10 minutes to remove dead cells and cell debris, respectively. We harvested exosomes by adding an exosome isolation reagent for 30 minutes (ExoQuick solution) before centrifugation at 1500 g for 30 minutes. We used a differential centrifugation protocol combined with an exosome isolation reagent where the exosome pellet was washed before RNA extraction, thus limiting potential contamination by cell-free or exosome-free miRNA.
Electron microscopy
Exosomes were characterized by electron microscopy using CD63 and CD81 as follows: pelleted exosomes were fixed in 0.1-M phosphate buffer (pH, 7.4) with 2% paraformaldehyde, then processed for ultrathin sectioning and immunogold labeling by using anti-CD63 and anti-CD81 antibodies and protein A coupled with 10- or 15-nm gold particles. Sections were observed at 80 kV on a TecnaiGβ Spirit BioTWIN transmission electron microscope (FEI), and images were recorded with an AMT 2k CCD camera. We confirmed the presence of exosomes by transmission electron microscopy and NanoSight analysis in MM samples before proceeding with the rest of the samples for exosome isolation.
RNA extraction and RNA sequencing
Total RNA was extracted from exosome pellets using the miRNeasy Micro Kit (Qiagen). Small RNA libraries were prepared and amplified using the NEBNext Small RNA Library Prep Set (New England BioLabs). Amplified libraries were resolved on a 10% polyacrylamide gel for size selection. The 140- to 160-nucleotide bands corresponding to adapter-ligated constructs derived from the 21- to 40-nucleotide RNA fragments were excised and recovered in a DNA elution buffer. The average size distribution of each library was determined using Agilent Bioanalyzer with High Sensitivity Chip Kit (Agilent) and quantified on an ABI 7900HT Fast reverse transcription PCR instrument using the KAPA Library Quantification Kit (Kapa Biosystems). Each library was adjusted to final concentration of 2 nM and pooled and sequenced on an Illumina HiSequation 2000 sequencer for single-read 50 cycles at the Center for Cancer Computational Biology at Dana-Farber Cancer Institute. The BCL files were demultiplexed using CASAVA 1.8.2 (Illumina) into FASTQ files. Raw sequencing reads were then analyzed using miRDeep2 to quantify known small RNA species. RNA sequencing was performed for circulating exosomes obtained from the serum of 10 patients with MM and 5 healthy individuals.
TaqMan Low-Density Array
For quantitative reverse transcription polymerase chain reaction (qRT-PCR), we designed a custom TaqMan low-density array (TLDA; Applied Biosystems). Twenty-two miRNAs were selected based on biological relevance in prior studies of tumor samples in MM.12,13 RNA concentrations were measured with a Qubit miRNA assay, and 5 ng of miRNA was reverse transcribed to complementary DNA using an miRNA reverse transcription kit (TaqMan; Applied Biosystems) and preamplified with a custom pool of primers and a PreAmp Master Mix (TaqMan; Applied Biosystems). qPCR reactions were performed with the TaqMan Universal Master Mix II reagent in Custom TaqMan Array Cards (384-well plate preloaded with 22 specific primers of interest) on a ViiA 7 reverse transcription PCR System (Applied Biosystems). TLDA quantification was performed based on the manufacturer’s protocol and based on prior publications.22 All cards were run on the Applied Biosystems 7900HT Fast reverse transcription PCR System using the AgPath-ID One-Step Kit (Applied Biosystems). All assays were performed in duplicate in each card. A subset of samples was run in two different cards to test reproducibility. The gene expression levels were determined and relatively quantified by using the comparative cycle threshold (Ct) method. All Ct values >35 cycles were considered undetectable. We normalized qRT-PCR data using a robust global median normalization as described previously (supplemental Figure 2).23 Each plate was adjusted by a normalization factor as the difference between the global median Ct value and the plate median Ct value. We calculated the expression of miRNAs with ΔCt, in which the maximal Ct value for an miRNA was subtracted from the specific value for this miRNA. The average of the replicate expression values of the miRNAs was used in the analysis. In our experimental design, we controlled our data quality by having technical replicates of plates and technical replicates of each sample in each plate. Technical replicates of plates were used to assess the variability of measurement in two plates; we had 2 control plates in which the same set of samples was analyzed. We observed a high correlation with Ct values from the duplicate plate, which indicated good concordance between technical replicates of plates. We indeed observed that Ct values in some plates were higher, and this was corrected after global normalization. In addition, we calculated the Ct difference for all technical replicates for each sample in each plate.
Statistical analysis
The primary outcomes of interest were progression-free survival (PFS) and overall survival (OS). To illustrate the impact of the miRNAs graphically, the miRNAs were dichotomized at the median based on a low-versus-high expression. PFS and OS were plotted and compared using the Kaplan-Meier method and the log-rank test. A Cox proportional hazards model was employed to compute the hazard ratios and accompanying 95% confidence intervals. A multivariate Cox proportional hazards model was used to identify independent outcome predictors after adjustment for confounders, such as the ISS and cytogenetics.
We compared miRNA expressions between patients with MM and healthy adults by using the Mann-Whitney U test in the preliminary study, then further analyzed the effect of each miRNA expression in the serum samples of 156 newly diagnosed untreated patients with MM on outcomes. We dichotomized the expression levels of miRNAs according to the median level of all samples and further analyzed their clinical impact. We defined an miRNA as clinically significant when it significantly correlated with both PFS and OS in the multivariate analysis. Correction for multiple comparisons was performed by using Benjamini-Hochberg correction in the univariate analysis. The miRNAs that were significant after correction were adjusted for the ISS and cytogenetics in the multivariate analysis.
The area under the receiver operating characteristic curve (AUC) was used to evaluate the predictive values of miRNA expressions for the patients’ PFS and OS. We compared the curves between the miRNA expressions alone, the ISS with cytogenetics, and both to evaluate the predictive value of the miRNA expressions. To avoid overfitting, cross-validation was applied to this analysis. We evenly divided the study cohort into a training part and a validation part by random sampling. We used the Cox proportional hazards model to find the most significant two miRNAs in the training cohort. The two miRNAs were used to calculate the AUC in the validation cohort. We iterated 1000 times and reported the mean of the results.
All statistical analyses were performed in R (version 3.1.1), SAS 9.2 software (SAS Institute Inc., Cary, NC), and STATA statistical software (version 12.1; StataCorp). Those statistical analyses with P values < .05 in the univariate model were further analyzed in the multivariate analysis.
Results
Characterization of circulating exosomes in MM
We first characterized peripheral blood circulating exosomes from patients with MM and healthy adults. After isolation of circulating exosomes (supplemental Figure 1A), we confirmed the presence of exosomes by transmission electron microscopy with immunogold labeling for CD63 and CD81, which are specific markers of exosomes (supplemental Figure 1B).6 The diameter of isolated exosomes was confirmed to be ∼120 nm by NanoSight analysis (supplemental Figure 1C).
To define the content of exosomes in terms of small RNAs, we next performed a small RNA sequencing from circulating exosomes of 10 newly diagnosed patients with MM and 5 healthy individuals. A large majority of mappable RNAs were miRNAs (88.0% in MM samples and 86.7% in healthy donor samples). The rest of the RNAs were represented by small nuclear and nucleolar RNA, ribosomal RNA, mRNA, long intergenic noncoding RNA, and unclassified RNA. Importantly, we found no difference in distribution of small RNA between the exosomes of donors with MM and healthy donors. The distribution of small exosomal RNAs between samples from donors with MM and healthy donors is shown in Figure 1A and supplemental Figure 3.
Circulating exosome characterization in patients with MM. (A) Distribution of mappable small RNAs by next-generation sequencing in circulating exosomes from 10 patients with MM and 5 healthy donors. (B) qRT-PCR of circulating exosomal miRNAs in 10 patients with MM and 5 healthy donors. Box plots represent the medians and standard deviations of the normalized expression levels of 22 miRNAs. lincRNA, long intergenic noncoding RNA; misc_RNA, miscellaneous other RNA; mt-rRNA, ribosomal RNA located in mitochondrial genome; mt-tRNA, transfer RNA located in mitochondrial genome; rRNA, ribosomal RNA; snoRNA, small nucleolar RNA; snRNA, small nuclear RNA.
Circulating exosome characterization in patients with MM. (A) Distribution of mappable small RNAs by next-generation sequencing in circulating exosomes from 10 patients with MM and 5 healthy donors. (B) qRT-PCR of circulating exosomal miRNAs in 10 patients with MM and 5 healthy donors. Box plots represent the medians and standard deviations of the normalized expression levels of 22 miRNAs. lincRNA, long intergenic noncoding RNA; misc_RNA, miscellaneous other RNA; mt-rRNA, ribosomal RNA located in mitochondrial genome; mt-tRNA, transfer RNA located in mitochondrial genome; rRNA, ribosomal RNA; snoRNA, small nucleolar RNA; snRNA, small nuclear RNA.
Exosomal miRNAs and survival in MM
Given that the majority of the RNA content was miRNAs, we focused on miRNAs that could be differentially expressed between patients with MM and could be predictive of prognosis. We sought to design a panel of miRNAs that was specific for MM pathogenesis based on prior studies in tumor cells and thereby restricted our prognostic studies of exosomal miRNAs to those that should be critical for MM tumor–derived miRNAs. This helped us avoid detecting miRNAs that were derived from other nontumor cells as well as having to perform comparisons of circulating exosomal miRNAs with tumor-derived miRNAs, given that we did not have access to matched tumor cells from this cohort of patients. In this respect, we focused on 22 miRNAs that were selected based on their biological relevance in prior studies of tumor samples in MM and their presence in the RNA sequencing study performed (supplemental Figure 4).12,13 We developed a custom-made TaqMan assay using these miRNAs to perform a screening of the prognostic relevance of exosomal miRNAs in patients with MM. The expression levels of these 22 miRNAs were further confirmed using RT-PCR in the same samples that were profiled by RNA sequencing. The expression levels of these 22 miRNAs were significantly lower in patients with MM compared with healthy individuals (Figure 1B).
We aimed to identify whether circulating exosomal miRNAs were of clinical prognostic significance in patients with newly diagnosed MM. Therefore, we obtained serum samples of 156 patients with newly diagnosed MM who were uniformly treated with bortezomib and dexamethasone (from the Intergroupe Francophone du Myélome group). The clinical characteristics of the patients are listed in Table 1. All serum samples were harvested at diagnosis, before the initiation of therapy. The median follow-up of the cohort was 5.4 years (interquartile range, 4.6-5.8 years). We performed a custom qRT-PCR TLDA to assess the clinical significance of the 22 selected miRNAs. The association between the miRNAs and patient outcomes was analyzed in the univariate and multivariate Cox regression models. We performed Benjamini-Hochberg correction for multiple comparisons. After correction, let-7b, let-7e, miR-106a, miR-106b, miR-155, miR-16, miR-17, miR-18a, and miR-20a were significant risk factors for PFS in the univariate analyses. All of these miRNAs were still significant after adjustment for the ISS and specific cytogenetic abnormalities. After Benjamini-Hochberg correction, let-7b, miR-155, and miR-18a were significantly associated with patient survival in the univariate analyses. However, only let-7b and miR-18a were significant predictors for OS in the multivariate models (Table 2). We further analyzed the association between let-7b and miR-18a and patient characteristics, because only these two exosomal miRNAs were demonstrated to be independent predictors for both PFS and OS in the univariate and multivariate analyses. Lower expression of let-7b or miR-18a was significantly associated with a high ISS stage (supplemental Tables 1 and 2). However, both let-7b and miR-18a were independent predictors after adjusting for the ISS and specific cytogenetic abnormalities. The effect of the two miRNAs on PFS and OS was illustrated by Kaplan-Meier curves with dichotomized miRNAs at the median (Figure 2). These data indicate that specific miRNAs can be critical in defining worse prognosis in patients with newly diagnosed MM.
Clinical characteristics of patients (N = 156)
| Characteristic . | No. . | % . |
|---|---|---|
| Age, y | ||
| Median | 56 | |
| Range | 34-73 | |
| Male sex | 89 | 57 |
| IgH | ||
| IgG | 81 | 52 |
| IgA | 36 | 23 |
| IgD | 2 | 1 |
| No heavy chain | 22 | 14 |
| No data | 15 | 10 |
| IgL | ||
| κ | 94 | 60 |
| Λ | 41 | 26 |
| No data | 21 | 14 |
| ISS score | ||
| I | 63 | 40 |
| II | 56 | 36 |
| III | 33 | 21 |
| No data | 4 | 3 |
| FISH* | ||
| del(13q) | 59 | 40 |
| t(4;14) | 14 | 10 |
| del(17p) | 5 | 3 |
| High risk† | 17 | 12 |
| PFS | ||
| Relapse or death | 111 | 71 |
| 3 years | 50 | |
| 95% CI | 42-28 | |
| OS | ||
| Death | 27 | 17 |
| 3 years | 97 | |
| 95% CI | 95-99 | |
| Follow-up, years | ||
| Median | 5.6 | |
| IQR | 5.4-5.9 | |
| Characteristic . | No. . | % . |
|---|---|---|
| Age, y | ||
| Median | 56 | |
| Range | 34-73 | |
| Male sex | 89 | 57 |
| IgH | ||
| IgG | 81 | 52 |
| IgA | 36 | 23 |
| IgD | 2 | 1 |
| No heavy chain | 22 | 14 |
| No data | 15 | 10 |
| IgL | ||
| κ | 94 | 60 |
| Λ | 41 | 26 |
| No data | 21 | 14 |
| ISS score | ||
| I | 63 | 40 |
| II | 56 | 36 |
| III | 33 | 21 |
| No data | 4 | 3 |
| FISH* | ||
| del(13q) | 59 | 40 |
| t(4;14) | 14 | 10 |
| del(17p) | 5 | 3 |
| High risk† | 17 | 12 |
| PFS | ||
| Relapse or death | 111 | 71 |
| 3 years | 50 | |
| 95% CI | 42-28 | |
| OS | ||
| Death | 27 | 17 |
| 3 years | 97 | |
| 95% CI | 95-99 | |
| Follow-up, years | ||
| Median | 5.6 | |
| IQR | 5.4-5.9 | |
CI, confidence interval; FISH, fluorescence in situ hybridization; IgH, immunoglobulin heavy chain; IgL, immunoglobulin light chain; IQR, interquartile range.
del(13q): unknown for 7 participants; t(4;14): unknown for 13 participants; del(17p): unknown for 8 participants.
High-risk cytogenetics include t(4;14) and/or del(17p).
Risk factors for mortality and progression among patients with MM
| . | PFS . | OS . | ||||||
|---|---|---|---|---|---|---|---|---|
| Crude HR (95% CI) . | P . | Adjusted* HR (95% CI) . | P . | Crude HR (95% CI) . | P . | Adjusted* HR (95% CI) . | P . | |
| Low let-7b | 1.78 (1.21-2.60) | .003† | 1.90 (1.22-2.94) | .004 | 4.57 (1.90-10.97) | .001† | 2.83 (1.07-7.50) | .036 |
| Low let-7c | 1.22 (0.84-1.78) | .302 | 2.28 (1.04-4.99) | .039 | ||||
| Low let-7e | 1.73 (1.19-2.53) | .005† | 2.01 (1.30-3.11) | .002 | 2.39 (1.09-5.24) | .030 | ||
| Low miR-106a | 1.90 (1.30-2.78) | .001† | 2.34 (1.52-3.61) | <.001 | 2.67 (1.21-5.88) | .015 | ||
| Low miR-106b | 2.52 (1.72-3.69) | <.001† | 3.54 (2.21-5.68) | <.001 | 2.76 (1.25-6.10) | .012 | ||
| Low miR-10b | 1.23 (0.85-1.80) | .273 | 1.82 (0.83-3.98) | .133 | ||||
| Low miR-125a | 1.28 (0.88-1.86) | .203 | 2.31 (1.05-5.08) | .037 | ||||
| Low miR-125b | 1.02 (0.70-1.49) | .906 | 1.27 (0.60-2.72) | .533 | ||||
| Low miR-155 | 1.64 (1.12-2.40) | .011† | 1.76 (1.15-2.69) | .009 | 3.19 (1.42-7.14) | .005† | 2.41 (0.96-6.05) | .061 |
| Low miR-15a | 1.37 (0.94-2.00) | .101 | 2.27 (1.02-5.06) | .046 | ||||
| Low miR-16 | 1.86 (1.27-2.72) | .001† | 2.21 (1.41-3.47) | .001 | 2.37 (1.09-5.17) | .030 | ||
| Low miR-17 | 1.83 (1.25-2.67) | .002† | 2.29 (1.48-3.55) | <.001 | 2.21 (1.01-4.83) | .046 | ||
| Low miR-181a | 1.45 (0.99-2.12) | .054 | 2.15 (1.00-4.63) | .051 | ||||
| Low miR-18a | 2.01 (1.37-2.94) | <.001† | 2.76 (1.79-4.26) | <.001 | 4.62 (1.85-11.50) | .001† | 4.52 (1.57-12.98) | .005 |
| Low miR-19a | 0.13 (0.02-0.99) | .049 | ‡ | ‡ | .990 | |||
| Low miR-19b | 0.83 (0.56-1.22) | .335 | 0.48 (0.22-1.03) | .060 | ||||
| Low miR-20a | 2.00 (1.37-2.92) | <.001† | 2.31 (1.52-3.53) | <.001 | 2.91 (1.29-6.54) | .010 | ||
| Low miR-21 | 1.53 (1.05-2.23) | .028 | 1.88 (0.88-4.06) | .106 | ||||
| Low miR-223 | 1.33 (0.91-1.94) | .147 | 1.77 (0.83-3.76) | .140 | ||||
| Low miR-25 | 1.20 (0.82-1.76) | .344 | 2.56 (1.16-5.65) | .020 | ||||
| Low miR-744 | 1.32 (0.91-1.93) | .144 | 2.10 (0.97-4.53) | .059 | ||||
| Low miR-92a | 1.39 (0.95-2.02) | .089 | 2.15 (1.00-4.65) | .051 | ||||
| . | PFS . | OS . | ||||||
|---|---|---|---|---|---|---|---|---|
| Crude HR (95% CI) . | P . | Adjusted* HR (95% CI) . | P . | Crude HR (95% CI) . | P . | Adjusted* HR (95% CI) . | P . | |
| Low let-7b | 1.78 (1.21-2.60) | .003† | 1.90 (1.22-2.94) | .004 | 4.57 (1.90-10.97) | .001† | 2.83 (1.07-7.50) | .036 |
| Low let-7c | 1.22 (0.84-1.78) | .302 | 2.28 (1.04-4.99) | .039 | ||||
| Low let-7e | 1.73 (1.19-2.53) | .005† | 2.01 (1.30-3.11) | .002 | 2.39 (1.09-5.24) | .030 | ||
| Low miR-106a | 1.90 (1.30-2.78) | .001† | 2.34 (1.52-3.61) | <.001 | 2.67 (1.21-5.88) | .015 | ||
| Low miR-106b | 2.52 (1.72-3.69) | <.001† | 3.54 (2.21-5.68) | <.001 | 2.76 (1.25-6.10) | .012 | ||
| Low miR-10b | 1.23 (0.85-1.80) | .273 | 1.82 (0.83-3.98) | .133 | ||||
| Low miR-125a | 1.28 (0.88-1.86) | .203 | 2.31 (1.05-5.08) | .037 | ||||
| Low miR-125b | 1.02 (0.70-1.49) | .906 | 1.27 (0.60-2.72) | .533 | ||||
| Low miR-155 | 1.64 (1.12-2.40) | .011† | 1.76 (1.15-2.69) | .009 | 3.19 (1.42-7.14) | .005† | 2.41 (0.96-6.05) | .061 |
| Low miR-15a | 1.37 (0.94-2.00) | .101 | 2.27 (1.02-5.06) | .046 | ||||
| Low miR-16 | 1.86 (1.27-2.72) | .001† | 2.21 (1.41-3.47) | .001 | 2.37 (1.09-5.17) | .030 | ||
| Low miR-17 | 1.83 (1.25-2.67) | .002† | 2.29 (1.48-3.55) | <.001 | 2.21 (1.01-4.83) | .046 | ||
| Low miR-181a | 1.45 (0.99-2.12) | .054 | 2.15 (1.00-4.63) | .051 | ||||
| Low miR-18a | 2.01 (1.37-2.94) | <.001† | 2.76 (1.79-4.26) | <.001 | 4.62 (1.85-11.50) | .001† | 4.52 (1.57-12.98) | .005 |
| Low miR-19a | 0.13 (0.02-0.99) | .049 | ‡ | ‡ | .990 | |||
| Low miR-19b | 0.83 (0.56-1.22) | .335 | 0.48 (0.22-1.03) | .060 | ||||
| Low miR-20a | 2.00 (1.37-2.92) | <.001† | 2.31 (1.52-3.53) | <.001 | 2.91 (1.29-6.54) | .010 | ||
| Low miR-21 | 1.53 (1.05-2.23) | .028 | 1.88 (0.88-4.06) | .106 | ||||
| Low miR-223 | 1.33 (0.91-1.94) | .147 | 1.77 (0.83-3.76) | .140 | ||||
| Low miR-25 | 1.20 (0.82-1.76) | .344 | 2.56 (1.16-5.65) | .020 | ||||
| Low miR-744 | 1.32 (0.91-1.93) | .144 | 2.10 (0.97-4.53) | .059 | ||||
| Low miR-92a | 1.39 (0.95-2.02) | .089 | 2.15 (1.00-4.65) | .051 | ||||
CI, confidence interval; HR, hazard ratio.
Adjusted for ISS stage, del(17q), and t(4;14).
Significance by using Benjamini-Hochberg correction (corrected significance level = .020 for PFS and .006 for OS).
Data did not converge.
Kaplan-Meier survival curves according to different levels of circulating exosomal miRNAs in MM. (A) PFS and (B) OS in patients with MM. The miRNAs were dichotomized at the median based on a low-versus-high expression.
Kaplan-Meier survival curves according to different levels of circulating exosomal miRNAs in MM. (A) PFS and (B) OS in patients with MM. The miRNAs were dichotomized at the median based on a low-versus-high expression.
AUC
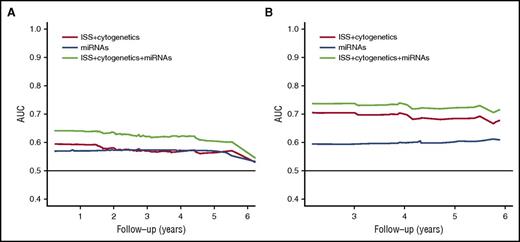
To further assess the prognostic value of the signature of the miRNAs in addition to their clinical value, we performed an AUC analysis with cross-validation. The combination of the signature of the two miRNAs with ISS and cytogenetic status had better prediction for PFS and OS than when the miRNA signature was excluded (Figure 3). Together, these data indicate that circulating exosomal miRNAs could improve the prognostic stratification of patients with MM, in addition to the ISS and cytogenetics.
The AUC. Comparisons between the AUC curves of the ISS and cytogenetics, the signature of the two miRNAs, and the combination for (A) PFS and (B) OS. Cross-validation was applied to this analysis to avoid overfitting.
The AUC. Comparisons between the AUC curves of the ISS and cytogenetics, the signature of the two miRNAs, and the combination for (A) PFS and (B) OS. Cross-validation was applied to this analysis to avoid overfitting.
Discussion
To our knowledge, this is the first large study that proves the clinical significance of circulating exosomal miRNAs in a uniformly treated newly diagnosed MM cohort. Indeed, miRNAs have recently emerged as promising biomarkers for different pathological conditions, including cancer, and their inherent stability in the serum and exosomes and reproducible levels across many individuals make miRNAs attractive biomarkers. Unlike current conventional genomic approaches that require a bone marrow biopsy, exosomal miRNAs can be obtained from the peripheral blood, making them candidates for use as noninvasive biomarkers. In this study, we examined a novel prognostic biomarker from patients’ serum samples to improve the prediction of PFS and OS in patients with MM. Our results show that two miRNAs, let-7b and miR-18a, from circulating exosomes can predict PFS and OS in patients with newly diagnosed MM in an independent manner and improve the prognostic value of ISS and cytogenetic status in MM.
Cell-free miRNAs have emerged as appealing biomarkers because they are noninvasive and have been reported as prognostic tools in many cancer types,24 including MM.25-27 However, many circulating miRNAs are passively released from apoptotic and necrotic cells19,24 and therefore may not truly reflect the biological changes that occur in these tumor cells. In contrast, exosomes are actively secreted in the peripheral blood by different cell types, including cancer cells, and are biologically relevant because they promote tumorigenesis through miRNA transfer.19 Recent studies have shown that cancer exosomes are capable of cell-independent miRNA processing and transfer of mature miRNAs into recipient cells; they thus mediate significant transcriptome alterations in target cells and lead to the induction of proliferation and conversion of nontumorigenic cells into tumor-forming cells.9 This indicates that exosomes carry specifically selected miRNAs as well as their own miRNA biogenesis machinery.9 Therefore, exosomal miRNAs may truly represent specific molecular biomarkers, in contrast with cell-free miRNAs.
Peripheral blood exosomes have been reported to be strong diagnostic and prognostic markers in cancer. Indeed, patients with stage IV melanoma and a high protein concentration in circulating exosomes have shorter survival.7 Specifically, markers of melanoma, such as TYRP2, VLA-4, and HSP70, are significantly higher in patients with stage IV disease compared with those at other stages. More recently, circulating exosomal miRNAs were studied in castration-resistant prostate cancer, and two (miR-1290 and miR-375) were shown to be independent predictive markers of OS in patients with this disease, improving the predictive value of the standard clinical staging system.28 Melo et al29 identified the presence of glypican-1 on circulating exosomes in patients with pancreatic cancer, which could serve as a powerful marker in both the diagnosis and prognosis of this disease. Future studies to determine the source of exosomes present in the peripheral blood and whether the level of miRNAs in circulating exosomes correlates with tumor-derived miRNAs should be performed. However, it is well known that circulating exosomal miRNAs can be prognostic in many cancer types without the need for profiling of the tumor cellular counterparts.7,28,29
In this study, we used the mean expression value for normalization instead of RNU6B normalization. Xiang et al30 reported that U6 was not a suitable endogenous control for the quantification of circulating miRNAs, especially in frozen samples. Mestdagh et al31 demonstrated that using the mean expression value was better than other small noncoding RNAs for normalization. Cui et al32 reported that normalization based on the global mean of each plate was superior to normalization based on mammalian U6 or spiked-in miR159. Our study shows high concordance across plates and for replicates of each sample, and the interplate variability was corrected by global normalization.
Established markers of prognosis in MM include the ISS and cytogenetics.3 The ISS classification is based on the nonclonal markers albumin and B2M. Although B2M is a useful marker of the tumor burden, it may not be specific enough to define the clinical and biological heterogeneities of patients with MM.2 Cytogenetics and several gene expression signatures are truly reflective of the molecular and biological characteristics of the tumor clone. However, these are performed only on tumor cells obtained from bone marrow biopsies. Therefore, there is a need to develop noninvasive biomarkers that reflect the molecular aspect of the disease.
We identified two exosomal miRNA signatures, let-7b and miR-18a, that were associated with poor outcomes with regard to PFS and OS. The let-7 family is one of the most studied groups of miRNAs in cancer.33 We and others have recently reported that there was a significant decrease in the expression of let-7 family members in MM tumor cells compared with normal plasma cells.12,13,34 Indeed, we demonstrated that let-7 acted as a tumor-suppressor miRNA in MM.34 This miRNA family is notably processed by the RNA-binding protein LIN28B, which is often deregulated in cancer.35,36 Low levels of let-7 induce cell proliferation and growth by derepressing oncogenes such as CCND1, MYC, and RAS.37 MiR-18a is a component of the miR-17-92 cluster on chromosome 13q31.3, which undergoes amplification in lymphomas and solid tumors.38 Krutilina et al39 reported that miR-18 inhibited hypoxia-inducible factor 1α activity in breast cancer and repressed tumor dissemination through a hypoxia-inducible factor 1α–dependent pathway. Teng et al40 found that miR-18a mediated induction of M1 macrophages by directly targeting IRF2. Natural killer cells and natural killer T cells activated by miR-18a play a critical role in the inhibition of tumor metastasis.
The results of this study need to be confirmed in large series of patients with MM. In addition, the prognostic relevance of these miRNAs in patients treated with other therapeutic agents may be different, indicating the need for larger studies to be performed in different therapeutic settings. However, this large study provides evidence of the association between exosomal miRNAs and outcomes in newly diagnosed patients with MM. We have identified an unprecedented prognostic significance for circulating exosomal miRNAs in patients with MM, which requires further validation in other independent prospective MM cohorts.
The RNA sequencing data reported in this article are publicly available at http://www.ncbi.nlm.nih.gov/geo/ (Gene Expression Omnibus accession number GSE94564).
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Sonal Jhaveri-Schneider (Office for Postdoctoral Training & Career Development, Dana-Farber Cancer Institute) for the English editing of the manuscript.
This work was supported by a grant from the National Institutes of Health, National Cancer Institute (R01CA154648).
Authorship
Contribution: S. Manier and I.M.G. designed the study; S. Manier, K.Z.S., D.H., S.V.G., B.R., A.S., and J.B. obtained and assembled the data; H.A.-L., S. Minvielle, P.M., T.F., and X.L. collected the samples and clinical information; S. Manier, C.-J.L., J.P., J.S., F.C., K.Z.S., A.M.R., E.W., L.T., and I.M.G. analyzed and interpreted the data; S. Manier, C.-J.L., and I.M.G. wrote the report; all authors revised the report and approved the final version.
Conflict-of-interest disclosure: The authors have no conflicts of interest to declare.
Correspondence: Irene M. Ghobrial, Dana-Farber Cancer Institute, 450 Brookline Ave, Boston, MA 02215; e-mail: irene_ghobrial@dfci.harvard.edu.
References
Author notes
S. Manier and C.-J.L. contributed equally to this study.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal