Key Points
miR-125b overexpression accelerates MLL-AF9–driven AML and endows partial addiction to its overexpression.
A miR-125b-TET2-VEGFA pathway promotes leukemogenesis involving a non–cell-intrinsic mechanism.
Abstract
The hematopoietic stem cell–enriched miR-125 family microRNAs (miRNAs) are critical regulators of hematopoiesis. Overexpression of miR-125a or miR-125b is frequent in human acute myeloid leukemia (AML), and the overexpression of these miRNAs in mice leads to expansion of hematopoietic stem cells accompanied by perturbed hematopoiesis with mostly myeloproliferative phenotypes. However, whether and how miR-125 family miRNAs cooperate with known AML oncogenes in vivo, and how the resultant leukemia is dependent on miR-125 overexpression, are not well understood. We modeled the frequent co-occurrence of miR-125b overexpression and MLL translocations by examining functional cooperation between miR-125b and MLL-AF9. By generating a knock-in mouse model in which miR-125b overexpression is controlled by doxycycline induction, we demonstrated that miR-125b significantly enhances MLL-AF9–driven AML in vivo, and the resultant leukemia is partially dependent on continued overexpression of miR-125b. Surprisingly, miR-125b promotes AML cell expansion and suppresses apoptosis involving a non–cell-intrinsic mechanism. MiR-125b expression enhances VEGFA expression and production from leukemia cells, in part by suppressing TET2. Recombinant VEGFA recapitulates the leukemia-promoting effects of miR-125b, whereas knockdown of VEGFA or inhibition of VEGF receptor 2 abolishes the effects of miR-125b. In addition, significant correlation between miR-125b and VEGFA expression is observed in human AMLs. Our data reveal cooperative and dependent relationships between miR-125b and the MLL oncogene in AML leukemogenesis, and demonstrate a miR-125b-TET2-VEGFA pathway in mediating non–cell-intrinsic leukemia-promoting effects by an oncogenic miRNA.
Introduction
The mammalian miR-125 family microRNAs (miRNAs) are important regulators in hematopoiesis and consist of miR-125a and miR-125b, which reside at 3 genomic loci.1-4 Expression of miR-125a and miR-125b is enriched in both mouse and human hematopoietic stem cells (HSCs) and decreases in more mature cells.4-7 Overexpression of miR-125a and miR-125b is often observed in myeloid and lymphoid malignancy specimens.3,8-14 miR-125b has been estimated to be overexpressed in ∼15% to 25% of human acute myeloid leukemias (AMLs)3,12,15-17 (supplemental Figure 1, available on the Blood Web site; Figure 7). Mechanisms of such deregulation are likely diverse, including rare translocations, chromosomal amplification,8-10,15,18 and transcriptional regulation.14,19
Consistent with their expression pattern, overexpression of miR-125a or miR-125b in HSCs leads to expansion of HSC number and/or function in vivo.4-7,20 At the same time, miR-125a or miR-125b overexpression results in a plethora of perturbations in normal hematopoiesis; most studies report myeloproliferative phenotypes,4,7,12,21 such as a chronic myelomonocytic leukemia–like condition.3,21 Occasionally, lymphoid-biased differentiation has also been reported.22,23 The myeloproliferative conditions in mice overexpressing miR-125a are addicted to its overexpression; phenotypes are largely corrected upon overexpression termination.21 Overexpression of miR-125b enhances GATA1s-induced serial replating activity of megakaryocyte colony–forming units in vitro.8 In a BCR-ABL–driven chronic myeloid leukemia model, coexpression of miR-125b and BCR-ABL accelerated chronic myeloid leukemia development in vivo.12 However, whether and how miR-125 miRNAs synergize with known oncogenes in AML pathogenesis in vivo have not been well explored, and whether AMLs with miRNA overexpression are dependent on such overexpression in vivo remains largely unknown.
VEGFA and VEGFC are VEGF family members that are often overexpressed in AML.24-27 VEGFA signals through vascular endothelial growth factor receptor 1 (VEGFR1) or VEGFR2,25,28-30 activating downstream pathways such as extracellular signal-regulated kinase (ERK) and elevating B-cell lymphoma 2 (BCL2).25 ,29,30 VEGFR2 is often overexpressed in human AMLs, including those with MLL translocation.31 High VEGFA levels have been associated with poor prognosis in AML,32-34 and clinical trials that therapeutically target VEGFA or VEGFR signaling have been actively pursued.23,30,31,35,36 VEGFA and VEGFR signaling has been reported to play an important role in regulating both normal hematopoiesis and AML.27,35,37-39 The mechanisms by which VEGFA is overexpressed in AML specimens are poorly understood. One study has found that RUNX1 can suppress VEGFA transcription, and may explain VEGFA overexpression in RUNX1-ETO AML cases.40 However, the mechanisms of VEGFA overexpression in other AML subtypes remain largely unknown.
In this work, by generating a new doxycycline (Dox)-inducible miR-125b knock-in mouse model, we demonstrate that miR-125b promotes MLL-AF9–driven AML leukemogenesis in vivo, and the resultant AML is partially dependent on continued miR-125b overexpression. Mechanistically, miR-125b overexpression promotes leukemia cell expansion and suppresses apoptosis involving a non–cell-intrinsic mechanism. MiR-125b upregulates VEGFA production partially through suppressing TET2, and the secreted VEGFA promotes leukemia cell expansion. These data indicate a novel mechanism by which oncogenic miRNAs promote AML, and elucidate a miR-125b-TET2-VEGFA pathway in leukemogenesis.
Materials and methods
Genetic mouse models
This study was regulated by the Yale University Institutional Animal Care and Use Committee. All mice were maintained at the Yale Animal Resource Center. C57BL/6 mice and Rosa26-rtTA-M2 mice41 were from the Jackson Laboratory. The i125b allele was generated in this study. Mouse A2Lox.cre embryonic stem cells42 were used with a miR-125b–containing targeting vector (see “Constructs”) to generate the knock-in line 818-7, which generated knock-in mice through blastocyst injection performed by Yale Animal Genomics Services. Germ line transmitted mice were crossed with Rosa26-rtTAm2 mice and backcrossed for 6 generations onto the C57BL/6 background (National Cancer Institute strain no. 01B96) to generate Ri125b mice. Male Ri125b mice have a genotype of rtTA/rtTA, i125b/y, and female Ri125b mice have a genotype of rtTA/rtTA, i125b/i125b/i125b.
Constructs
pMSCV-hMLL-AF9-ires-green fluorescent protein (GFP) was kindly provided by Krivtsov et al.43 To construct pMSCV-hMLL-AF9-ires-ΔNGFR, the EcoRI fragment from pMSCV-hMLL-AF9-ires-GFP was inserted into the EcoRI site of the pMSCV-ires-ΔNGFR vector.44 pMSCV-mmu-miR-125a-phosphoglycerate kinase (PGK)-DsRED-Express vector was cloned using the pMIRWAY-GFP vector7 by first replacing GFP with DsRED-Express, and then recombining mmu-miR-125a in front of the PGK promoter. Short hairpin TET2 (shTET2) and shCtrl were from a published study.45 shp53 and shScr were previously described.46 The cloning of shVEGFA_1, shVEGFA_2, and shCtrl are detailed in the supplemental Materials and methods. The mouse TET2 complementary DNA (cDNA) expression construct was generated by cloning the cDNA into a pMSCV-based pMIRWAY-DsRED-Express vector through Gateway (Invitrogen) cloning.45,47 The targeting vector for knock-in mice was cloned based on P2Lox plasmid.42 Details are in the supplemental Materials and methods.
Retroviral transduction and bone marrow transplantation
Genotyping, cell culture, flow cytometry, cell sorting, Giemsa staining, RNA, and protein analyses
Details are in the supplemental Materials and methods.
Statistical tests
Data are presented as mean values ± standard deviations. Two-tailed Student t test was used to determine P values (unequal variance), unless otherwise stated. For Kaplan-Meier analyses, log-rank tests were performed. For evaluating the outcome of the chimeric leukemia recipients, statistical comparisons were performed between the +Dox and −Dox groups; the P value reflected the probability of the null hypothesis that the +Dox group would not have a higher leukemia cell level than the −Dox group.
Results
Overexpression of miR-125a regulates MLL-AF9–induced AML leukemogenesis
Using data from a published study,48 we observed that both miR-125a and miR-125b were overexpressed in a subset of MLL-translocation AMLs with an outlier expression pattern (supplemental Figure 1). We thus used MLL-AF9 as a representative MLL-translocation oncogene to study the cooperation with miR-125 miRNAs. We initially used a viral model for miR-125a overexpression and followed an established protocol49 in which granulocyte-monocyte progenitors (GMPs) from wild-type mice were transduced with an MLL-AF9 virus containing a GFP marker (supplemental Figure 2A). It has been recently demonstrated that GMPs can be the natural target cells of MLL fusion oncogene–based transformation.50,51 For comparison, GMPs were cotransduced with MLL-AF9 and a miR-125a–coding virus marked by DsRED-Express, or with miR-125a alone (supplemental Figure 2A). MLL-AF9 led to transformation of GMP cells, resulting in a lethal AML with a median survival of 87 days after transplantation (supplemental Figure 2B). miR-125a alone did not result in hematopoietic malignancy. Mice that were recipients of miR-125a and MLL-AF9 cotransduction showed significantly shortened survival compared with mice with MLL-AF9 transduction, with a median survival of 47.5 days (supplemental Figure 2B). These data suggest that overexpression of miR-125a accelerates MLL-AF9–driven leukemogenesis.
Generation of i125b, an inducible miR-125b knock-in mouse model
To avoid uncontrolled integration sites by viral miR-125 delivery, we generated a knock-in mouse model with inducible expression of the miRNA at a defined genetic locus. We focused on miR-125b, because miR-125b is often overexpressed to a higher level than miR-125a in human AML samples, including MLL-translocation AMLs (supplemental Figure 1).
We used a previously published embryonic stem cell line for genetic targeting at a defined locus on the X chromosome to achieve inducible gene expression.42 A targeting vector was cloned with MIR125B1 inserted between 2 splicing sites in the 3′ untranslated region of GFP (Figure 1A), and recombined into the X chromosome locus,42 resulting in the GFP-miR-125b cassette under the control of tet-responsive promoter. The design of splicing sites is to avoid destabilization of GFP RNA by miRNA hairpin52 (the benefits of this design will be described in a future manuscript). We refer to this knock-in allele as “i125b.” Knock-in mice (Figure 1B) were crossed with the ROSA26-rtTAm2 alleles.41 The resulting mice carried both the i125b allele and the rtTA allele; we refer to such mice in this study as “Ri125b” mice.
Generation and confirmation of Dox-inducible miR-125b knock-in mouse. (A) Schematic of knock-in design, with miR-125b precursor sequence flanked by splicing sites inserted in the 3′ untranslated region of enhanced green fluorescent protein (EGFP). The targeting vector was knocked into a Dox-inducible locus on the X chromosome, creating GFP-miR-125b cassette under the control of the tet response element (TRE) promoter. Neomycin (neo), under the control of phosphoglycerate kinase (PGK), is a selection marker for successfully recombined events. (B) Representative genotyping results from wild-type (WT), heterozygous (Het), and homozygous (Homo) i125b knock-in (K/I) mice. (C) Representative flow cytometry to show inducible GFP expression in peripheral blood and bone marrow cells from Ri125b mice with or without Dox water for 7 days. WT mice treated with Dox water were used as controls. (D) WT Mac-1+ or Ri125b GFP+/Mac-1+ myeloid cells were sorted from bone marrow after 7 days of Dox induction. Relative miR-125b expression was determined by quantitative reverse transcription polymerase chain reaction. U6 was used as a control. (E) miR-125b expression in Ficoll-purified bone marrow cells from 6 cytogenetically normal AML patients were determined. Relative expression to U6 small RNA are plotted. Data were aligned from low to high expression of miR-125b. Error bars represent standard deviations; n = 3 for technical replications.
Generation and confirmation of Dox-inducible miR-125b knock-in mouse. (A) Schematic of knock-in design, with miR-125b precursor sequence flanked by splicing sites inserted in the 3′ untranslated region of enhanced green fluorescent protein (EGFP). The targeting vector was knocked into a Dox-inducible locus on the X chromosome, creating GFP-miR-125b cassette under the control of the tet response element (TRE) promoter. Neomycin (neo), under the control of phosphoglycerate kinase (PGK), is a selection marker for successfully recombined events. (B) Representative genotyping results from wild-type (WT), heterozygous (Het), and homozygous (Homo) i125b knock-in (K/I) mice. (C) Representative flow cytometry to show inducible GFP expression in peripheral blood and bone marrow cells from Ri125b mice with or without Dox water for 7 days. WT mice treated with Dox water were used as controls. (D) WT Mac-1+ or Ri125b GFP+/Mac-1+ myeloid cells were sorted from bone marrow after 7 days of Dox induction. Relative miR-125b expression was determined by quantitative reverse transcription polymerase chain reaction. U6 was used as a control. (E) miR-125b expression in Ficoll-purified bone marrow cells from 6 cytogenetically normal AML patients were determined. Relative expression to U6 small RNA are plotted. Data were aligned from low to high expression of miR-125b. Error bars represent standard deviations; n = 3 for technical replications.
To determine transgene inducibility, we induced the mice with Dox via drinking water for 1 week. Most bone marrow cells (98%) became GFP positive (Figure 1C), and bone marrow Mac-1+/GFP+ cells showed ∼1000-fold miR-125b overexpression as compared with Mac-1+ bone marrow cells from wild-type controls (Figure 1D), supporting robust transgene induction. We further show data in later sections (supplemental Figure 5D) that indicate that miR-125b overexpression was ∼25-fold higher in AML cells in Dox-treated mice. A survey of 6 human AML specimens demonstrated an even larger range of differential miR-125b expression; the highest sample was ∼10 000-fold higher than the lowest sample (Figure 1E). These 6 samples were selected to represent the highest and lowest and the intermediate levels of miRNA-125b expression in a cohort of 67 cytogenetically normal AMLs.45 These data suggest that the inducible Ri125b mice can model miR-125b overexpression in human AML.
Continued Dox induction of miR-125b expression in Ri125b mice led to the development of myeloproliferative neoplasm–like symptoms (supplemental Figure 3A-C); these mice showed an increased mortality rate compared with Ri125b−Dox mice after long-term miR-125b induction (supplemental Figure 3A), with a median survival of 370 days after Dox induction. These phenotypes were similar to those in previous reports of miR-125b viral overexpression models,12,21 but we did not observe them to develop full-blown AML in this cohort (eg, supplemental Figure 3B-C). Taken together, Ri125b mice support robust inducible transgene expression, providing a mouse model that will enable analysis of the cooperative role of miR-125b with other known AML oncogenes.
miR-125b accelerates MLL-AF9–induced leukemogenesis
To test whether miR-125b cooperates with MLL-AF9 in leukemogenesis, we used a low dose of MLL-AF9 virus to infect GMPs from uninduced Ri125b mice, so that mice without miR-125b induction would have a relatively low probability of developing leukemia (Figure 2A). The MLL-AF9–encoding retrovirus also contains a ΔNGFR marker that could be used to distinguish transduced cells. Cells were transplanted into wild-type recipients, which were then randomly divided into 2 cohorts; 1 cohort was immediately given Dox water and the other was given regular water (Figure 2A). This design restricts the miR-125b transgene overexpression in GMP-derived cells within the recipients. For simplicity, mice with and without Dox are referred to as AF9+Ri125b+Dox and AF9+Ri125b−Dox, respectively.
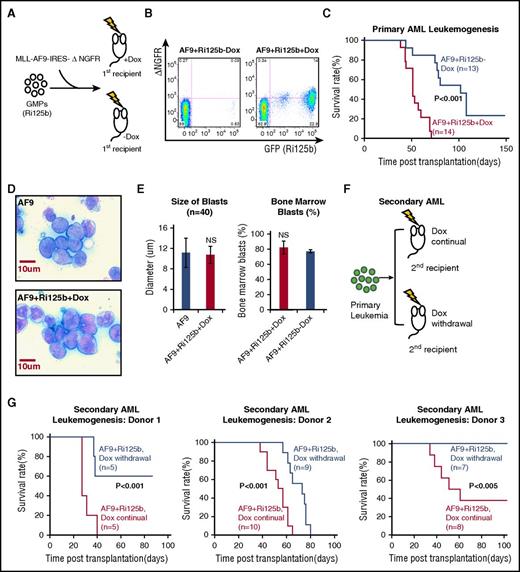
miR-125b accelerates leukemogenesis induced by MLL-AF9. (A) Schematics of the experiment, in which GMPs from Ri125b mice were transduced with MLL-AF9-ΔNGFR (AF9) and injected into wild-type recipients. Recipients were randomly assigned to the Dox treatment group or the −Dox group. (B) Representative flow cytometry plots demonstrating the detection of GFP+ΔNGFR+ cells in peripheral blood 4 weeks after transplantation in +Dox and −Dox mice. GFP marks miR-125b induction and ΔNGFR marks MLL-AF9–transduced cells. (C) Kaplan-Meier survival curve of primary AF9+Ri125b+Dox and AF9+Ri125b−Dox cohorts (P < .001). (D) Representative Giemsa staining of bone marrow cells from MLL-AF9–only or AF9+Ri125b+Dox leukemia mice, showing similar blast morphology. Images were captured by an Olympus BX51 (Olympus) using a SensiCam QE camera (PCO) and IPLab 4.0.8 software (BioVision Technologies). (E) Left: bone marrow blast sizes of MLL-AF9–only and AF9+Ri125b+Dox leukemia (n = 40 cells). Right: bone marrow blast percentages quantified from AF9+Ri125b+Dox and AF9+Ri125b−Dox leukemia-bearing mice (n = 3). (F) Schematics of experiment to analyze secondary leukemogenesis for dependence on miR-125b overexpression. Primary AML cells from AF9+Ri125b+Dox mice were transplanted into lethally irradiated wild-type mice. Secondary recipients were randomized into cohorts for continuation or withdrawal of Dox. (G) Kaplan-Meier survival curves for AF9+Ri125b secondary AML cohorts with continued Dox and Dox withdrawal, with results from 3 primary AML donors. NS, not significant.
miR-125b accelerates leukemogenesis induced by MLL-AF9. (A) Schematics of the experiment, in which GMPs from Ri125b mice were transduced with MLL-AF9-ΔNGFR (AF9) and injected into wild-type recipients. Recipients were randomly assigned to the Dox treatment group or the −Dox group. (B) Representative flow cytometry plots demonstrating the detection of GFP+ΔNGFR+ cells in peripheral blood 4 weeks after transplantation in +Dox and −Dox mice. GFP marks miR-125b induction and ΔNGFR marks MLL-AF9–transduced cells. (C) Kaplan-Meier survival curve of primary AF9+Ri125b+Dox and AF9+Ri125b−Dox cohorts (P < .001). (D) Representative Giemsa staining of bone marrow cells from MLL-AF9–only or AF9+Ri125b+Dox leukemia mice, showing similar blast morphology. Images were captured by an Olympus BX51 (Olympus) using a SensiCam QE camera (PCO) and IPLab 4.0.8 software (BioVision Technologies). (E) Left: bone marrow blast sizes of MLL-AF9–only and AF9+Ri125b+Dox leukemia (n = 40 cells). Right: bone marrow blast percentages quantified from AF9+Ri125b+Dox and AF9+Ri125b−Dox leukemia-bearing mice (n = 3). (F) Schematics of experiment to analyze secondary leukemogenesis for dependence on miR-125b overexpression. Primary AML cells from AF9+Ri125b+Dox mice were transplanted into lethally irradiated wild-type mice. Secondary recipients were randomized into cohorts for continuation or withdrawal of Dox. (G) Kaplan-Meier survival curves for AF9+Ri125b secondary AML cohorts with continued Dox and Dox withdrawal, with results from 3 primary AML donors. NS, not significant.
Examination of the peripheral blood of the AF9+Ri125b+Dox mice, at 4 weeks after transplantation and Dox induction, showed GFP and ΔNGFR double-positive cells (Figure 2B), indicating successful transgene induction in cells derived from MLL-AF9–transduced GMPs. In contrast, in AF9+Ri125b−Dox mice, there were much lower levels of detectable GFP+ or ΔNGFR+ cells at this time point (Figure 2B). Consistent with our hypothesis, AF9+Ri125b+Dox mice showed significantly earlier mortality than AF9+Ri125b−Dox mice (Figure 2C). Moribund mice from both AF9+Ri125b+Dox and AF9+Ri125b−Dox cohorts had enlarged spleens and higher-than-normal peripheral white blood cell counts, and similar bone marrow blast percentages (Figure 2E; supplemental Figure 4A-B). Bone marrow leukemia blast cells from AF9+Ri125b+Dox mice showed similar morphology as leukemia with only the MLL-AF9 oncogene (Figure 2D-E), and were positive for Mac-1 (supplemental Figure 4D), consistent with a myeloid leukemia. The accelerated leukemogenesis is not simply a side effect of Dox treatment, given that MLL-AF9–transduced wild-type GMP cells gave rise to lethal leukemia (referred to as “AF9-only” AML) with a similar time course in the presence and absence of Dox (supplemental Figure 5A). In addition, this acceleration effect cannot be due to GMPs being transformed by miR-125b alone in the absence of MLL-AF9. This is because Ri125b mice treated with Dox developed myeloproliferation (supplemental Figure 3A-C), but did not develop AML, and had a much longer survival than the MLL-AF9 AML models (supplemental Figure 3A). Furthermore, our previous study has shown that miR-125 is incapable of endowing GMPs with self-renewal capacity.7 Of note, the AF9-only AML cells had similar miR-125b expression as normal bone marrow mononuclear cells (supplemental Figure 4C), suggesting that MLL-AF9 does not upregulate endogenous murine miR-125b expression. Taken together, these data indicate that miR-125b accelerates MLL-AF9–induced leukemogenesis.
Partial addiction of AML to continued miR-125b overexpression
To determine whether leukemia that develops in the presence of miR-125b overexpression is dependent on the continued expression of this miRNA, we transplanted primary leukemia cells from AF9+Ri125b+Dox mice into wild-type secondary recipients, and randomly assigned recipients to continued Dox and Dox withdrawal cohorts (Figure 2F). We performed 3 independent experiments using primary AML cells from 3 different donor AML mice. Compared with secondary AF9+Ri125b mice with continued Dox treatment, secondary mice in the Dox withdrawal cohorts showed significantly delayed lethality in all 3 experiments (Figure 2G). For the first donor, 3 of 5 mice in the Dox withdrawal cohort became AML free, whereas all mice with continued Dox treatment died. For the second donor, all mice within the Dox withdrawal cohort died with a significant extension of survival compared with mice with continued Dox treatment. For the third donor, all mice with Dox withdrawal became AML free, whereas 5 of 8 mice with continued Dox died. The differences between the 3 experiments could be due to differences in the primary AML donor cells used in these experiments. Examination of bone marrow cells from moribund leukemic mice revealed that miR-125b was overexpressed at ∼25-fold in AF9+Ri125b+Dox mice when compared with AF9-only AML mice (supplemental Figure 5D). In mice moribund after Dox withdrawal (AF9+Ri125b−Dox), miR-125b expression returned to a level similar to that of AF9-only mice (supplemental Figure 5D), supporting effective shutoff of miR-125b overexpression. Further quantification revealed that AF9+Ri125b+Dox AML cells contained on average 10 939 ± 2750 molecules of miR-125b per cell, whereas AF9+Ri125b−Dox AML cells had 435 ± 34 molecules per cell. Taken together, these experiments support that AML that develops in the presence of miR-125b overexpression is partially dependent on the continued overexpression of miR-125b.
miR-125b regulates MLL-AF9 AML cell expansion and survival involving a non–cell-intrinsic mechanism
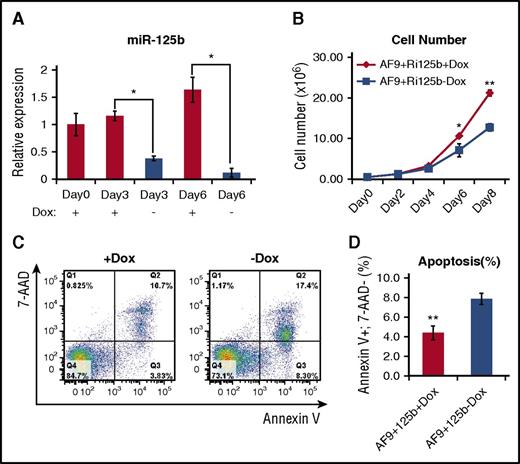
To facilitate the elucidation of the mechanism of miR-125b, we first determined if we could recapitulate the in vivo leukemia-promoting effect of miR-125b in in vitro experiments. Primary AML bone marrow cells from AF9+Ri125b+Dox mice were cultured in the presence and absence of Dox. Dox withdrawal led to a decrease in miR-125b expression (Figure 3A); miR-125b expression levels decreased to 7% of those of +Dox controls after 6 days. We observed a significant decrease in cell numbers after 6 days of Dox withdrawal (Figure 3B). This decrease in cell expansion upon Dox withdrawal was accompanied by an increase in apoptosis, quantified by the annexin V+, 7AAD− population (Figure 3C-D). These data support a role of continued miR-125b overexpression in promoting the expansion of the leukemia cells in vitro, which is consistent with delayed leukemogenesis upon miR-125b withdrawal in vivo.
Withdrawal of miR-125b expression leads to decreased cell number and increased apoptosis. (A) Primary AF9+Ri125b+Dox AML cells were cultured in vitro in the presence or absence of Dox. Relative miR-125b expression levels were determined at the indicated days. U6 was used as a control (n = 3; *P < .01). (B) Viable cell counts for +Dox and −Dox cells at indicated days, for experiment described in (A) (n = 3; *P < .05; **P < .001). (C) Cells in experiment described in (A) were harvested on day 3 and stained with annexin V and 7-aminoactinomycin D (7-AAD). Representative flow cytometry plots (of 3 replicates) are shown. (D) Quantification of annexin V+ 7-AAD− cells for the experiment described in (C) (n = 3; error bars represent standard deviation; **P < .01).
Withdrawal of miR-125b expression leads to decreased cell number and increased apoptosis. (A) Primary AF9+Ri125b+Dox AML cells were cultured in vitro in the presence or absence of Dox. Relative miR-125b expression levels were determined at the indicated days. U6 was used as a control (n = 3; *P < .01). (B) Viable cell counts for +Dox and −Dox cells at indicated days, for experiment described in (A) (n = 3; *P < .05; **P < .001). (C) Cells in experiment described in (A) were harvested on day 3 and stained with annexin V and 7-aminoactinomycin D (7-AAD). Representative flow cytometry plots (of 3 replicates) are shown. (D) Quantification of annexin V+ 7-AAD− cells for the experiment described in (C) (n = 3; error bars represent standard deviation; **P < .01).
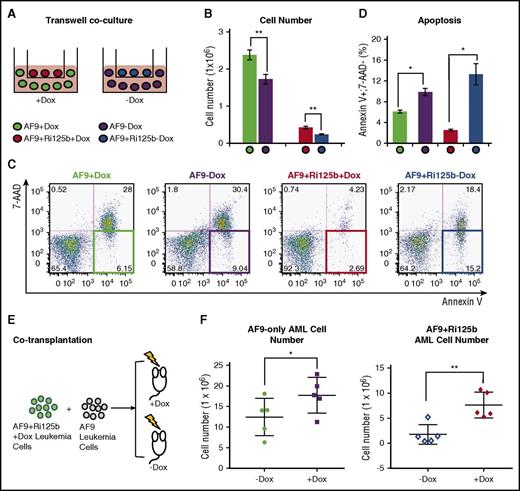
We next asked whether the effect of miR-125b on MLL-AF9 AML cells is through a cell-intrinsic effect or a non–cell-intrinsic effect. We performed a transwell coculture experiment, as illustrated in Figure 4A. Specifically, AF9+Ri125b primary AML cells were cultured in the transwell insert in the presence or absence of Dox, and AF9-only primary AML cells were cultured in the bottom of the transwell. The pore size of the transwell plate is 0.4 μm, thus preventing cellular migration through the membrane. AF9+Ri125b primary AML cells expanded better in the presence of Dox, but unexpectedly, a significant increase in expansion was also observed for the AF9-only AML cells within this coculture system, in the presence of Dox (Figure 4B). This increased expansion in AF9-only AML cells was accompanied by a significant decrease in apoptosis (Figure 4C-D). In contrast, without transwell coculture, AF9-only AML cells cultured in the presence or absence of Dox had similar levels of expansion and apoptosis (supplemental Figure 5B-C), indicating that the observed effect was not simply due to Dox treatment. We also noticed that Kit+ normal bone marrow cells cocultured in the transwell with AF9+Ri125b AML cells expanded better in the +Dox condition than the −Dox condition (supplemental Figure 6E). These data support that a non–cell-intrinsic mechanism was involved in the leukemia-promoting effect of miR-125b in vitro.
Non–cell-intrinsic effects of miR-125b in AF9+Ri125b leukemia. (A) Schematics of the transwell coculture experiment, in which AF9-only leukemia cells were cocultured with AF9+Ri125b AML leukemia cells in the presence or absence of Dox. (B) Cell numbers and (C-D) apoptosis levels were determined for the described experiment in (A) 2 days after plating cells (n = 3; *P < .01; **P < .001). Representative flow cytometry plots are shown in (C). (E) Schematic of cotransplantation of AF9-only AML cells and AF9+Ri125b AML cells. Wild-type recipient mice were treated with or without Dox. All mice were analyzed on day 28 for leukemia cells in right femur bone. (F) Total AF9-only leukemia cells or AF9+Ri125b leukemia cells in the right femur bones were plotted, with each dot representing 1 mouse (n = 5; error bars represent standard deviations; *P < .05; **P < .01).
Non–cell-intrinsic effects of miR-125b in AF9+Ri125b leukemia. (A) Schematics of the transwell coculture experiment, in which AF9-only leukemia cells were cocultured with AF9+Ri125b AML leukemia cells in the presence or absence of Dox. (B) Cell numbers and (C-D) apoptosis levels were determined for the described experiment in (A) 2 days after plating cells (n = 3; *P < .01; **P < .001). Representative flow cytometry plots are shown in (C). (E) Schematic of cotransplantation of AF9-only AML cells and AF9+Ri125b AML cells. Wild-type recipient mice were treated with or without Dox. All mice were analyzed on day 28 for leukemia cells in right femur bone. (F) Total AF9-only leukemia cells or AF9+Ri125b leukemia cells in the right femur bones were plotted, with each dot representing 1 mouse (n = 5; error bars represent standard deviations; *P < .05; **P < .01).
To determine whether a non–cell-intrinsic leukemia-promoting mechanism exists in vivo, we cotransplanted AF9+Ri125b AML cells with AF9-only AML cells into the same wild-type recipients (Figure 4E), and randomly assigned recipients to +Dox and −Dox groups. Bone marrow cells were then analyzed after 28 days, before any mice died. In addition to an increased level of total AF9+Ri125b AML cells in +Dox mice, we also observed a significant increase of total AF9-only AML cells in the femur bones of +Dox mice compared with −Dox mice (Figure 4F). Combined with the fact that Dox did not affect AF9-only AML cells in vitro or in vivo (supplemental Figure 5A-C), these data support a non–cell-intrinsic effect of miR-125b in vivo. Of note, the differences between the absolute numbers of AF9-only AML cells and AF9+Ri125b cells in vivo could be a result of differences in primary AML donor properties, similar to what we observed in Figure 2G. Together, our experiments suggest that a soluble factor was actively secreted by miR-125b–expressing AML cells to influence other AML cells.
miR-125b upregulates VEGFA production in AML cells, which accounts for the majority of leukemia-promoting effects of miR-125b in vitro
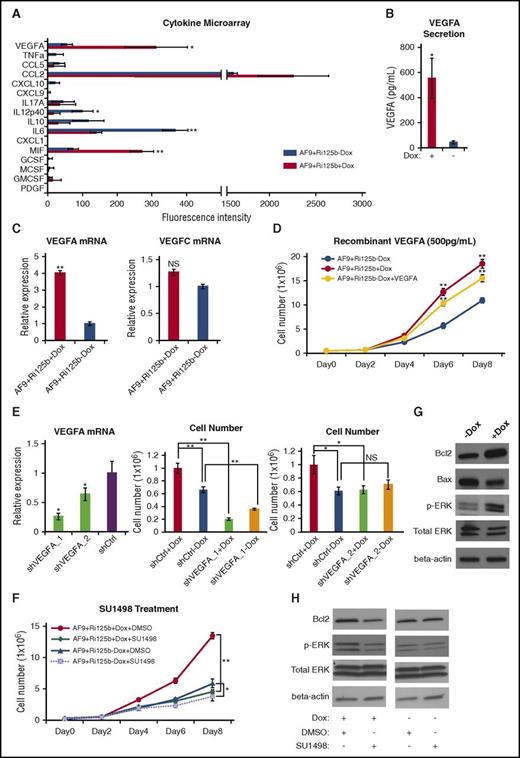
To search for the potential secretory factor that mediates the non–cell-intrinsic mechanism, we used a microchamber-based cytokine microarray53 that was designed for multiplex detection of 16 cytokines by enzyme-linked immunosorbent assay (Figure 5A). Comparing the supernatants of AF9+Ri125b+Dox AML cells to those without Dox, we noticed a significant, >12-fold increase in the level of secreted VEGFA (Figure 5B). An increase in the VEGFA RNA level could also be detected in AF9+Ri125b+Dox AML cells (Figure 5C). In contrast, a related VEGF family member, VEGFC, was not altered (Figure 5C). These data indicate that miR-125b expression in MLL-AF9 AML cells upregulates VEGFA production on both RNA and protein levels.
miR-125b upregulates VEGFA production and signaling to promote leukemia cell expansion. (A) Cytokine microarray analyses were performed for the culture supernatants from AF9+Ri125b AML cells in +Dox or −Dox conditions. The names of the 16 different cytokines are indicated. The fluorescence intensities from the cytokine microarray assay are plotted (n = 3; *P < .05; **P < .001). (B) The absolute concentrations of VEGFA in the culture supernatants from AF9+Ri125b AML cells in +Dox or −Dox conditions were determined by recombinant VEGFA standards (n = 3; *P < .05). (C) VEGFA and VEGFC messenger RNA (mRNA) expressions were determined by quantitative reverse transcription polymerase chain reaction in AF9+Ri125b+Dox and AF9+Ri125b−Dox cells on day 6 after Dox withdrawal. 18S ribosomal RNA was used as a control (n = 3; **P < .01). (D) Primary AF9+Ri125b leukemia cells were cultured in the presence or absence of Dox. Recombinant VEGFA (500 pg/mL) was added to AF9+Ri125b−Dox cells. Cell numbers were counted on indicated days (n = 3; **P < .001). (E) AF9+Ri125b+Dox cells were transduced with control shRNA (shCtrl) or shRNAs against VEGFA (shVEGFA_1, shVEGFA_2). VEGFA RNA levels were determined by quantitative reverse transcription polymerase chain reaction after 3 days (n = 3; *P < .05). Dox was withdrawn from shRNA-transduced AF9+Ri125b AML cells for 5 days, and AF9+Ri125b+Dox and AF9+Ri125b−Dox cells were then plated. Cell numbers were counted 2.5 days after plating (n = 3; *P < .05; **P < .01). (F) AF9+Ri125b+Dox and AF9+Ri125b−Dox cells were treated with dimethyl sulfoxide (DMSO; vehicle) or 3 nM SU1498. Cell numbers were plotted on indicated days (n = 3; *P < .05; **P < .001). (G) VEGFR2 downstream proteins Bcl2, Bax, phosphorylated ERK (pERK), and total ERK were measured in AF9+Ri125b leukemia cells cultured with or without Dox for 6 days. β-actin was used as a loading control. (H) AF9+Ri125b+Dox and AF9+Ri125b−Dox cells were treated with DMSO (vehicle) or 3 nM SU1498. Protein expression levels were determined by western blot after 3 days. Representative blots are shown. For all panels, error bars represent standard deviations.
miR-125b upregulates VEGFA production and signaling to promote leukemia cell expansion. (A) Cytokine microarray analyses were performed for the culture supernatants from AF9+Ri125b AML cells in +Dox or −Dox conditions. The names of the 16 different cytokines are indicated. The fluorescence intensities from the cytokine microarray assay are plotted (n = 3; *P < .05; **P < .001). (B) The absolute concentrations of VEGFA in the culture supernatants from AF9+Ri125b AML cells in +Dox or −Dox conditions were determined by recombinant VEGFA standards (n = 3; *P < .05). (C) VEGFA and VEGFC messenger RNA (mRNA) expressions were determined by quantitative reverse transcription polymerase chain reaction in AF9+Ri125b+Dox and AF9+Ri125b−Dox cells on day 6 after Dox withdrawal. 18S ribosomal RNA was used as a control (n = 3; **P < .01). (D) Primary AF9+Ri125b leukemia cells were cultured in the presence or absence of Dox. Recombinant VEGFA (500 pg/mL) was added to AF9+Ri125b−Dox cells. Cell numbers were counted on indicated days (n = 3; **P < .001). (E) AF9+Ri125b+Dox cells were transduced with control shRNA (shCtrl) or shRNAs against VEGFA (shVEGFA_1, shVEGFA_2). VEGFA RNA levels were determined by quantitative reverse transcription polymerase chain reaction after 3 days (n = 3; *P < .05). Dox was withdrawn from shRNA-transduced AF9+Ri125b AML cells for 5 days, and AF9+Ri125b+Dox and AF9+Ri125b−Dox cells were then plated. Cell numbers were counted 2.5 days after plating (n = 3; *P < .05; **P < .01). (F) AF9+Ri125b+Dox and AF9+Ri125b−Dox cells were treated with dimethyl sulfoxide (DMSO; vehicle) or 3 nM SU1498. Cell numbers were plotted on indicated days (n = 3; *P < .05; **P < .001). (G) VEGFR2 downstream proteins Bcl2, Bax, phosphorylated ERK (pERK), and total ERK were measured in AF9+Ri125b leukemia cells cultured with or without Dox for 6 days. β-actin was used as a loading control. (H) AF9+Ri125b+Dox and AF9+Ri125b−Dox cells were treated with DMSO (vehicle) or 3 nM SU1498. Protein expression levels were determined by western blot after 3 days. Representative blots are shown. For all panels, error bars represent standard deviations.
To determine whether the level of upregulated VEGFA could account for a significant portion of the miR-125b–mediated leukemia-promoting effect, we quantified the concentration of VEGFA in the supernatant of AF9+Ri125b+Dox AML cells, which was 552 ± 160 pg/mL, by using recombinant VEGFA as a standard (supplemental Figure 6A). In contrast, the concentration of VEGFA levels in the −Dox culture was 44 ± 13 pg/mL. We then treated AF9+Ri125b cells in the −Dox condition with 500 pg/mL recombinant VEGFA, which led to an increase in cell expansion compared with −Dox control cells (Figure 5D). Compared with cells treated with Dox, the increase in cell expansion by recombinant VEGFA was ∼70% of that in +Dox condition, indicating that miR-125b–induced VEGFA production accounts for the majority of the leukemia-promoting effect of miR-125b. Titration experiments showed that at concentrations above 500 pg/mL, the biological effects of VEGFA were saturated, whereas a concentration of 100 pg/mL did not have a significant effect (supplemental Figure 6B), suggesting that the level of VEGFA produced from miR-125b–expressing AML cells is over a threshold level. To further evaluate the requirement of VEGFA in this biology, we used 2 independent shRNAs that knocked down VEGFA expression, and observed that both VEGFA shRNAs abolished miR-125b–induced expansion of the AF9+Ri125b AML cell culture (Figure 5E).
The biological effect of VEGFA suggests that VEGFA-mediated signaling in leukemia cells is important for the expansion of MLL-AF9 AML cells. Given that VEGFR2 is often overexpressed in AML with MLL translocation,30,31 we tested whether inhibiting VEGFR2 could inhibit miR-125b–induced AML cell expansion by using a specific VEGFR2 inhibitor, SU1498.54 SU1498 treatment led to a significant decrease in the expansion of the +Dox AML cells to a level similar to that of −Dox cells (Figure 5F), whereas drug treatment of −Dox AF9+Ri125b AML cells only yielded a mild suppressive effect (Figure 5F). The suppression of AF9+Ri125b+Dox cells by SU1498 was accompanied by an increase in cell death (supplemental Figure 6C-D). Consistent with an effect of SU1498, AF9+Ri125b cells with Dox treatment had increased phosphorylated ERK and increased BCL2 (Figure 5G), both of which are downstream of VEGFR2 signaling.25,28-30 Treatment of SU1498 reduced phosphor-ERK and BCL2 levels in +Dox cells, but not in −Dox cells (Figure 5H). Taken together, the data above support that miR-125b–regulated VEGFA overproduction is a key mechanism for the leukemia-promoting effect of miR-125b.
miR-125b upregulates VEGFA partially by targeting TET2
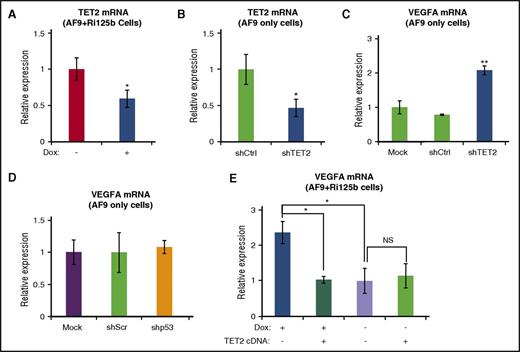
We have previously shown that TET2 is a direct target of miR-125 miRNAs.45 We tested the possibility that miR-125b upregulates VEGFA through targeting TET2. We first examined the RNA level of TET2 in AF9+Ri125b+Dox cells, and observed a decrease in TET2 expression in comparison with −Dox cells (Figure 6A). We observed similar results when examining AML cells directly isolated from AF9+Ri125b+Dox mice and AF9+Ri125b−Dox mice (supplemental Figure 6F). We next asked whether the downregulation of TET2 was sufficient to lead to increased VEGFA expression. AF9-only AML cells were transduced with a published shRNA against TET2 (shTET2), a shRNA against p53 (shp53), or their corresponding controls. shTET2 knocked down TET2 expression (Figure 6B) by ∼50%, which is comparable to miR-125b–induced TET2 downregulation (Figure 6A). shTET2 resulted in a ∼twofold increase in VEGFA RNA expression, whereas shp53 did not (Figure 6C-D). We further asked whether increasing TET2 levels could inhibit miR-125b–induced VEGFA upregulation. We expressed a 3′ untranslated region–less TET2 that was not under the control of miR-125b45 into AF9+Ri125b AML cells. The expression of TET2 cDNA significantly suppressed the upregulation of VEGFA in +Dox cells, but not in −Dox cells (Figure 6E). Taken together, these experiments support that miR-125b upregulates VEGFA expression, at least partially, by targeting and downregulating TET2 expression.
miR-125b upregulates VEGFA expression in part through suppression of TET2. (A) TET2 messenger RNA (mRNA) levels were assessed by quantitative reverse transcription polymerase chain reaction (qRT-PCR) in AF9+Ri125b+Dox and AF9+Ri125b−Dox cells at day 6. 18S ribosomal RNA (rRNA) was used as an internal control (n = 3). (B) shRNA against TET2 or a control shRNA was transduced into primary MLL-AF9–only AML cells. TET2 expression levels were determined by qRT-PCR 7 days after transduction (n = 3). (C) MLL-AF9–only leukemia cells were infected with shTET2, shCtrl, or a mock infection. After 7 days of puromycin selection, total mRNAs were harvested for TET2 expression analysis. 18S rRNA was used as an internal control (n = 3). (D) VEGFA mRNA levels were analyzed in MLL-AF9–only AML cells that were infected with shp53 or a control shScr vector. 18s was used as an internal control (n = 3). (E) TET2 cDNA or a control vector was transduced into AF9+Ri125b AML cells in the presence or absence of Dox treatment. Transduced cells were fluorescence-activated cell sorted 36 hours post infection, and the levels of VEGFA expression were determined by qRT-PCR and normalized to 18S rRNA (n = 3; error bars represent standard deviations; *P < .01; **P < .001 in all panels). NS, not significant.
miR-125b upregulates VEGFA expression in part through suppression of TET2. (A) TET2 messenger RNA (mRNA) levels were assessed by quantitative reverse transcription polymerase chain reaction (qRT-PCR) in AF9+Ri125b+Dox and AF9+Ri125b−Dox cells at day 6. 18S ribosomal RNA (rRNA) was used as an internal control (n = 3). (B) shRNA against TET2 or a control shRNA was transduced into primary MLL-AF9–only AML cells. TET2 expression levels were determined by qRT-PCR 7 days after transduction (n = 3). (C) MLL-AF9–only leukemia cells were infected with shTET2, shCtrl, or a mock infection. After 7 days of puromycin selection, total mRNAs were harvested for TET2 expression analysis. 18S rRNA was used as an internal control (n = 3). (D) VEGFA mRNA levels were analyzed in MLL-AF9–only AML cells that were infected with shp53 or a control shScr vector. 18s was used as an internal control (n = 3). (E) TET2 cDNA or a control vector was transduced into AF9+Ri125b AML cells in the presence or absence of Dox treatment. Transduced cells were fluorescence-activated cell sorted 36 hours post infection, and the levels of VEGFA expression were determined by qRT-PCR and normalized to 18S rRNA (n = 3; error bars represent standard deviations; *P < .01; **P < .001 in all panels). NS, not significant.
miR-125b and VEGFA expression levels are significantly correlated in human AML
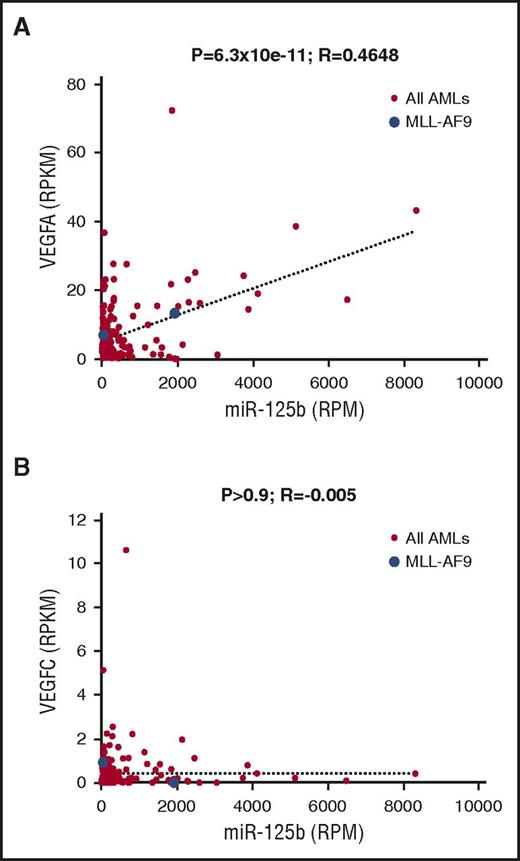
To evaluate the relevance of VEGFA upregulation by miR-125b in human AML, we examined the published data set on AML from The Cancer Genome Atlas, in which 178 samples have public RNA sequencing and small RNA sequencing data. We observed a significant positive correlation between miR-125b RNA expression and VEGFA RNA expression (Figure 7A; P < 1 × 10−10). In contrast, we did not observe any significant correlation between miR-125b expression and VEGFC expression (Figure 7B; P > .9). Of note, the correlation between miR-125b and VEGFA expression was not perfect, which is expected, given that VEGFA is likely under additional mechanisms of regulation. We also observed a weak but significant negative correlation between TET2 expression and VEGFA expression (P < .05), but not between TET2 and VEGFC (supplemental Figure 6G), consistent with our model that TET2 suppresses VEGFA expression in AML cells. These data suggest that the mechanism of miR-125b–mediated VEGFA upregulation is not simply a mouse model phenomenon, but rather is relevant to human AML.
miR-125b correlates with VEGFA mRNA expression in samples from human patients with AML. (A) Correlation analysis between miR-125b expression and VEGFA mRNA expression levels in human patients with AML from The Cancer Genome Atlas data, which contain a total of 178 samples. Expression levels were expressed in reads per million (RPM) or reads per million per kilobase (RPKM). Each dot represents 1 sample. Blue dots: patient samples with MLL-AF9 translocation. Red dots: patients with AML without MLL-AF9 translocation. (B) Similar analysis as in (A) between miR-125b and VEGFC. P values and R values are indicated.
miR-125b correlates with VEGFA mRNA expression in samples from human patients with AML. (A) Correlation analysis between miR-125b expression and VEGFA mRNA expression levels in human patients with AML from The Cancer Genome Atlas data, which contain a total of 178 samples. Expression levels were expressed in reads per million (RPM) or reads per million per kilobase (RPKM). Each dot represents 1 sample. Blue dots: patient samples with MLL-AF9 translocation. Red dots: patients with AML without MLL-AF9 translocation. (B) Similar analysis as in (A) between miR-125b and VEGFC. P values and R values are indicated.
Discussion
In this study, we examined the synergy and dependence of miR-125b overexpression in MLL-AF9–driven leukemogenesis. Functional experiments in vivo indicate that miR-125b enhances AML leukemogenesis. Upon withdrawal of miR-125b overexpression, secondary leukemogenesis is delayed, and in some cases, completely stopped. One of the unexpected findings from this study is that miR-125b promotes MLL-AF9–driven leukemogenesis involving a non–cell-intrinsic mechanism. The role of the miR-125 family miRNAs in inhibiting hematopoietic stem and progenitor cell apoptosis has been reported by multiple laboratories,4-7 with a common, yet unproven, assumption that miR-125 miRNAs inhibit apoptosis through a cell-intrinsic mechanism, such as targeting of Bak1, Bmf, Puma, and the p53 pathway.3,55 However, evidence proving the cell-intrinsic nature of the antiapoptotic effect of miR-125b remains largely absent. It is possible that similar non–cell-intrinsic mechanisms may be involved in the regulation of hematopoietic stem and progenitor cells.
Our study indicates a new mechanism by which VEGFA can be overproduced in AML. We have shown that miR-125b strongly upregulates VEGFA on the levels of both RNA and secreted proteins in mouse AML cells. A significant correlation also exists between miR-125b expression and VEGFA RNA in human AML samples. This mechanism is unlikely to be the only mechanism governing VEGFA overproduction, given that there are some human AML samples (Figure 7A) with low miR-125b expression but high VEGFA expression, consistent with the previous finding that VEGFA overexpression in RUNX1-ETO AMLs may be regulated through RUNX1-mediated VEGFA suppression.40 We further demonstrated a link between the miR-125b–target TET2 and VEGFA expression. The mechanism by which TET2 suppresses VEGFA expression is currently unknown. TET2 is a frequently mutated hematopoietic tumor suppressor that can catalyze the first steps of DNA demethylation,56 and thus is associated with mechanisms of gene activation. Nevertheless, recent studies showed that TET2 can recruit histone deacetylases to function as suppressors of gene expression,57,58 a mechanism which may be at play in VEGFA regulation. It is also possible that TET2 indirectly regulates VEGFA expression, for example, through the hypoxia pathway, which is well known for VEGFA regulation.59,60 Our experiments also suggest that inhibiting VEGFR2 could be a potential approach to target miR-125b–overexpressing AML in vivo, a possibility that can be tested in the future.
Our finding that VEGFA-mediated non–cell-intrinsic effects are a mechanism for the leukemia-promoting effects of miR-125b does not exclude the possibility that other mechanisms, such as cell-intrinsic pathways, also contribute to enhanced leukemogenesis. In the in vivo competition experiment, we observed a larger-fold difference between +Dox and −Dox mice for AF9+Ri125b AML cells than for AF9-only AML cells, suggesting the existence of cell-intrinsic or VEGFA-mediated autocrine (in addition to paracrine) regulation in vivo. Furthermore, we and others have previously shown that protein phosphatases, many of which function to deactivate kinase signaling, can be directly targeted by miR-125 family miRNAs.21,61 These molecular interactions may augment VEGFA-mediated signaling. We also observed that the level of upregulation of secreted VEGFA by miR-125b is stronger than that on the RNA level, suggesting either that miR-125b affects posttranscriptional control of VEGFA production, or the existence of cellular heterogeneity that affects miR-125b function.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Ergin Beyret for help in culturing the knock-in embryonic stem cells.
This study was supported in part by National Institutes of Health, National Cancer Institute grant no. R01CA149109 (J. Lu) and National Institutes of Health, National Institute of General Medical Sciences grant no. R01GM099811 (J. Lu), Connecticut Regenerative Medicine Fund 15-RMB-YALE-06 (J. Lu), and a Leukemia and Lymphoma Society Fellowship (W.P.).
Authorship
Contribution: J. Lu, S.G. and J. Liu designed the research and analyzed data; J. Lu and J. Liu wrote the manuscript; J. Liu, B.G., Z.C., N.W., J.C., C.R., W.P., and S.G. performed the research; S.K. and R.F. participated in data analysis and interpretation; and M.I., S.C., M.K., and R.F. contributed vital reagents.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jun Lu, Yale Stem Cell Center, 10 Amistad St, Room 237C, New Haven, CT 06520-8005; e-mail: jun.lu@yale.edu.