Key Points
GCB DLBCLs are characterized by a defective IRE1-XBP1 pathway.
XBP1 expression reduces GCB DLBCL tumor growth in a mouse xenograft model.
Abstract
The endoplasmic reticulum kinase inositol-requiring enzyme 1 (IRE1) and its downstream target X-box–binding protein 1 (XBP1) drive B-cell differentiation toward plasma cells and have been shown to contribute to multiple myeloma development; yet, little is known of the role of this pathway in diffuse large B-cell lymphoma (DLBCL). Here, we show that in the germinal center B-cell–like (GCB) DLBCL subtype, IRE1 expression is reduced to a level that prevents XBP1 activation. Gene expression profiles indicated that, in GCB DLBCL cancer samples, expression of IRE1 messenger RNA was inversely correlated with the levels and activity of the epigenetic repressor, histone methyltransferase enhancer of zeste homolog 2 (EZH2). Correspondingly, in GCB-derived cell lines, the IRE1 promoter carried increased levels of the repressive epigenetic mark histone 3 lysine 27 trimethylation. Pharmacological inhibition of EZH2 erased those marks and restored IRE1 expression and function in vitro and in vivo. Moreover, reconstitution of the IRE1-signaling pathway, by expression of the XBP1-active form, compromised GCB DLBCL tumor growth in a mouse xenograft cancer model. These findings indicate that IRE1-XBP1 downregulation distinguishes GCB DLBCL from other DLBCL subtypes and contributes to tumor growth.
Introduction
Diffuse large B-cell lymphoma (DLBCL) represents the most common category of non-Hodgkin lymphomas and accounts for 30% to 40% of newly diagnosed patients.1 Gene expression profiling of patient-derived DLBCL tumors has uncovered 2 main and noticeably distinct DLBCL molecular subtypes.2,3 The activated B-cell–like (ABC) DLBCL subtype is characterized by expression of genes typically induced during in vitro activation of peripheral B cells. Most prominent oncogenic events that characterize ABC lymphomas include constitutive NF-κB signaling4-9 and frequent genetic inactivation of B-lymphocyte–induced maturation protein 1 (Blimp1), a master regulator of terminal plasma B-cell differentiation.10,11
The second main molecular subtype exhibits transcriptional programs characteristic of germinal center B cells and is named germinal center B-cell–like (GCB) DLBCL. This subtype is characterized by overexpression of B-cell lymphoma 6 (BCL6),12 a transcriptional suppressor that downregulates genes involved in plasma cell differentiation13,14 and contributes to disease onset by blocking B cells in the germinal center stage.15,16 Histone methyl-transferase enhancer of zeste homolog 2 (EZH2) is another frequently mutated17,18 transcriptional regulator that cooperates with aberrant BCL6 activity to promote GCB DLBCL pathology.19,20 EZH2, the catalytic subunit of the polycomb repressor complex 2 (PRC2), mediates histone 3 lysine 27 trimethylation (H3K27me3), leading to chromatin condensation and repression of its target genes.21,22 Similar to BCL6, mutated EZH2 permanently downregulates genes involved in plasma cell differentiation, including Blimp1 and interferon regulatory factor 4 (IRF4),23 and thereby blocks terminal differentiation to promote disease development. Therefore, constitutive inhibition of differentiation programs represents a general strategy that GCB DLBCLs deploy to promote tumor growth.
B-cell differentiation toward plasma cells is also highly dependent on functional inositol-requiring enzyme 1 (IRE1) signaling.24-26 IRE1 is a ubiquitously expressed transmembrane kinase and endoribonuclease localized within the endoplasmic reticulum (ER). IRE1 detects stress caused by misfolded proteins that accumulate within the ER lumen. Upon activation, IRE1 mediates X-box–binding protein 1 (XBP1) messenger RNA (mRNA) maturation, leading to production of the transcription factor XBP1 spliced (XBP1s) that translocates to the nucleus to trigger the expression of ER stress response genes such as DNAJB9, SEC23b, and SRPR. In addition to its main role in catalyzing XBP1 mRNA maturation, IRE1 can degrade several mRNA molecules in a process called regulated IRE1-dependent decay (RIDD) 27 . Upon exposure to stress, the signaling pathways steaming from IRE1 promote increased ER-folding capacity and restore homeostasis.28,29
In plasma cells, the IRE1-XBP1s signaling expands the ER network in order to equip these secretory cells with extensive ER-folding machinery competent enough to handle large amounts of immunoglobulins.30 In addition, XBP1s binds the interleukin 6 (IL6) promoter, leading to production of this cytokine that is required for appropriate plasma cell differentiation.24 Intriguingly, constitutive expression of the IRE1-downstream product XBP1s in murine B cells promotes a disease that resembles multiple myeloma (MM).31 Furthermore, pharmacological inhibition of IRE1 activity has been described as a promising therapeutic option in MM, indicating addiction of plasma cell–derived cancers to the IRE1-XBP1s signaling pathway.32,33
Although the importance of the IRE1-XBP1 pathway in MM is well established, the role of this pathway in DLBCL remains rather unclear. Comparison between molecular characteristics of ABC and GCB DLBCL subtypes indicated a dichotomy in expression of XBP1s target genes.34 However, the causes and therapeutic significance of this dichotomy was unknown. Here, we describe that specific downregulation of IRE1 expression impairs XBP1s production and renders GCB DLBCL more sensitive to ER stress–inducing agents. The analysis of IRE1 expression, in a cohort of 350 DLBCL patients, confirmed that IRE1 is specifically downregulated in GCB DLBCL, therefore suggesting that IRE1 levels could serve as a novel marker in DLBCL classification. We identified the epigenetic regulator EZH2 as a key factor that predefined basal levels of IRE1 expression. Pharmacological inhibition of EZH2 enzymatic activity restored IRE1 expression and function in GCB DLBCL. Moreover, reconstitution of the IRE1 pathway by ectopic XBP1s expression compromised tumor growth in a GCB DLBCL xenograft mouse model. This indicates that, in contrast to its tumor-promoting role in MM, the IRE1-XBP1s activity might negatively impact tumor growth in GCB DLBCL.
Materials and methods
Methods for cell culture and drug treatment, immunoblot analysis, chromatin immunoprecipitation (ChIP), reverse transcription polymerase chain reaction (RT-PCR), XBP1-splicing assay, and lentivirus production and cell line infection are detailed in supplemental Methods (available on the Blood Web site).
Mice
Animal experiments were approved by the Veterinary Office of the Canton de Vaud and the Animal Ethics Committee (authorization 2883). Immunocompromised AGR129 (interferon-α/β [IFN-α/β], IFN-γ receptor, and recombination-activating gene 2 [RAG-2]-deficient) mice were provided by M. Gilliet (Department of Dermatology, Centre Hospitalier Universitaire Vaudois, Lausanne, Switzerland) and housed at the University of Lausanne in accordance with local and national guidelines. Six- to 8-week-old females were randomly distributed in 2 groups and tumor cells were injected subcutaneously in a 200-μL volume of 1:1 phosphate-buffered saline to Matrigel (BD Biosciences). Each mouse was injected with 3 × 106 SUDHL4 cells in the left flank and the same number of SUDHL4-XBP1s cells (SUDHL4 infected with the vector-expressing XBP1s under doxycycline) was injected in the right flank. The same experimental procedure was followed to generate the ABC xenograft mouse model using 2.5 × 106 human DLBCL 1 (HBL1) or HBL1-XBP1s cells (HBL1 infected with the vector-expressing XBP1s under doxycycline). The control group was treated with vehicle (5% sucrose in H2O). The second group was treated with doxycycline (5% sucrose, 1 mg/mL doxycycline in H2O). Treatments were administered orally through water. Bottles with vehicle or doxycycline were replaced every 2 days. Tumors were measured 3 times per week and the volumes were calculated using the formula V = (length × width2)/2. Doxycycline was acquired from Sigma-Aldrich. To evaluate the in vivo effects of GSK126, the AGR129 mice were subcutaneously injected with 3 × 106 Karpas422 cells in a 200-μL volume of 1:1 phosphate-buffered saline to Matrigel. When palpable tumors were established, vehicle (20% sulfobutyl ether β-cyclodextrin adjusted to pH 4-4.5 with 1 N acetic acid) or GSK126 at a concentration of 150 mg/kg was injected daily (during 10 days) in the final volume of 0.2 mL per injection as previously described.35
Statistical analysis
All data are representative of at least 3 different experiments. Statistical significance was ascertained by performing the appropriate tests as described in the figure legends. Significant differences were indicated as follows: *P ≤ .05, **P ≤ .01, or ***P ≤ .001.
Data mining from published microarray data sets
Raw expression profiles (CEL files) were downloaded from the publicly available data sets (GSE10846,36 GSE23501,37 GSE40971,35 GSE5631538 ), imported, and normalized using the robust multiarray average algorithm in Partek Genomics Suite 6.4 (St. Louis, MO). Differences of expression between groups using the Wilcoxon rank-sum test and Pearson correlations were calculated using Stata/SE v.12.1 (StataCorp, College Station, TX); the Pearson correlations P values were calculated with the tool available at www.socscistatistics.com. Genes were considered differentially expressed between groups if bearing a value of P < .005, q < 0.05, and an absolute log ratio > 0.3 in both cohorts.
Results
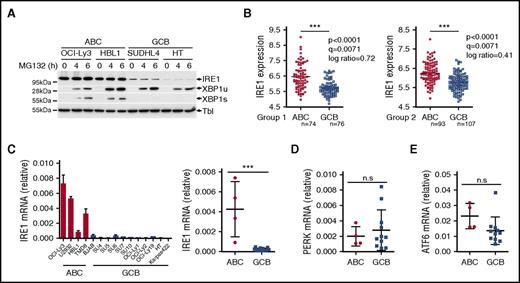
IRE1 signaling is defective in a subset of DLBCLs
In conditions of ER stress, IRE1 engages its endoribonuclease activity to promote unconventional processing of XBP1 mRNA leading to production of mature, spliced XBP1 mRNA. The spliced XBP1 mRNA product is consequently translated into the active transcription factor XBP1s. Gene expression profiles have indicated that ABC DLBCLs express XBP1 target genes at higher levels than GCB DLBCLs. This suggested that the IRE1/XBP1 pathway is differentially regulated between the 2 DLBCL subtypes and led to the hypothesis that ABC DLBCLs display enhanced basal XBP1s protein.34 To further evaluate this, we studied IRE1 activity in 4 representative DLBCL cell lines. To trigger IRE1 activation, we treated DLBCL cells with tunicamycin, an inhibitor of N-linked glycosylation that triggers robust ER stress, and measured gene expression. The induction of XBP1 target genes was impaired in the representative GCB DLBCL lines compared with ABC DLBCL cells (Figure 1A). Along these lines, XBP1 mRNA maturation and XBP1s protein expression were almost completely abolished in these 2 GCB DLBCL lines (Figure 1B). Inability of GCB DLCBL cells to splice XBP1 mRNA was combined with reduced levels of IRE1 protein (Figure 1B), indicating that IRE1 downregulation may impair the activation of a functional ER stress response in GCB DLBCL cells. To extend these findings further, we analyzed a larger panel of DLBCL cell lines and found that 9 of 11 GCB DLBCL cells had impaired IRE1 expression that correlated with defective production of active XBP1s upon 6 hours of treatment with tunicamycin (Figure 1C). In line with these observations, we found that ER stress–mediated induction of the XBP1 target gene DNAJB9 was impaired in GCB DLBCL compared with ABC DLBCL (supplemental Figure 1A) whereas expression of the RIDD target Bloc1S1 mRNA was not affected upon ER stress induction in GCB DLBCL (supplemental Figure 1B), further indicating that IRE1 functions are defective in GCB DLBCL. Importantly, other ER stress response pathways were similar in both subtypes, as measured by activating transcription factor 6 (ATF6) and protein kinase R-like ER kinase (PERK) levels (Figure 1C). Moreover, upon exposure to tunicamycin, PERK activation and production of its downstream effectors, phospho-eIF2α and ATF4, were unaffected in GCB DLBCL cells (Figure 1C; supplemental Figure 1C-D). This indicated that these cells can mount a response to tunicamycin and do not display a global ER stress response defect but rather a specific downmodulation of the IRE1/XBP1-signaling branch. Next, we treated DLBCL with various pharmacological inducers of ER stress pathways. Administration of proteasome inhibitor bortezomib led to specific activation of the PERK-ATF4 pathway without promoting robust IRE1-XBP1 activation in both subtypes. However, treatment with other stress inducers such as thapsigargin, dithiothreitol (DTT), brefeldin A, or nelfinavir induced XBP1s in representative ABC DLBCL while having no effect on its expression in GCB DLBCL lines (Figure 1D; supplemental Figure 1E). In stark contrast, IRE1 levels and response appeared functional in other experimental models, such as Jurkat T-cell leukemia cells, lymphoblastoid B-cell line CB33, human blood B cells, and in the panel of Burkitt lymphomas that, similar to GCB DLBCLs, are germinal center B-cell–derived cancers (supplemental Figure 2A-D).
GCB DLBCLs display impairment in IRE1 expression that affects the ER stress response. (A-B) Representative ABC DLBCL and GCB DLBCL cell lines were treated with 5 μg/mL tunicamycin (TM) for 6 hours and analyzed by quantitative real-time PCR for expression of XBP1 target genes Sec23b, DNAJB9, and SRPR mRNA levels relative to β-actin (mean and standard deviation [SD] of technical triplicates of 1 representative experiment of 3) (A). XBP1 mRNA splicing was measured by RT-PCR analysis of XBP1 mRNA maturation (B) DNA electrophoresis is shown in the top panel. Protein levels of XBP1s and IRE1 protein were analyzed by immunoblot. Tubulin (Tbl) is used as loading control (B, below). (C) DLBCL cell lines were treated for 6 hours with 5 μg/mL tunicamycin (TM) or vehicle and analyzed by immunoblot for XBP1s, IRE1, ATF4, PERK, ATF6, and tubulin (Tbl). (D) Representative ABC and GCB cell lines were treated with different ER stress inducers, tunicamycin (TM), thapsigargin (Tpg), DTT, brefeldin A (Bref A), and bortezomib (Btz) for 6 hours. IRE1, XBP1s, PERK, ATF4, and Tubulin (Tbl) expression was analyzed by immunoblot. (E) Viability of IRE1-high and IRE1-low DLBCL cells assessed by 3-(4,5-dimethylthiazol-2-yl)-2,5-dimethyltetrazolium bromide (MTS)/1-methoxy phenazine methosulfate (PMS) assay after treatment of the indicated time course with 0.05 μg/mL tunicamycin or 12.5 μM nelfinavir. P values were calculated using 2-way analysis of variance (ANOVA); *P ≤ .05; ***P ≤ .001.
GCB DLBCLs display impairment in IRE1 expression that affects the ER stress response. (A-B) Representative ABC DLBCL and GCB DLBCL cell lines were treated with 5 μg/mL tunicamycin (TM) for 6 hours and analyzed by quantitative real-time PCR for expression of XBP1 target genes Sec23b, DNAJB9, and SRPR mRNA levels relative to β-actin (mean and standard deviation [SD] of technical triplicates of 1 representative experiment of 3) (A). XBP1 mRNA splicing was measured by RT-PCR analysis of XBP1 mRNA maturation (B) DNA electrophoresis is shown in the top panel. Protein levels of XBP1s and IRE1 protein were analyzed by immunoblot. Tubulin (Tbl) is used as loading control (B, below). (C) DLBCL cell lines were treated for 6 hours with 5 μg/mL tunicamycin (TM) or vehicle and analyzed by immunoblot for XBP1s, IRE1, ATF4, PERK, ATF6, and tubulin (Tbl). (D) Representative ABC and GCB cell lines were treated with different ER stress inducers, tunicamycin (TM), thapsigargin (Tpg), DTT, brefeldin A (Bref A), and bortezomib (Btz) for 6 hours. IRE1, XBP1s, PERK, ATF4, and Tubulin (Tbl) expression was analyzed by immunoblot. (E) Viability of IRE1-high and IRE1-low DLBCL cells assessed by 3-(4,5-dimethylthiazol-2-yl)-2,5-dimethyltetrazolium bromide (MTS)/1-methoxy phenazine methosulfate (PMS) assay after treatment of the indicated time course with 0.05 μg/mL tunicamycin or 12.5 μM nelfinavir. P values were calculated using 2-way analysis of variance (ANOVA); *P ≤ .05; ***P ≤ .001.
To interrogate whether IRE1 expression modulated the ability of DLBCL to cope with stress, we monitored cell viability in a panel of IRE1-high and IRE1-low DLBCLs treated with the stress inducers tunicamycin or nelfinavir. Cells with reduced IRE1 displayed significantly impaired viability in the presence of stress (Figure 1E), indicating that decreased IRE1 expression affected optimal stress-adaption programs in GCB DLBCLs.
IRE1 expression is regulated at transcriptional level in GCB DLBCL
The identification of cell types that downregulate IRE1 to nonfunctional levels revealed an unanticipated mechanism of regulation of the ER stress response. IRE1 is expected to be expressed and functional in virtually all eukaryotic cells to elicit a swift response to homeostatic changes. To investigate protein degradation mechanisms that may regulate its expression, we monitored IRE1 protein levels in the presence of the proteasome inhibitor MG132. We found that proteasome inhibition stabilized XBP1 unspliced (XBP1u) protein39 in GCB DLCBL cells similar to the protein levels observed in ABC DLBCL (Figure 2A). This indicated that GCB DLBCL cells are capable of producing XBP1 mRNA. However, no recovery of IRE1 protein expression or XBP1s production was observed in GCB DLBCL cells (Figure 2A). This suggests that proteasomal degradation of IRE1 does not contribute to the observed IRE1 phenotype in this lymphoma subtype. To investigate whether IRE1 regulation occurred at the transcriptional level, we analyzed IRE1 mRNA expression in a data set profiling GCB DLBCL tumors and tonsil-derived B-cell subtypes38 and observed that IRE1 expression was reduced in GCB DLBCL compared with normal B cells, including germinal center B cells (supplemental Figure 2E). Then, we investigated IRE1 mRNA expression in 2 gene expression data sets profiling 167 ABC and 183 GCB DLBCL tumors.36 In both groups, clinical samples classified as GCB DLBCL had significantly lower IRE1 mRNA expression compared with ABC DLBCL tumors (Figure 2B). Similarly, IRE1 mRNA levels in DLBCL cell lines were significantly reduced in GCB DLBCL cells compared with ABC DLBCL cells (Figure 2C). In contrast, other stress-related genes, such as ATF4, Chop, PERK, and ATF6 were not significantly reduced in GCB DLBCL compared with ABC DLBCL (Figure 2D-E; supplemental Figure 3). Thus, IRE1 mRNA downregulation is a hallmark of GCB DLBCL tumors that occurs independently of the other branches of the ER stress response.
IRE1 mRNA levels are downregulated in GCB DLBCL. (A) Two ABC and 2 GCB DLBCL cell lines were treated for 4 or 6 hours with 10 μM proteasome inhibitor MG132. IRE1, XBP1u, and XBP1s protein expression was analyzed by immunoblot. Tubulin (Tbl) is used as loading control. (B) IRE1 mRNA levels in GCB-DLBCL (GCB) compared with ABC-DLBCL (ABC) molecular subtype in 2 independent gene expression–profiled DLBCL data sets GSE10846. Genes were considered differentially expressed between ABC and GCB if bearing a P < .005, q < 0.05, and an absolute log ratio > 0.3 in both cohorts. (C-E) Representative ABC and GCB DLBCL cell lines were analyzed for mRNA expression levels of IRE1 (C), PERK (D), and ATF6 (E) relative to β-actin. Relative IRE1 expression (mean and SD of technical triplicates of 1 representative experiment) for each cell line tested is shown in panel C (left panel). Statistical analyses were done using the unpaired Student t test to compare relative mRNA expression between ABC and GCB cells. ***P ≤ .001 (n.s.); P > .05. n.s., not significant.
IRE1 mRNA levels are downregulated in GCB DLBCL. (A) Two ABC and 2 GCB DLBCL cell lines were treated for 4 or 6 hours with 10 μM proteasome inhibitor MG132. IRE1, XBP1u, and XBP1s protein expression was analyzed by immunoblot. Tubulin (Tbl) is used as loading control. (B) IRE1 mRNA levels in GCB-DLBCL (GCB) compared with ABC-DLBCL (ABC) molecular subtype in 2 independent gene expression–profiled DLBCL data sets GSE10846. Genes were considered differentially expressed between ABC and GCB if bearing a P < .005, q < 0.05, and an absolute log ratio > 0.3 in both cohorts. (C-E) Representative ABC and GCB DLBCL cell lines were analyzed for mRNA expression levels of IRE1 (C), PERK (D), and ATF6 (E) relative to β-actin. Relative IRE1 expression (mean and SD of technical triplicates of 1 representative experiment) for each cell line tested is shown in panel C (left panel). Statistical analyses were done using the unpaired Student t test to compare relative mRNA expression between ABC and GCB cells. ***P ≤ .001 (n.s.); P > .05. n.s., not significant.
EZH2 regulates IRE1 expression in DLBCL
Gain-of-function EZH2 mutations are typical features that discriminate GCB from ABC DLBCL.17,40 EZH2 is the catalytic subunit of the PRC2 complex that catalyzes mono- through trimethylation of lysine 27 on histone H3 (H3K27). Trimethylation of H3K27 suppresses transcription of specific genes that are proximal to the site of histone modification, a mechanism that may contribute to tumor growth.41 Previously, it has been demonstrated that in GCB DLBCL, through the generation of de novo bivalent domains, EZH2 downregulates the genes involved in plasma cell differentiation and keeps the tumors arrested in the germinal center–like developmental stage.23 In the 2 available DLBCL gene expression profiling data sets,36 we observed that within the GCB DLBCL clinical samples EZH2 expression negatively correlated with IRE1 mRNA levels (Figure 3A). No such correlation was observed between EZH2 and PERK expression levels (supplemental Figure 4A). Moreover, analysis of gene expression profile data sets from GCB patients37 and GCB cell lines35 indicated that EZH2 gain-of-function mutations correlated with decreased IRE1 expression (Figure 3B; supplemental Figure 4B). To confirm that EZH2 modulates IRE1 expression and stress responses, we generated HeLa cells expressing the EZH2 gain-of-function mutant form (Y641F) that is typically observed in GCB DLBCL.18 Compared with control cells, EZH2Y641F-expressing cells showed increased levels of H3K27me3 whereas IRE1 expression was reduced (supplemental Figure 4C). Consistently, IRE1-mediated gene expression upon treatment with tunicamycin was impaired in EZH2Y641F-expressing cells (supplemental Figure 4D). This suggested that EZH2 could specifically silence IRE1 expression through epigenetic modification of its promoter. To test this hypothesis, we measured, by ChIP, H3K27me3 marks within the proximal promoter region of IRE1 and found that GCB cells displayed an increased enrichment of H3K27me3 marks compared with ABC cells (Figure 3C). As expected, IRF4, an EZH2-regulated gene in GCB19 showed a similar profile. In contrast, the promoters of the housekeeping gene GAPDH and of the constitutively expressed Bcl6 gene had no detectable H3K27me3 marks whereas the differentiation factors MyoD and HoxA7 harbored H3K27me3 marks in both ABC and GCB cells as previously reported.23 In addition to repressive marks, we noticed that the IRE1 promoter harbored H3K4me3-activating marks (supplemental Figure 4E), suggesting that its activity is poised and could potentially be restored by erasing the H3K27me3 marks.42 To test this, we treated cells with a cell-permeable EZH2 inhibitor GSK343.43 EZH2 inhibition decreased H3K27me3 marks on the IRE1 promoter (Figure 4A) and increased IRE1 mRNA (supplemental Figure 4F) and protein levels in the GCB subtypes (Figure 4B). Increased IRE1 expression correlated with decreased overall H3K27 trimethylation in GCB cells. In contrast, PERK expression was not significantly affected by EZH2 inhibition, further demonstrating that the 2 ER stress signaling branches are controlled by different mechanisms. Finally, EZH2 inhibition restored IRE1 functionality in GCB cells as demonstrated by the production of active XBP1s in the presence of tunicamycin (Figure 4C). To rule out the possibility that the effects on IRE1 expression are exclusive to GSK343, we treated the cells with GSK126, another selective EZH2 inhibitor.35 We demonstrated that, similar to GSK343, GSK126 increased IRE1 levels in various GCB DLBCL lines (supplemental Figure 4G). Then, we analyzed whether this effect could be reproduced in vivo. We implanted Karpas422 GCB DLBCLs into immunodeficient AGR129 (IFN-α/β, IFN-γ receptor, and RAG-2–deficient) mice44 and treated the animals with GSK126 for 10 days. Analysis of postmortem tumor biopsies showed that pharmacological EZH2 inhibition in vivo promoted IRE1 protein and mRNA expression and increased the levels of the XBP1 target gene DNAJB9 (Figure 4D-E). Altogether these data show that increased EZH2 activity in DLBCL contributes to decreased IRE1 levels and that pharmacological inhibition of H3K27 trimethylation can restore a functional IRE1 pathway.
IRE1 promoter in GCB DLBCL carries high levels of repressive histone mark H3K27me3. (A) Correlations as measured by the Pearson correlation coefficient r between EZH2 expression and IRE1 expression in GCB DLBCL clinical samples from 2 data sets extracted from GSE10846. (B) IRE1 mRNA levels in EZH2 wild-type (wt) and EZH2-mutated (mt) GCB DLBCL patients. Data extracted from gene expression data sets GSE23501. Wilcoxon signed-rank test, *P ≤ .05. (C) ChIP was performed on ABC and GCB DLBCL cell lines using H3K27me3-specific antibody. Enrichments of indicated promoters were probed by real-time PCR (mean and SD of technical triplicates of 1 representative experiment of 3). MyoD and Hoxa7 regions are positive controls for H3K27me3-mediated repression. GAPDH and BCL6 promoters are negative controls for H3K27me3-mediated repression.
IRE1 promoter in GCB DLBCL carries high levels of repressive histone mark H3K27me3. (A) Correlations as measured by the Pearson correlation coefficient r between EZH2 expression and IRE1 expression in GCB DLBCL clinical samples from 2 data sets extracted from GSE10846. (B) IRE1 mRNA levels in EZH2 wild-type (wt) and EZH2-mutated (mt) GCB DLBCL patients. Data extracted from gene expression data sets GSE23501. Wilcoxon signed-rank test, *P ≤ .05. (C) ChIP was performed on ABC and GCB DLBCL cell lines using H3K27me3-specific antibody. Enrichments of indicated promoters were probed by real-time PCR (mean and SD of technical triplicates of 1 representative experiment of 3). MyoD and Hoxa7 regions are positive controls for H3K27me3-mediated repression. GAPDH and BCL6 promoters are negative controls for H3K27me3-mediated repression.
IRE1 expression levels and activity are regulated by histone methyltransferase EZH2. (A) Karpas422 cells treated for 5 days with 2.5 μM of the EZH2 inhibitor GSK343 or vehicle were analyzed by ChIP for H3K27me3 promoter marks on MyoD and IRF4 (EZH2-dependent), GAPDH (negative control), and IRE1 (mean and SD of technical triplicates of 1 representative experiment of 3). (B) Representative ABC and GCB DLBCL cell lines treated with GSK343 or vehicle were probed by immunoblot for IRE1, PERK, H3K27me3, H3 total, and tubulin (Tbl). IRE1 protein levels were quantified and plotted (right panel). Paired Student t test (nontreated vs treated); *P ≤ .05. (C) Representative GCB DLBCL cell lines were treated for 5 days with 2.5 μM EZH2 inhibitor GSK343. Then, 5 μg/mL tunicamycin (TM) was added to the cells for 6 hours. Immunoblots were revealed using antibodies against XBP1s, IRE1, PERK, ATF4, H3K27me3, and tubulin (experiment representative of 3). (D) Karpas422 cells were subcutaneously injected in immunodeficient AGR129 mice. Mice were treated during 10 days with vehicle or 150 mg/kg GSK126 as described in “Materials and methods.” IRE1, H3K27me3, H3 total, and tubulin levels were analyzed by immunoblot. (E) Human IRE1 and DNAJB9 relative mRNA values were assessed in postmortem biopsies. (F) Parental and IRE1-deficient SUDHL4 cells were treated for 4 days with vehicle or 5 μM GSK343. IRE1, H3K27me3, H3 total, and tubulin levels were assessed by immunoblot. (G) Viability of IRE1-proficient and IRE1-deficient SUDHL4 cells was assessed by MTS/PMS assay after treatment with 10 μM GSK343 for 48 hours. P values were obtained using the unpaired Student t test; **P ≤ .01.
IRE1 expression levels and activity are regulated by histone methyltransferase EZH2. (A) Karpas422 cells treated for 5 days with 2.5 μM of the EZH2 inhibitor GSK343 or vehicle were analyzed by ChIP for H3K27me3 promoter marks on MyoD and IRF4 (EZH2-dependent), GAPDH (negative control), and IRE1 (mean and SD of technical triplicates of 1 representative experiment of 3). (B) Representative ABC and GCB DLBCL cell lines treated with GSK343 or vehicle were probed by immunoblot for IRE1, PERK, H3K27me3, H3 total, and tubulin (Tbl). IRE1 protein levels were quantified and plotted (right panel). Paired Student t test (nontreated vs treated); *P ≤ .05. (C) Representative GCB DLBCL cell lines were treated for 5 days with 2.5 μM EZH2 inhibitor GSK343. Then, 5 μg/mL tunicamycin (TM) was added to the cells for 6 hours. Immunoblots were revealed using antibodies against XBP1s, IRE1, PERK, ATF4, H3K27me3, and tubulin (experiment representative of 3). (D) Karpas422 cells were subcutaneously injected in immunodeficient AGR129 mice. Mice were treated during 10 days with vehicle or 150 mg/kg GSK126 as described in “Materials and methods.” IRE1, H3K27me3, H3 total, and tubulin levels were analyzed by immunoblot. (E) Human IRE1 and DNAJB9 relative mRNA values were assessed in postmortem biopsies. (F) Parental and IRE1-deficient SUDHL4 cells were treated for 4 days with vehicle or 5 μM GSK343. IRE1, H3K27me3, H3 total, and tubulin levels were assessed by immunoblot. (G) Viability of IRE1-proficient and IRE1-deficient SUDHL4 cells was assessed by MTS/PMS assay after treatment with 10 μM GSK343 for 48 hours. P values were obtained using the unpaired Student t test; **P ≤ .01.
Enhanced EZH2 activity regulates IRE1 levels in different tumor types
EZH2 function is predominant during embryonic development and in differentiating tissues, however, its activity decreases in the majority of differentiated cells.45 Nevertheless, aberrantly increased EZH2 activity has been reported in multiple malignancies including cervical cancer.41,46,47 To assess the epigenetic status of the IRE1 promoter in the context of another malignancy characterized by increased EZH2 activity, we quantified IRE1 histone modifications in cervical cancer–derived HeLa cells (supplemental Figure 5A). We observed that repressive (H3K27me3) and activating (H3K4me3) marks coexisted at the IRE1 promoter, indicating promoter bivalency in this cancer cell line. In contrast to GCB DLBCL and HeLa cells, the IRE1 promoter from nontumoral mouse embryonic fibroblasts was labeled only with the activating H3K4me3 epigenetic mark (supplemental Figure 5B). Next, we tested whether EZH2 inhibition affected IRE1 expression in HeLa cells. These cells have detectable IRE1 levels, yet in the presence of the EZH2 inhibitor, we observed an increase in IRE1 protein that correlated with a decrease in H3K27 trimethylation (supplemental Figure 5C). Similarly, IRE1 mRNA was augmented upon EZH2 inhibition (supplemental Figure 5D). Consistently, upon deletion of the EZH2 gene by CRISPR/Cas9, IRE1 expression was increased, enhancing XBP1s protein production in presence of mild ER stress (supplemental Figure 5E). These data indicate that in different tumors characterized by enhanced EZH2 function, IRE1 expression levels are predefined by aberrant epigenetic activity that controls its signaling outputs.
Reactivation of the IRE1 pathway contributes to toxic effects of EZH2 inhibitors in GCB DLBCL
EZH2 inhibition was demonstrated to be specifically toxic to GCB lymphomas.23,35 Similar to previous reports, we observed that EZH2 inhibition with GSK343 predominantly affected GCB growth in vitro (supplemental Figure 6). To evaluate the contribution of IRE1, we generated IRE1-deficient SUDHL4 (GCB DLBCL) cells (Figure 4F) and analyzed cell viability upon treatment with GSK343. We found that EZH2 inhibition-mediated toxicity was reduced in IRE1-deficient clones compared with controls (Figure 4G). These data indicate that reactivation of the IRE1 pathway contributes, at least partially, to the antitumoral effects of EZH2 inhibitors.
Expression of XBP1s impairs tumor growth in a GCB DLBCL xenograft model
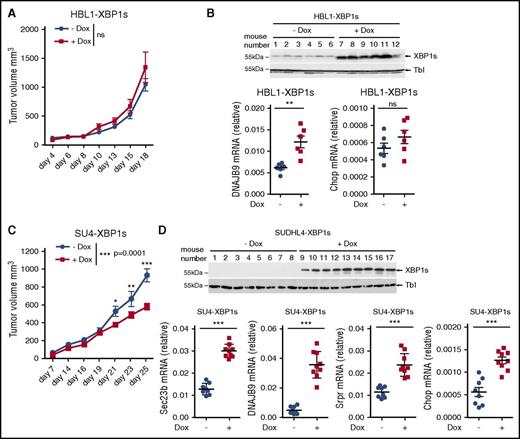
Because the aberrantly decreased IRE1-XBP1s pathway is a feature of most GCB DLBCLs, we interrogated the contribution of this pathway to tumor growth. We generated HBL1 (ABC DLBCL) and SUDHL4 (GCB DLBCL) cells that produce a doxycycline-inducible XBP1s protein. These cells were transplanted into the flanks of immunodeficient AGR129 mice,44 and tumor growth was monitored over time. We found that doxycycline administration did not affect growth of the ABC tumors (Figure 5A). On the other hand, treatment with doxycycline resulted in impaired growth of the grafted GCB tumors (Figure 5C). As expected, analysis of postmortem tumor biopsies showed that, in both models, doxycycline administration increased XBP1s expression and its downstream target DNAJB9 (Figure 5B,D). Interestingly, in the GCB xenograft model, XBP1s induced expression of the proapoptotic gene Chop (Figure 5D). Doxycycline per se did not impair the growth of tumors originating from parental HBL1 and SUDHL4 cells (supplemental Figure 7A,C). Similarly, this treatment did not promote the transcription of DNAJB9 as measured in the tumor biopsies (supplemental Figure 7B,D). These data indicate that XBP1s can decrease tumor growth in GCB DLBCLs, further suggesting that decreased IRE1 activation in particular cancer types may contribute to tumorigenesis.48
XBP1 signaling impairs GCB DLBCL tumor growth. (A-D) Immunodeficient AGR129 mice were injected subcutaneously with 2.5 × 106 HBL1 (A-B) or 3 × 106 SUDHL4 cells (C-D) that express XBP1s under treatment with doxycycline. Mice were daily-administered vehicle or doxycycline at 1 mg/mL concentration through water as described in “Materials and methods.” Mean tumor volume measurements were represented against the days of treatment (A,C). P values were calculated using 2-way ANOVA followed by Bonferroni posttest; n.s., P > .05, *P ≤ .05, **P ≤ .01, ***P ≤ .001. Efficiency of doxycycline-induced XBP1s expression and its target genes was validated by immunoblot and quantitative real-time PCR for the mice carrying HBL1-XBP1s (B) or SUDHL4-XBP1s tumors. (D) Each dot in the graph represents relative gene expression for a single mouse (B and D bottom panels). P values were calculated using the unpaired Student t test to compare nontreated vs treated groups; ***P ≤ .001.
XBP1 signaling impairs GCB DLBCL tumor growth. (A-D) Immunodeficient AGR129 mice were injected subcutaneously with 2.5 × 106 HBL1 (A-B) or 3 × 106 SUDHL4 cells (C-D) that express XBP1s under treatment with doxycycline. Mice were daily-administered vehicle or doxycycline at 1 mg/mL concentration through water as described in “Materials and methods.” Mean tumor volume measurements were represented against the days of treatment (A,C). P values were calculated using 2-way ANOVA followed by Bonferroni posttest; n.s., P > .05, *P ≤ .05, **P ≤ .01, ***P ≤ .001. Efficiency of doxycycline-induced XBP1s expression and its target genes was validated by immunoblot and quantitative real-time PCR for the mice carrying HBL1-XBP1s (B) or SUDHL4-XBP1s tumors. (D) Each dot in the graph represents relative gene expression for a single mouse (B and D bottom panels). P values were calculated using the unpaired Student t test to compare nontreated vs treated groups; ***P ≤ .001.
Discussion
Here, we demonstrate that EZH2-mediated epigenetic regulation predefines basal IRE1 expression levels. In GCB DLBCL, where EZH2 frequently carries gain-of-function mutations, the IRE1 promoter is labeled by high amounts of H3K27me3-repressive marks. This promotes IRE1 downregulation and blunts ER stress responses as demonstrated by inability of GCB DLBCL to activate XBP1 or downregulate the targets of IRE1-mediated mRNA decay. IRE1 expression and function could be restored upon EZH2 inhibition, demonstrating the role of this regulation in DLBCL. Additionally, in a cervical cancer cell line, characterized by enhanced EZH2 activity, we observed a similar mode of epigenetic downregulation of the IRE1 promoter, indicating that this regulatory mechanism is conserved among different tumors.
The IRE1-XBP1 pathway plays an important role in secretory cell differentiation and maintenance.30,49 It is therefore conceivable that differentiation programs may engage epigenetic modifiers to establish IRE1 protein levels and therefore couple ER stress response capacity to the physiological requirements. In line with this hypothesis, EZH2 has been involved in the differentiation and maintenance of secretory cells including pancreatic β cells.50 Similarly, EZH2 regulates the genetic programs associated with differentiation of germinal center B cells to plasma cells or memory B cells,19,23 a process that is also dependent on the IRE1-XBP1 pathway.24,26 The mechanisms involved in IRE1 activation during B-cell differentiation are still poorly understood. Studies have shown that XBP1s production in this context precedes ER stress.51 Our findings indicating that EZH2 status regulates IRE1 expression raise the question of whether, during B-cell differentiation, increased IRE1 expression could be mediated by decreased EZH2 activity.52 It was shown that overexpression of IRE1 promotes its own activation,53 thus enhanced IRE1 expression during differentiation could be sufficient to initiate the pathway and trigger XBP1s production.
In MM, activation of the IRE1-XBP1s signaling pathway has been considered as a tumor-promoting event.31,33 In contrast, here we found that in GCB DLBCL this pathway negatively affects tumor growth and is therefore downregulated. Reconstitution of XBP1s signaling specifically impaired the growth of GCB DLBCL tumors in vivo. These data highlight the key role of the IRE1-XBP1 pathway in malignancies and suggest that it can contribute to both tumor promotion and tumor repression, depending on the tumor type.
Although our results indicate that XBP1s is an important regulator of GCB DLBCL growth downstream of IRE1, we cannot exclude that other pathways stemming from IRE1, such as RIDD, could also play a role and further affect GCB tumor growth. XBP1 may regulate GCB DLBCL tumors by different mechanisms. It drives expression of the unfolded protein response network genes that is a hallmark of plasma cell differentiation.30 Therefore, decreasing XBP1s production, by means of EZH2-mediated inhibition of IRE1 may contribute to differentiation arrest and thereby promote tumorigenesis in GCB DLBCL. In addition, we observed that expression of the proapoptotic gene Chop was induced downstream of XBP1s exclusively in GCB DLBCLs. It is therefore possible that XBP1s directly engages proapoptotic programs that may contribute to decreased tumor growth in this lymphoma subtype.
The identification of mechanisms controlling IRE1 expression in DLBCLs suggests that differences in the XBP1 signatures between ABC and GCB DLBCL34 could be a consequence of the specific downregulation of IRE1 signaling observed in GCB DLBCL. Despite a possible scenario where, due to an advanced differentiation state, IRE1 activation takes place in some ABC tumors, we clearly demonstrated that GCB cells suffer an obvious shortage in IRE1 expression and downstream XBP1 signaling. Therefore, we suggest that IRE1 expression level could be a robust biomarker of DLBCL classification.
Although we cannot exclude that additional mechanisms may contribute to IRE1 downregulation in GCB DLBCL, the fact that the pathway could be restored in vivo upon treatment with EZH2 inhibitors is quite remarkable. This indicates that these drugs that have been developed as cancer therapeutics41 could be positioned to modulate ER stress responses in human diseases characterized by epigenetic deregulation of the IRE1/XBP1 pathway.54 Because IRE1 plays a role in tumorigenesis,33,55,56 it would be important to interrogate how these drugs affect ER stress responses and what consequences this may have on tumor progression in treated patients.
IRE1 is a key pathway that determines cellular fate in various tissues and cancers. Emerging regulatory mechanisms that tune IRE1 expression and function, such as EZH2, could play an important role in various pathologies and conditions. It is therefore likely that further studies of these mechanisms will uncover their importance beyond their role in DLBCL, including during differentiation programs, in the course of stress responses and in other tumors.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank L. H. Glimcher, A. Bruhat, Margot Thome, Nicolas Gestermann, Elisa Oricchio, Dinis Calado, Dong Eun Kim, Paolo Dotto, and Steve Elledge for sharing key reagents. The authors thank Margot Thome and Jerome Lugrin for comments and discussions. The authors thank Melanie Op and Emanuele Bulla for the technical help with in vivo experiments.
This work was supported by European Research Council starting grant 281996. F.B. was supported by a research grant of the Gelu Foundation. B.B. was supported by a fellowship of the Swiss Institute for Experimental Cancer Research Foundation.
Authorship
Contribution: F.M. and B.B. conceptualized the study, developed the study methodology, wrote the original draft of the manuscript, and acquired funding for the study; B.B., A.D.G., R.T., S.C., F.B., O.D., and L.Z. performed experiments; F.B. and M.G. provided resources; and F.M. supervised the study.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Fabio Martinon, Department of Biochemistry, University of Lausanne, 155 Ch. des Boveresses, Epalinges 1066, Switzerland; e-mail: fabio.martinon@unil.ch.

![Figure 1. GCB DLBCLs display impairment in IRE1 expression that affects the ER stress response. (A-B) Representative ABC DLBCL and GCB DLBCL cell lines were treated with 5 μg/mL tunicamycin (TM) for 6 hours and analyzed by quantitative real-time PCR for expression of XBP1 target genes Sec23b, DNAJB9, and SRPR mRNA levels relative to β-actin (mean and standard deviation [SD] of technical triplicates of 1 representative experiment of 3) (A). XBP1 mRNA splicing was measured by RT-PCR analysis of XBP1 mRNA maturation (B) DNA electrophoresis is shown in the top panel. Protein levels of XBP1s and IRE1 protein were analyzed by immunoblot. Tubulin (Tbl) is used as loading control (B, below). (C) DLBCL cell lines were treated for 6 hours with 5 μg/mL tunicamycin (TM) or vehicle and analyzed by immunoblot for XBP1s, IRE1, ATF4, PERK, ATF6, and tubulin (Tbl). (D) Representative ABC and GCB cell lines were treated with different ER stress inducers, tunicamycin (TM), thapsigargin (Tpg), DTT, brefeldin A (Bref A), and bortezomib (Btz) for 6 hours. IRE1, XBP1s, PERK, ATF4, and Tubulin (Tbl) expression was analyzed by immunoblot. (E) Viability of IRE1-high and IRE1-low DLBCL cells assessed by 3-(4,5-dimethylthiazol-2-yl)-2,5-dimethyltetrazolium bromide (MTS)/1-methoxy phenazine methosulfate (PMS) assay after treatment of the indicated time course with 0.05 μg/mL tunicamycin or 12.5 μM nelfinavir. P values were calculated using 2-way analysis of variance (ANOVA); *P ≤ .05; ***P ≤ .001.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/129/17/10.1182_blood-2016-09-741348/4/m_blood741348f1.jpeg?Expires=1767705248&Signature=q0FbqkentAqLZ8mnXl~yhN6QrDCezkX2Z7oTqS~nVDF3HKik1ltgd29eOyGpacHxiXgcOlqVKF8WePV0lAt0B0lFFDwhuefCkWoSGi12liatTh28sbXz9kySWXaxxJW-nLP7sFziGi6Khvg8zkxzhwZPxxLi3uQ5cTyckkgGd4pf-2jMnpTA7NYnU2-u7rbqyScXaDdYVc7XWRT5Vnjy~fk-c03XsZA4277zCu5WI0at~VMYnKsj~v2QO4lPaP8dlZAHYyFUDdwRW-p-S3881O9BDRP490iiALEoStiGTPmq23ry7CvopBl6cPae~E7KZb2E3rW6KpXAxfN9kZ1v9g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal