Key Points
CMV is prevalent in pretreatment bone marrow from childhood ALL and not in acute myeloid leukemia.
In utero infection with CMV is a risk factor for ALL (OR = 3.71, P = .0016) and is more pronounced in Hispanics (OR = 5.90, P = .006).
Abstract
It is widely suspected, yet controversial, that infection plays an etiologic role in the development of acute lymphoblastic leukemia (ALL), the most common childhood cancer and a disease with a confirmed prenatal origin in most cases. We investigated infections at diagnosis and then assessed the timing of infection at birth in children with ALL and age, gender, and ethnicity matched controls to identify potential causal initiating infections. Comprehensive untargeted virome and bacterial analyses of pretreatment bone marrow specimens (n = 127 ALL in comparison with 38 acute myeloid leukemia cases in a comparison group) revealed prevalent cytomegalovirus (CMV) infection at diagnosis in childhood ALL, demonstrating active viral transcription in leukemia blasts as well as intact virions in serum. Screening of newborn blood samples revealed a significantly higher prevalence of in utero CMV infection in ALL cases (n = 268) than healthy controls (n = 270) (odds ratio [OR], 3.71, confidence interval [CI], 1.56-7.92, P = .0016). Risk was more pronounced in Hispanics (OR=5.90, CI=1.89-25.96) than in non-Hispanic whites (OR=2.10 CI= 0.69-7.13). This is the first study to suggest that congenital CMV infection is a risk factor for childhood ALL and is more prominent in Hispanic children. Further investigation of CMV as an etiologic agent for ALL is warranted.
Introduction
A role for infection in the etiology of childhood acute lymphoblastic leukemia (ALL) has long been hypothesized.1-3 Although many individual infectious agents and proxies have been investigated,4-8 a comprehensive untargeted study of possible etiologic viruses and bacteria has not previously been attempted.
Studies of tumor genetic changes in newborn blood spots (NBS) provide unambiguous evidence of the presence of ALL initiation in utero for some subtypes of the disease.9-11 Decreased interleukin-10 levels at birth in children who later develop ALL12 and more medically diagnosed infections early in life7,13 suggest that dysregulated immunity plays an etiologic role. Hispanics have the highest risk of ALL, which appears to be increasing.14 In this study we seek to answer fundamental etiologic questions: (1) What infections are unique to childhood ALL in comparison with acute myeloid leukemia (AML, used here as an immune suppressed control group at the time of diagnosis)? (2) Are such infections acquired in utero, and are they risk factors for ALL?
Study design
We conducted two complementary studies, briefly described below (details are available in the supplemental Materials section, available on the Blood Web site).
Viral and bacterial discovery at diagnosis
We conducted a comprehensive next-generation sequencing-based virome and bacterial metagenomic analysis in pretreatment diagnostic bone marrow of both childhood ALL and AML, drawn from the California Childhood Leukemia Study (CCLS).15 (All data and samples have been approved for use by the University of California, Berkeley, institutional review board [California Childhood Leukemia Study/Protocol 2010-10-2438] and California State institutional review board [Childhood Cancer Record Linkage Project/Protocol 12-07-0529 and California Childhood Leukemia Study/Protocol 2010-10-2438].)
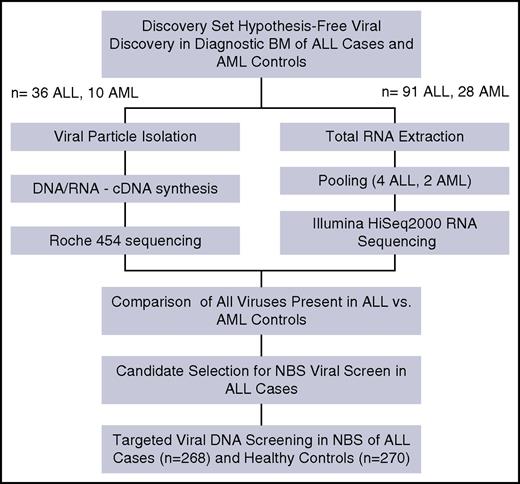
Two independent patient sample sets were selected from the CCLS (see the supplemental Methods section) and interrogated using two separate metagenomic methods (Figure 1). The first method used 36 diagnostic bone marrow (BM) samples from children with ALL and 10 diagnostic BM samples from children with AML and were examined using virus isolation by ultracentrifugation and 454 sequencing. The second method used total RNA extracted from 91 ALL BM pooled into 4 groups and 28 AML BM pooled into 2 groups, then sequenced by Illumina Hi-Seq2500. Data generated from each method were analyzed using separate custom bioinformatics pipelines that categorized all nonhuman reads using BLAST (see the supplemental Methods section). Additionally, we used 16s-based bacterial metagenomics on 91 ALL BM and 28 AML BM as a control for bacterial diversity and differential bacterial contamination of the BM samples (see the supplemental Materials section). Sample size was determined on the basis of sample availability and budget constraints.
Viral screen at birth
NBS from 268 ALL cases and 270 cancer-free controls, randomly drawn from the California Department of Public Health (see the supplemental Methods section, supplemental Table 2), were screened for 2 herpes viruses identified in the discovery set at diagnosis (ie, cytomegalovirus [CMV] and Epstein-Barr virus [EBV]). Viral screen was done by a droplet digital polymerase chain reaction PCR-based direct-detection method, specifically, ultrasensitive “third-generation” ddPCR (Figure 1).
Results and discussion
In general, ALL patients were younger than AML patients but were otherwise demographically similar (supplemental Table 1). A limitation of our methods is the ability to primarily detect active infections, although evidence of viruses and bacteria was detected in all pretreatment bone marrow specimens from both ALL and AML groups by both Illumina sequencing (supplemental Figures 1-7) and virus enrichment isolation/454 sequencing. Only herpes viruses distinguished ALL from AML in both analysis sets; all 4 ALL Illumina pools expressed CMV transcripts, whereas no CMV transcripts were detected in AML pools (Figure 2A). Particle isolation and 454 sequencing showed a greater prevalence of CMV (OR = 18, P = .003) in individual ALL bone marrow specimens in comparison with AML specimens (Figure 2B).
Nonhuman sequence search identification (“blast hits”) from pretreatment bone marrow. (A) Results of deep-sequencing RNA pools (73 ALLs in 4 pools and 28 AMLs in 2 pools) after ribosomal RNA depletion (RiboZERO). Raw Illumina reads were aggressively quality filtered, then aligned to hg19 using BowTie2; nonaligning reads were categorized using an E value less than or equal to 1 × E−10 with blastn and the nt database. Cytomegalovirus hits were normalized by sequencing effort per pool (blast hits/quality reads × 1.0 × E8 ). Odds ratios were calculated by comparing presence/absence of virus in ALL versus AML pools; P values were computed by using Fisher’s exact test (for additional details, see the supplemental Methods section). (B) Results of particle isolation and 454 sequencing from an independent study of 36 ALL and 10 AML patients. Virus was isolated using centrifugation and Millipore filter, and then viral nucleic acids were extracted and sequenced using Roche 454 (for additional details, see the supplemental Methods section). Recovered sequences were assembled into contigs in which alignments to hg19 were removed using Bowtie2. Contigs were then categorized with a cutoff of E value less than or equal to 1.0 × E−10 using blastn and the refseq viral database. Cytomegalovirus contigs were normalized by recovered contigs per patient (blast hits/contigs assembled × 1 × E3 ); odds ratios were calculated comparing presence/absence of virus in ALL versus AML patients; P values were computed using Fisher’s exact test (for additional details, see the supplemental Methods section). CMV was found to be the only virus that showed statistically significant variation between ALL and AML in the Illumina and 454 groups. CI, confidence interval; NA, not applicable; OR, odds ratio.
Nonhuman sequence search identification (“blast hits”) from pretreatment bone marrow. (A) Results of deep-sequencing RNA pools (73 ALLs in 4 pools and 28 AMLs in 2 pools) after ribosomal RNA depletion (RiboZERO). Raw Illumina reads were aggressively quality filtered, then aligned to hg19 using BowTie2; nonaligning reads were categorized using an E value less than or equal to 1 × E−10 with blastn and the nt database. Cytomegalovirus hits were normalized by sequencing effort per pool (blast hits/quality reads × 1.0 × E8 ). Odds ratios were calculated by comparing presence/absence of virus in ALL versus AML pools; P values were computed by using Fisher’s exact test (for additional details, see the supplemental Methods section). (B) Results of particle isolation and 454 sequencing from an independent study of 36 ALL and 10 AML patients. Virus was isolated using centrifugation and Millipore filter, and then viral nucleic acids were extracted and sequenced using Roche 454 (for additional details, see the supplemental Methods section). Recovered sequences were assembled into contigs in which alignments to hg19 were removed using Bowtie2. Contigs were then categorized with a cutoff of E value less than or equal to 1.0 × E−10 using blastn and the refseq viral database. Cytomegalovirus contigs were normalized by recovered contigs per patient (blast hits/contigs assembled × 1 × E3 ); odds ratios were calculated comparing presence/absence of virus in ALL versus AML patients; P values were computed using Fisher’s exact test (for additional details, see the supplemental Methods section). CMV was found to be the only virus that showed statistically significant variation between ALL and AML in the Illumina and 454 groups. CI, confidence interval; NA, not applicable; OR, odds ratio.
We initiated this study with no a priori candidate, following identification of CMV at diagnosis and given the in utero origins of ALL. Children who went on to develop ALL were 3.71 times more likely to be CMV positive at birth (P = .0016) (Table 1). We found suggestive evidence for statistical interaction between CMV positivity and Hispanic ethnicity (logistic interaction term P = .227). Stratification by Hispanic ethnicity showed 5.9-fold increased risk of ALL in Hispanics infected perinatally with CMV (CI = 1.9, 26.0). The prevalence of CMV infection was higher in whites overall, and an increased risk for ALL was also demonstrated in this group; however, the result was not statistically significant (OR = 2.10; CI = 0.69, 7.13). We observed no difference in EBV prevalence at birth between cases and controls (11 positive cases and 11 positive controls; OR = 1.01, P = 1.0). After stratifying by ethnicity, no significant effect modification was observed (Table 1). CMV viral load among positive case samples averaged 0.214 copies/ul and was higher than the CMV viral load among positive control samples (0.071 copies/ul; supplemental Table 2; P = .003). There was no significant difference in the age at diagnosis between CMV-positive (mean age, 4.8 years) and CMV-negative (mean age, 5.15) ALLs at birth.
Results of neonatal blood spot screen: cytomegalovirus and Epstein Barr virus
| . | CMV− . | CMV+ . | OR (CI) . | P value . | EBV− . | EBV+ . | OR (CI) . | P value . |
|---|---|---|---|---|---|---|---|---|
| Overall* | ||||||||
| ALL | 242 | 26 | 3.71 (1.71-8.95) | 0.0016 | 257 | 11 | 1.01 (0.42-2.42) | 1 |
| Controls | 262 | 8 | — | — | 259 | 11 | — | — |
| By race† | ||||||||
| Hispanic ALL | 134 | 15 | 5.90 (1.89-25.96) | 0.006 | 141 | 8 | 1.38 (0.46-4.35) | 0.56 |
| Hispanic controls | 152 | 3 | — | — | 149 | 6 | — | — |
| White ALL | 71 | 10 | 2.10 (0.69-7.13) | 0.204 | 79 | 2 | 0.53 (0.09-2.94) | 0.68 |
| White controls | 82 | 5 | — | — | 83 | 4 | — | — |
| Asian/PI ALL | 25 | 1 | N/A | 1 | 25 | 1 | N/A | 1 |
| Asian/PI controls | 21 | 0 | — | — | 21 | 0 | — | — |
| Black ALL | 10 | 0 | — | — | 10 | 0 | — | — |
| Black controls | 4 | 0 | — | — | 4 | 0 | — | |
| Other ALL | 2 | 0 | — | — | 2 | 0 | — | 1 |
| Other controls | 3 | 0 | — | — | 2 | 1 | — | — |
| . | CMV− . | CMV+ . | OR (CI) . | P value . | EBV− . | EBV+ . | OR (CI) . | P value . |
|---|---|---|---|---|---|---|---|---|
| Overall* | ||||||||
| ALL | 242 | 26 | 3.71 (1.71-8.95) | 0.0016 | 257 | 11 | 1.01 (0.42-2.42) | 1 |
| Controls | 262 | 8 | — | — | 259 | 11 | — | — |
| By race† | ||||||||
| Hispanic ALL | 134 | 15 | 5.90 (1.89-25.96) | 0.006 | 141 | 8 | 1.38 (0.46-4.35) | 0.56 |
| Hispanic controls | 152 | 3 | — | — | 149 | 6 | — | — |
| White ALL | 71 | 10 | 2.10 (0.69-7.13) | 0.204 | 79 | 2 | 0.53 (0.09-2.94) | 0.68 |
| White controls | 82 | 5 | — | — | 83 | 4 | — | — |
| Asian/PI ALL | 25 | 1 | N/A | 1 | 25 | 1 | N/A | 1 |
| Asian/PI controls | 21 | 0 | — | — | 21 | 0 | — | — |
| Black ALL | 10 | 0 | — | — | 10 | 0 | — | — |
| Black controls | 4 | 0 | — | — | 4 | 0 | — | |
| Other ALL | 2 | 0 | — | — | 2 | 0 | — | 1 |
| Other controls | 3 | 0 | — | — | 2 | 1 | — | — |
ALL, acute lymphoblastic leukemia; CI, confidence interval; CMV, cytomegalovirus; EBV, Epstein Barr virus; OR, odds ratio; PI, Pacific Islander.
Logistic regression adjusted for race, age, and sex.
†Logistic regression adjusted for age and sex.
Several studies of childhood ALL have examined herpes viruses at diagnosis with conflicting results.5,6 Two previous studies examined herpes viruses in NBS from children who developed ALL. The first study examined EBV and HHV-6, in which no association was found, but did not assess CMV.16 Another screened NBS for CMV from 48 cases and 46 controls yet found no infected children in either group.17 With a significantly larger sample size and a more sensitive assay, we could detect extremely low quantities of CMV DNA that may otherwise have been missed. Further replication of our findings is warranted using higher input volumes of NBS DNA, protein-based detection methods, and samples from geographically diverse locations.
CMV is a common virus that infects over 90% of adults worldwide. In the United States, seroprevalence of adult CMV ranges from ∼50% to 80% and is highest in Hispanics and blacks,18 which is particularly interesting given the high rate of ALL in Hispanics.14 Timing of infection has important implications. In utero infection with CMV is a leading cause of birth defects, specifically hearing loss. Although primary CMV infection during pregnancy yields the greatest risk for vertical transmission, most women of childbearing age are previously infected with CMV, and therefore vertical transmission of reactivated virus accounts for the majority of congenital CMV infections.19 The presence of CMV in the child prior to birth may have important immune control implications. Infection prior to the development of a robust adaptive immune response in the fetus and neonate may affect central tolerance and enable CMV to persist in an infectious course, unlike the majority of children who are infected after birth, thus leading to increased risk of developing ALL.
Two features of CMV infection support a role in oncogenesis in ALL. First, congenital CMV infection has been noted to cause chromosomal instability, which is suspected to be related to its teratogenic properties.19 This is of interest in leukemia, because specific chromosomal lesions are present at birth in children with ALL.20 Second, CMV has the largest genome of any known human viral pathogen and harbors many immune evasion genes, indicating that host immune dysregulation is a critical aspect of the CMV lifecycle. The functions of many CMV genes remain poorly understood, and it is likely that their genomic complexity is the direct result of the evolution of latency. Along these lines, CMV has been hypothesized to be an “oncomodulator” in adult gliomas.21 A delicate balance has evolved between CMV and the immune system, which has led to a high population prevalence of the virus with relatively low morbidity. Nevertheless, disruptions to this balance can have serious and complex pathologic consequences for the host.
This study provides a putative candidate for future investigations. The comparison of ALLs with AMLs was designed solely to generate a putative agent in ALL and was not matched to race, ethnicity, or age. Furthermore, the preponderance of lymphoid versus myeloid cells in the BM samples may bias the results in relation to viruses with a tropism for B cells. A major limitation in the dried blood spots (DBS) CMV screen is our lack of information on ALL subtype and cytogenetic characteristics. Additionally, the low input quantity of DBS DNA may have resulted in false negatives, though this may drive results toward the null. Additional studies defining in utero CMV infection in terms of ALL subtype, cytogenetics, and overall prevalence are needed.
Our findings lead us to hypothesize that in utero or perinatal CMV infection initiates immune dysregulation during the critical period of fetal immune development, allowing a greater number of and more fulminant infections later in life, as observed in epidemiologic studies.7,13 Further validation of our findings is warranted to establish ALL as an additional reason to develop an effective CMV vaccine.22
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the families that participate in the California Childhood Leukemia Study. Without their time and effort none of our studies would be possible. The authors also thank our clinical collaborators throughout California for their continued support of our research and commitment to their patients. Finally, the authors thank the California Department of Public Health Genetic Disease Screening Program.
This work was supported by grants from the National Institutes of Health (NIH), National Institute of Environmental Health Sciences (R01ES09137); the NIH, National Heart, Lung, and Blood Institute (R01HL105770); and the NIH, National Cancer Institute (1T32CA151022-01, R01CA155461, and R01CA185058).
Authorship
Contribution: S.S.F. conceived and designed the study, performed laboratory experiments, analyzed data, and wrote the primary and subsequent versions of the manuscript; C.M. and G.V.D. enrolled participants and obtained samples; A.D.W. and L.L. helped to design and conduct laboratory experiments; G.A.W. provided bioinformatic and data analysis support; S.K., X.M., and K.M.W. aided in refining the study and helped prepare the manuscript; E.D., F.L., X.M., and L.W.R. helped to design the study; J.L.W. helped to design and conduct the study and prepare the manuscript; all coauthors have reviewed and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Stephen Starko Francis, 1450 3rd St, HD442, University of California, San Francisco, CA 94158; e-mail: ssfrancis@unr.edu or stephen.francis@ucsf.edu.