Key Points
New approaches to identifying functionally relevant mutations in CTLA-4 deficiency syndromes.
Measuring responses to stimulation and degradation distinguishes between CTLA-4 and LRBA mutations.
Abstract
Heterozygous CTLA-4 deficiency has been reported as a monogenic cause of common variable immune deficiency with features of immune dysregulation. Direct mutation in CTLA-4 leads to defective regulatory T-cell (Treg) function associated with impaired ability to control levels of the CTLA-4 ligands, CD80 and CD86. However, additional mutations affecting the CTLA-4 pathway, such as those recently reported for LRBA, indirectly affect CTLA-4 expression, resulting in clinically similar disorders. Robust phenotyping approaches sensitive to defects in the CTLA-4 pathway are therefore required to inform understanding of such immune dysregulation syndromes. Here, we describe assays capable of distinguishing a variety of defects in the CTLA-4 pathway. Assessing total CTLA-4 expression levels was found to be optimal when restricting analysis to the CD45RA−Foxp3+ fraction. CTLA-4 induction following stimulation, and the use of lysosomal-blocking compounds, distinguished CTLA-4 from LRBA mutations. Short-term T-cell stimulation improved the capacity for discriminating the Foxp3+ Treg compartment, clearly revealing Treg expansions in these disorders. Finally, we developed a functionally orientated assay to measure ligand uptake by CTLA-4, which is sensitive to ligand-binding or -trafficking mutations, that would otherwise be difficult to detect and that is appropriate for testing novel mutations in CTLA-4 pathway genes. These approaches are likely to be of value in interpreting the functional significance of mutations in the CTLA-4 pathway identified by gene-sequencing approaches.
Introduction
Common variable immune deficiency (CVID) is a heterogeneous group of primary immune deficiencies, containing of a number of different genetic etiologies. Although diagnosis is characterized by low levels of immunoglobulins, a significant fraction of patients suffer from complications, some of which are autoimmune in nature including enteropathy and cytopenias.1,2 The use of exome and genome sequencing has identified an increasing number of genes that are associated with CVID,3,4 however, this raises the issue of determining whether individual mutations in such genes are functionally significant. Accordingly, functional dissection is required in order to validate the impact of gene mutations. Recently, heterozygous mutations in the CTLA-4 gene have been reported in humans with features of CVID with autoimmune complications.5,6 In addition, biallelic mutations in a second gene, LRBA, also affect the CTLA-4 pathway,7,8 resulting in a similar disease phenotype, which, in contrast to CTLA-4 mutation, has nearly complete penetrance.9,10 In both conditions, insufficient functionally active CTLA-4 is produced to permit the proper functioning of regulatory T cells (Tregs), giving rise to immunodysregulation polyendocrinopathy enteropathy X-linked (IPEX)-like disorders. It is also likely that additional mutations affecting the function of the CTLA-4 pathway will be identified in the future, which will require robust functional assays. Treg testing in vitro is notoriously difficult and in vitro assays are frequently performed in ways that are uninformative for investigating CTLA-4 function.11
Despite an understanding of the general principles of CD28 and CTLA-4 in T-cell biology,12 the precise physiological mechanisms behind CTLA-4 function are still debated,13-15 hampering the design of functional tests. Much of the biology of CTLA-4 concerns Foxp3+ Tregs,16 although it is also induced upon activation of Foxp3− conventional T cells (Tcons). Accordingly, mice completely deficient in CTLA-4, and those conditionally deficient only in Tregs, develop wide-ranging and typically fatal autoimmunity17-19 but with some variation.20,21 We recently identified a mechanism of action whereby CTLA-4 acts to capture and remove its ligands from antigen-presenting cells by a process known as transendocytosis.22 Because T-cell costimulation via CD28 is triggered by these same ligands (CD80 and CD86), CTLA-4 therefore acts to regulate CD28 stimulation. Accordingly, uptake of ligands by CTLA-4 represents a measure of its functional capacity. Indeed, the principle of controlling availability of CD28 ligands has been used to generate soluble forms of CTLA-4 (abatacept and its high-affinity derivative belatacept) for use as immune-suppressive agents,23 which are increasingly being evaluated in immune deficiencies with immune dysregulation.7
In addition to ligand binding, the cell biology of CTLA-4 is unusual and requires consideration. Although ∼10% of CTLA-4 protein is typically found at the plasma membrane, the majority of CTLA-4 is actually located intracellularly as a result of rapid internalization by clathrin-mediated endocytosis.24 Subsequently, trafficking of CTLA-4–containing vesicles through the cell involves both recycling to the plasma membrane and degradation in lysosomes.25 Accordingly, disturbances in trafficking can result in defective CTLA-4 expression. This issue has been recently highlighted by the discovery that LRBA affects CTLA-4 trafficking and lysosomal degradation. Consequently, individuals with defective LRBA have low levels of CTLA-4, but in the absence of CTLA-4 mutations.7
Assessing CTLA-4 and LRBA mutations and the pathway in general therefore requires a number of approaches at the intersection of CTLA-4 and Treg biology to determine functional significance. Such methodologies should be capable of reliably detecting heterozygous (ie, incomplete) loss of CTLA-4 expression in the presence of the remaining unaffected allele. Moreover, assays are needed that detect the impacts of different mutations as well as distinguishing between direct causes (eg, CTLA-4 mutation) and indirect causes (eg, LRBA mutation). Here, we describe a number of approaches that, when used together, provide detailed assessment of the likely functional significance of mutations in this pathway as well as highlight the differences between LRBA and CTLA-4 deficiency and their impact on CTLA-4 expression.
Methods
PBMC isolation
Blood was diluted at 1/1 with phosphate-buffered saline, layered on Ficoll-Paque PLUS (GE Healthcare), and centrifuged at 1060g for 25 minutes. Peripheral blood mononuclear cells (PBMCs) were resuspended in phosphate-buffered saline containing 2 mM EDTA/0.5% bovine serum albumin for T-cell purification using a CD4+ T-cell enrichment kit (StemCell). Patient samples were submitted for the purpose of diagnostic evaluation and processed under institutional approval for the investigation of immunodeficiency diseases.
T-cell stimulation
CD4 T cells were resuspended at 1 × 106/mL in RPMI 1640 with 10% fetal bovine serum, 2 mM l-glutamine, 1% penicillin, and 1% streptomycin. Cells were stimulated with anti-CD3/CD28 T-cell expander dynabeads (Invitrogen) at a ratio of 1 bead to 2 T cells for 16 hours. To inhibit lysosomal degradation, bafilomycin A (BafA; Sigma-Aldrich) was added at 50 nM. Cells were cultured in a 96-well round-bottomed plate at 37°C, 95% humidity, and 5% CO2.
Flow cytometry
For surface staining, cells were incubated with CD25 BV605 (clone 2A3; BD), CD4 Alexa Fluor 700 (clone RPA-T4; BD), CD45RA peridinin chlorophyll–Cy5.5 (clone HI100; eBioscience) at 4°C for 30 minutes. For analysis of total CTLA-4 and Foxp3 expression, cells were fixed and permeabilized with Foxp3 staining buffer (eBioscience) and incubated with Foxp3 allophycocyanin (clone 236A-E7; eBioscience) and CTLA-4 phycoerythrin (clone BNI3; BD). Cells were acquired on a BD LSRII cytometer and the data analyzed using FlowJo software (TreeStar).
Ligand uptake assay
CD4 T cells were incubated with recombinant human CD80-Ig (R&D Systems) at 2 μg/mL in the presence of CD3/CD28 bead stimulation for 16 hours. To block ligand uptake, abatacept (Bristol-Myers Squibb) was added at 10 μg/mL. Cells were then labeled for CD4, CD25, and CD45RA as described in the previous section. For intracellular staining, cells were fixed and permeabilized with Foxp3 staining buffer (eBioscience) and stained for Foxp3, total CTLA-4, and CD80-Ig uptake. For Foxp3, anti-Foxp3 eFluor 450 (clone 236A-E7; eBioscience) was used. Total CTLA-4 was stained using a CTLA-4 C-terminal antibody (C-19; Santa Cruz Biotechnology) and detected with anti-goat immunoglobulin G (IgG) Alexa Fluor 647. CD80-Ig was detected with rabbit anti-human IgG phycoerythrin (SouthernBiotech). The efficiency of ligand uptake (ligand uptake/ CTLA-4) was calculated by extracting CD80-Ig and CTLA-4 mean fluorescence intensity (MFI) values and determining the slope of the line of best fit using linear regression.
Results
CTLA-4 deficiency is most robustly detected in memory Tregs
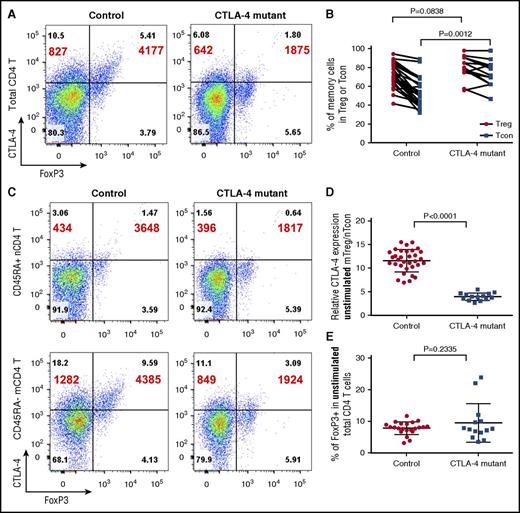
CTLA-4 is expressed in both activated Tcons and Foxp3+ Tregs. We therefore performed flow cytometric staining using a multiplex panel to examine CTLA-4 and Foxp3 in both naive and memory T cells. Total CTLA-4 stains where cells were fixed and permeabilized were used to determine overall deficits in expression. However, it should be appreciated that CTLA-4 trafficking is dynamic and can give rise to specific defects that are not detected in total stains. As shown in Figure 1A, analysis of peripheral blood CD4+ T cells revealed that Foxp3+ Tregs expressed higher CTLA-4 compared with Foxp3− cells as expected. On average, the MFI of Tregs was approximately fivefold brighter than Foxp3− T cells, however, this value was influenced by the numbers of naive and memory T cells in the Foxp3− populations as well as their activation state. To account for variability in naive and memory T-cell fractions, we analyzed naive and memory subsets in both Foxp3+ and Foxp3− compartments independently. This revealed a number of features. First, as expected, the fraction of naive or memory T cells varied considerably between individuals and we observed higher numbers of CD4+ memory Tcons in CTLA-4 deficiency (Figure 1B). Second, when gating on the naive compartment, it was more difficult to detect CTLA-4 deficiency even among Foxp3+ Tregs as CTLA-4 had lower expression (Figure 1C top panels). In contrast, differences in CTLA-4 expression between individuals with CTLA-4 mutations and control individuals were readily detected in the memory (CD45RA−Foxp3+) Treg population (Figure 1C bottom panels). Therefore, analyzing memory Tregs was useful because it prevented incorrect identification of low CTLA-4 expression simply due to high numbers of naive Tregs and instead focused analysis on cells expressing the highest levels of CTLA-4, thereby making detection of CTLA-4 deficiency more robust (Figure 1C bottom panels).
Reduced CTLA-4 expression in memory Tregs in individuals with CTLA-4 mutations. (A) Expression of Foxp3 and total CTLA-4 in unstimulated CD4 T cells. CTLA-4 MFI (large font) is shown for the total Foxp3+ and Foxp3− populations. Percentage values are shown in quadrants. (B) Comparison of percentage of memory CD4 T cells (CD45RA−) in Foxp3+ (Treg) and Foxp3− (Tcon) compartments in CTLA-4–deficient individuals (n = 14) and controls (n = 22). (C) Representative expression of Foxp3 and total CTLA-4 in unstimulated CD4 T cells gated on CD45RA+ naive (top) or CD45RA− memory subsets (bottom). CTLA-4 MFI (large font) is shown for total Foxp3+ cells and Foxp3− cells. Percentages are shown in quadrants. (D) Relative CTLA-4 expression in healthy controls (n = 33) and individuals with CTLA-4 heterozygous mutations (n = 14). Relative expression is calculated as the fold CTLA-4 MFI change between of nTcons and mTregs. (E) Foxp3+ Treg percentage in unstimulated CD4 T cells comparing CTLA-4 mutation carriers and controls.
Reduced CTLA-4 expression in memory Tregs in individuals with CTLA-4 mutations. (A) Expression of Foxp3 and total CTLA-4 in unstimulated CD4 T cells. CTLA-4 MFI (large font) is shown for the total Foxp3+ and Foxp3− populations. Percentage values are shown in quadrants. (B) Comparison of percentage of memory CD4 T cells (CD45RA−) in Foxp3+ (Treg) and Foxp3− (Tcon) compartments in CTLA-4–deficient individuals (n = 14) and controls (n = 22). (C) Representative expression of Foxp3 and total CTLA-4 in unstimulated CD4 T cells gated on CD45RA+ naive (top) or CD45RA− memory subsets (bottom). CTLA-4 MFI (large font) is shown for total Foxp3+ cells and Foxp3− cells. Percentages are shown in quadrants. (D) Relative CTLA-4 expression in healthy controls (n = 33) and individuals with CTLA-4 heterozygous mutations (n = 14). Relative expression is calculated as the fold CTLA-4 MFI change between of nTcons and mTregs. (E) Foxp3+ Treg percentage in unstimulated CD4 T cells comparing CTLA-4 mutation carriers and controls.
Because unstimulated (CD45RA+Foxp3−) CD4+ naive Tcons (nTcons) do not express CTLA-4, we used this population as an internal control with which to compare CTLA-4 expression between individuals. Using this approach, memory Tregs (mTregs) from healthy controls expressed on average 10-fold higher CTLA-4 (MFI) than naive CD4 T cells (Figure 1D). In contrast, patients with CTLA-4 deficiency generally had less than fivefold increase (Figure 1D). Thus, the fold change in CTLA-4 MFI between naive CD4 T cells and mTregs is a robust indicator of CTLA-4 deficiency, which can be used to compare between individuals. Finally, because CTLA-4 affects Treg homeostasis, we also examined the percentage of Tregs as a fraction of CD4+ cells in individuals with CTLA-4 deficiency (Figure 1E). This revealed some heterogeneity with marked expansions in some individuals but not others. Thus, although expansions of Tregs are a feature of CTLA-4 deficiency, they are not observed in all individuals, suggesting they may be mutation specific.
Defective CTLA-4 expression remains after T-cell stimulation
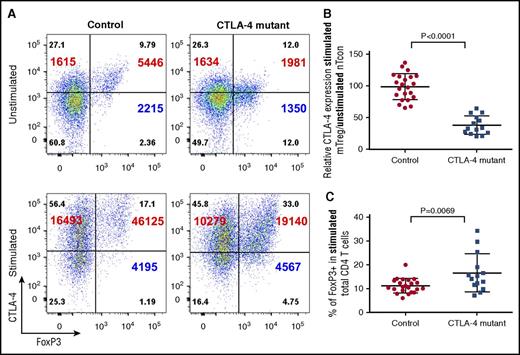
Given that CTLA-4 expression is induced upon activation of Tcons, we measured its induction in individuals with CTLA-4 mutations following stimulation. CTLA-4 expression was substantially increased upon stimulation in both Tcons as well as in Tregs (Figure 2A) with ∼10-fold increase in MFI over the unstimulated levels, in both Treg and non-Treg populations. This upregulation occurred in both healthy controls and in individuals carrying CTLA-4 mutations, suggesting that mutation did not alter the response to stimulation. However, despite the ability to upregulate CTLA-4, the fold change in CTLA-4 mutation carriers (relative to naive T cells) remained approximately half that of healthy individuals (Figure 2B). Stimulation therefore provides important additional verification that reduced CTLA-4 expression due to genetic deficiency cannot be corrected by T-cell activation. During the stimulation process, we also noted that stimulation revealed increased percentages of Foxp3+ T cells which was particularly evident in individuals with CTLA-4 mutation. This suggested that brief T-cell activation enhanced detection of Tregs that were otherwise missed, possibly due to low levels of Foxp3 expression in the ex vivo state (Figure 2C).
CTLA-4 deficiency persists after stimulation. (A) CD4 T cells were stimulated with anti-CD3/anti-CD28 beads for 16 hours to stimulate CTLA-4 expression. Foxp3 and total CTLA-4 (BN13) staining are compared between unstimulated (top panels) or stimulated T cells (bottom panels). Cells were gated on CD45RA− memory CD4 T cells. CTLA-4 MFI (large font) is shown for total Foxp3+ cells (right) and Foxp3− cells (left). Percentages are shown in quadrants and Foxp3 MFI on Tregs (bottom right). (B) Relative CTLA-4 expression in healthy controls and individuals with CTLA-4 heterozygous mutations after stimulation. Relative expression is calculated as in Figure 1. (C) Foxp3+ percentage in stimulated CD4 T cells comparing CTLA-4 mutation (n = 14) and control (n = 22).
CTLA-4 deficiency persists after stimulation. (A) CD4 T cells were stimulated with anti-CD3/anti-CD28 beads for 16 hours to stimulate CTLA-4 expression. Foxp3 and total CTLA-4 (BN13) staining are compared between unstimulated (top panels) or stimulated T cells (bottom panels). Cells were gated on CD45RA− memory CD4 T cells. CTLA-4 MFI (large font) is shown for total Foxp3+ cells (right) and Foxp3− cells (left). Percentages are shown in quadrants and Foxp3 MFI on Tregs (bottom right). (B) Relative CTLA-4 expression in healthy controls and individuals with CTLA-4 heterozygous mutations after stimulation. Relative expression is calculated as in Figure 1. (C) Foxp3+ percentage in stimulated CD4 T cells comparing CTLA-4 mutation (n = 14) and control (n = 22).
T-cell stimulation upregulates both CTLA-4 and Foxp3 expression
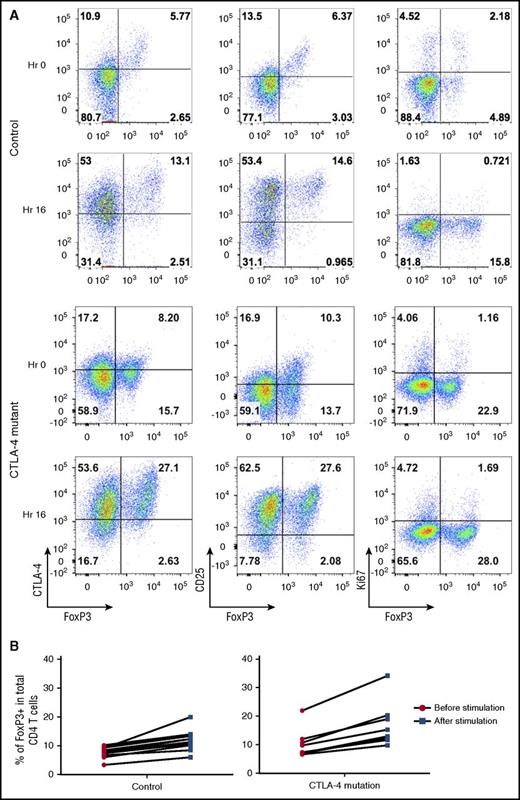
Following stimulation, the increase in the percentage of Foxp3+ cells was accompanied by upregulation of CD25 and CTLA-4 but occurred in the absence of increased proliferation as measured by Ki67 upregulation (Figure 3A). These data, along with the short time period of stimulation, indicated that the increase was not due to an outgrowth of induced Tregs. The fold increase in the percentage of Foxp3-expressing T cells was consistent between individuals and seen in both control and CTLA-4 mutation carriers (Figure 3B). Thus, we concluded that brief stimulation helped to enhance both Foxp3+ and CD25 staining and provided a more distinct population on which to base CTLA-4 analysis and to assess Treg percentages.
T-cell stimulation increases Treg detection by upregulating Foxp3 expression. (A) CD4 T cells were analyzed for Foxp3, CTLA-4, and CD25, Ki67 in a healthy control and a CTLA-4–deficient patient at 0 hours (Hr) and 16 hours after CD3/28 bead stimulation. (B) Percentage of Tregs before or after bead stimulation in controls and individuals with CTLA-4 mutations.
T-cell stimulation increases Treg detection by upregulating Foxp3 expression. (A) CD4 T cells were analyzed for Foxp3, CTLA-4, and CD25, Ki67 in a healthy control and a CTLA-4–deficient patient at 0 hours (Hr) and 16 hours after CD3/28 bead stimulation. (B) Percentage of Tregs before or after bead stimulation in controls and individuals with CTLA-4 mutations.
Assessing functional capacity in CTLA-4 deficiency
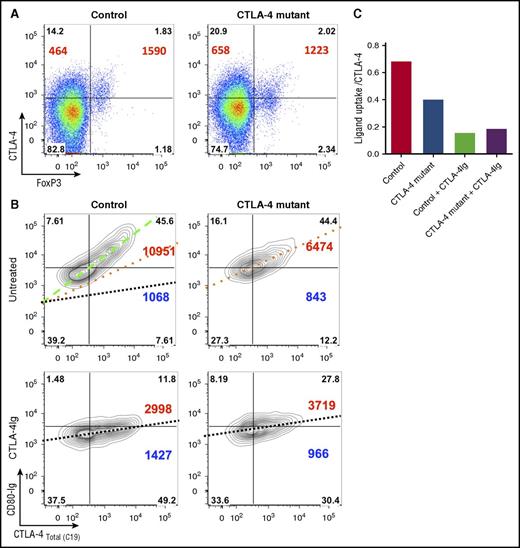
Although some mutations (eg, stop mutations) may cause true haploinsufficiency, missense mutations in CTLA-4 can have a range of effects which require further dissection. For example, some mutations may result in proteins that do not bind CTLA-4 antibodies, although others may have a limited impact on antibody binding but still affect the ability to bind ligands. As shown in Figure 4A, cells from an individual harboring a mutation in the CTLA-4 ligand-binding site revealed antibody staining similar to a healthy control. To account for such issues, we established an assay that measures soluble ligand uptake by CTLA-4 as a surrogate for normal ligand capture and effector function. Previously, we have used assays which rely on uptake of green fluorescent protein–tagged ligands from transfected cells, however, this requires specialized cellular reagents and is strongly influenced by cell numbers and cell-cell contact. We therefore developed an assay monitoring the uptake of soluble ligands by stimulated Tregs from patients carrying CTLA-4 mutations. Using this approach, ligand uptake at 37°C can be compared with the total amount of CTLA-4 protein per cell. Importantly, ligand uptake requires both effective CTLA-4 trafficking to the cell surface as well as ligand-binding capacity so the assay is therefore capable of probing a number of functional defects.
CTLA-4 ligand uptake reveals defects in patients with CTLA-4 deficiency. (A) Expression of Foxp3 and total CTLA-4 (BN13) on unstimulated CD45RA− memory CD4 T cells were compared between a ligand-binding mutant (P137R) and healthy control. CTLA-4 MFI in Foxp3+ and Foxp3− populations is shown in large font. Percentages are shown in quadrants. (B) Impaired ligand uptake by CTLA-4–deficient patient. CD4 T cells were stimulated with CD3/CD28 beads and gated on Foxp3+ cells. Total CTLA-4 staining (C19 antibody) is plotted against ligand uptake (CD80-Ig). Changes in slope reflect alterations in ligand uptake efficiency and are overlaid in control plot. CD80-Ig MFI (top right) and CTLA-4 MFI (bottom right) are shown in large font. (C) Comparison of the slope of the line of best fit from the data in panel B.
CTLA-4 ligand uptake reveals defects in patients with CTLA-4 deficiency. (A) Expression of Foxp3 and total CTLA-4 (BN13) on unstimulated CD45RA− memory CD4 T cells were compared between a ligand-binding mutant (P137R) and healthy control. CTLA-4 MFI in Foxp3+ and Foxp3− populations is shown in large font. Percentages are shown in quadrants. (B) Impaired ligand uptake by CTLA-4–deficient patient. CD4 T cells were stimulated with CD3/CD28 beads and gated on Foxp3+ cells. Total CTLA-4 staining (C19 antibody) is plotted against ligand uptake (CD80-Ig). Changes in slope reflect alterations in ligand uptake efficiency and are overlaid in control plot. CD80-Ig MFI (top right) and CTLA-4 MFI (bottom right) are shown in large font. (C) Comparison of the slope of the line of best fit from the data in panel B.
As shown in Figure 4B, in healthy controls the ability of CTLA-4 to capture ligands was proportional to its expression level. However, a much reduced slope was obtained with Tregs from a patient with a known ligand-binding defect (P137R). This indicated the presence of CTLA-4 protein that was impaired in its ligand capture ability. As a control, abatacept (CTLA-4-Ig) was used to block ligand uptake. Therefore, the decreased slope in these plots reflects lower ligand uptake per CTLA-4 molecule (Figure 4B). The quantification of this decrease in slope (CTLA-4 functional efficacy) is shown in Figure 4C, providing an integrated assessment of level of expression, the ability of CTLA-4 to traffic to the membrane and to bind its ligands. Thus, functionally significant mutations affecting the amount of CTLA-4, the quality of ligand binding, or its trafficking can be detected using this assay.
Distinguishing CTLA-4 mutations from LRBA deficiency
Recently, mutations in the protein LRBA have been shown to impact on CTLA-4 expression. In LRBA deficiency, CTLA-4 is synthesized normally, but appears aberrantly trafficked, resulting in enhanced degradation in lysosomes. Because both CTLA-4 and LRBA mutations result in reduced CTLA-4 expression, we attempted to distinguish between these conditions.
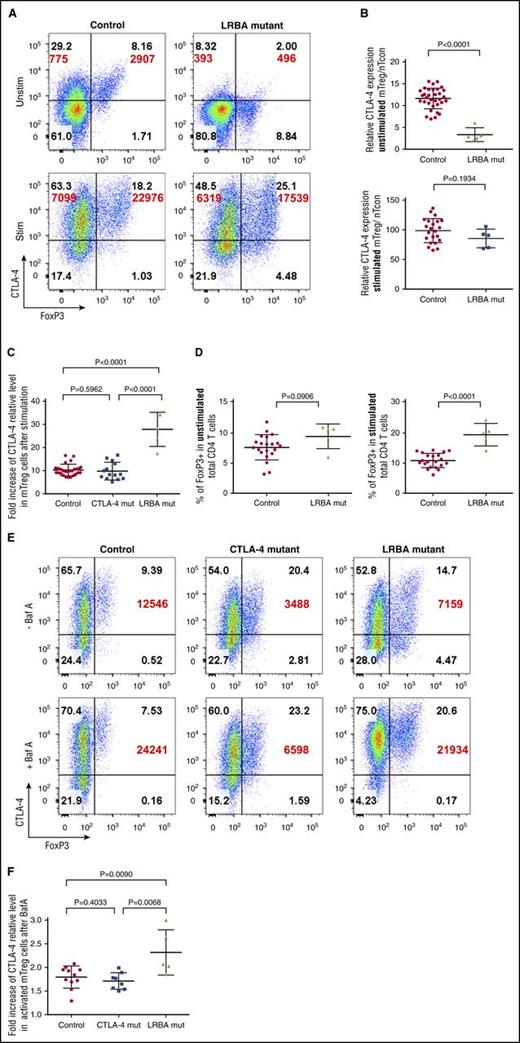
As shown in Figure 5A-B, levels of CTLA-4 in LRBA-deficient mTregs were even lower than those bearing CTLA-4 mutations. However, in contrast to T cells from CTLA-4–deficient individuals (see Figure 2B), in response to stimulation, the levels of CTLA-4 expression seen in stimulated LRBA T cells recovered to levels similar to controls (Figure 5B), representing a 20- to 30-fold upregulation from baseline (Figure 5C). Thus, following anti-CD3/anti-CD28 stimulation, CTLA-4 gene expression in LRBA patients results in strong induction of CTLA-4 and a higher fold change from baseline levels.
LRBA deficiency and CTLA-4 deficiency have different patterns of expression. (A) Representative expression of Foxp3 and total CTLA-4 (BN13) on unstimulated (Unstim) and stimulated (Stim) memory CD4 T cells from control or LRBA mutations. CD4 T cells were stimulated with CD3/CD28 beads; CTLA-4 MFI is shown in large font. (B) Relative expression of CTLA-4 in healthy controls and LRBA-deficient patients (n = 5) in unstimulated or stimulated conditions. Relative expression is calculated as the fold change in CTLA-4 MFI between nTcon and mTreg. (C) Fold increase in CTLA-4 MFI between Foxp3+ memory CD4 T cells before and after stimulation with CD3/CD28 beads. (D) Foxp3+ Treg percentage in unstimulated or stimulated LRBA-deficient and control CD4 T cells. (E) CD4 T cells were stimulated with CD3/CD28 beads in the presence of absence of BafA and stained for Foxp3 and total CTLA-4 (BN13) expression. CTLA-4 MFI in Foxp3+ mTregs is shown in large font. (F) Collated BafA data for healthy controls, CTLA-4, or LRBA mutations. Fold increase is the change in CTLA-4 MFI in mTregs before and after BafA treatment.
LRBA deficiency and CTLA-4 deficiency have different patterns of expression. (A) Representative expression of Foxp3 and total CTLA-4 (BN13) on unstimulated (Unstim) and stimulated (Stim) memory CD4 T cells from control or LRBA mutations. CD4 T cells were stimulated with CD3/CD28 beads; CTLA-4 MFI is shown in large font. (B) Relative expression of CTLA-4 in healthy controls and LRBA-deficient patients (n = 5) in unstimulated or stimulated conditions. Relative expression is calculated as the fold change in CTLA-4 MFI between nTcon and mTreg. (C) Fold increase in CTLA-4 MFI between Foxp3+ memory CD4 T cells before and after stimulation with CD3/CD28 beads. (D) Foxp3+ Treg percentage in unstimulated or stimulated LRBA-deficient and control CD4 T cells. (E) CD4 T cells were stimulated with CD3/CD28 beads in the presence of absence of BafA and stained for Foxp3 and total CTLA-4 (BN13) expression. CTLA-4 MFI in Foxp3+ mTregs is shown in large font. (F) Collated BafA data for healthy controls, CTLA-4, or LRBA mutations. Fold increase is the change in CTLA-4 MFI in mTregs before and after BafA treatment.
In addition, we also noted that although the percentage Foxp3+ as a fraction of CD4+ T cells was not obviously different between unstimulated LRBA samples and controls, brief stimulation revealed significantly higher Treg percentages in LRBA patients (Figure 5D), suggesting that stimulation preferentially helps detect Tregs in conditions associated with CTLA-4 deficiency. This expanded Treg compartment is highly consistent with the known impact of CTLA-4 deficiency on Treg homeostasis in mice.20,21,26
Because in LRBA deficiency CTLA-4 protein is incorrectly trafficked to lysosomes, we also assessed CTLA-4 expression in the presence of BafA to prevent lysosomal degradation. As shown in Figure 5E, both control individuals and those carrying CTLA-4 mutations showed a 1.5-fold to twofold increase in CTLA-4 in response to BafA. In contrast, in patients with LRBA deficiency, T cells stimulated in the presence of BafA displayed between a twofold and threefold increase in CTLA-4 expression and recovered expression to levels similar to control values (Figure 5F). Thus, although there was variation between individuals, increased responses to stimulation and enhanced BafA sensitivity appear to be useful in distinguishing between low CTLA-4 expression due to genetic CTLA-4 deficiency and that as a result of aberrant handling of CTLA-4 due to LRBA deficiency.
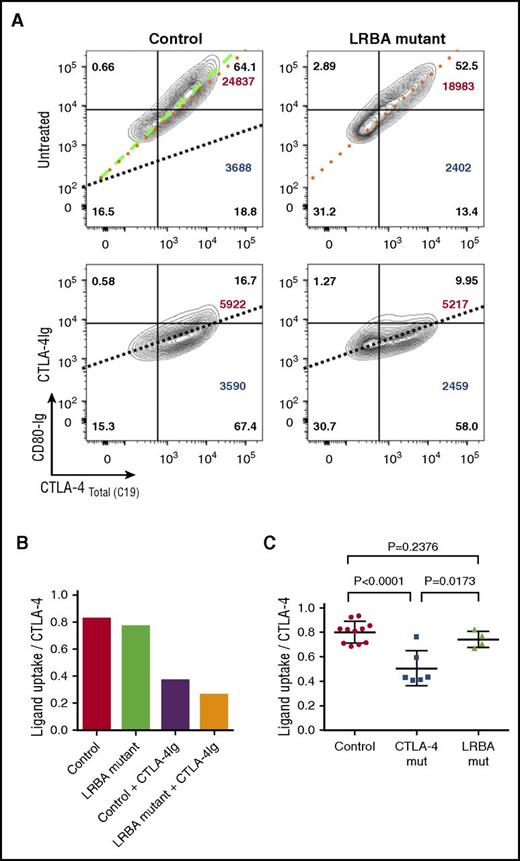
Finally, we also compared ligand uptake in patients with LRBA mutations, using CD80-Ig. In keeping with the fact that CTLA-4 expression is reasonably well corrected by transient stimulation and the CTLA-4 is qualitatively normal, we observed that the slope of ligand binding against CTLA-4 expression in stimulated cells was very similar to controls (Figure 6A-B). Thus, in patients with LRBA mutations, ligand uptake efficiency is much less affected in comparison with CTLA-4 mutations and may be useful in distinguishing LRBA from CTLA-4 defects (Figure 6C).
Ligand uptake is relatively unaffected in LRBA-deficient patients. (A) CD4 T cells were stimulated with CD3/CD28 beads and total CTLA-4 (C19) plotted against CD80-Ig uptake gating on CD4+ memory Tregs. Slope of the line represents efficiency of CD80 uptake. Dotted lines are overlaid in the control plot (top left) for comparison. CTLA-4-Ig treatment (bottom panels) provides a negative control by blocking ligand uptake. CD80-Ig MFI is shown in large font (top right) and CTLA-4 MFI in large font (bottom right). Percentages are shown in all quadrants. (B) Graph is generated using the slope of the line of best fit from the data in panel A. (C) Collated ligand uptake efficiency data are shown for CTLA-4 and LRBA mutations.
Ligand uptake is relatively unaffected in LRBA-deficient patients. (A) CD4 T cells were stimulated with CD3/CD28 beads and total CTLA-4 (C19) plotted against CD80-Ig uptake gating on CD4+ memory Tregs. Slope of the line represents efficiency of CD80 uptake. Dotted lines are overlaid in the control plot (top left) for comparison. CTLA-4-Ig treatment (bottom panels) provides a negative control by blocking ligand uptake. CD80-Ig MFI is shown in large font (top right) and CTLA-4 MFI in large font (bottom right). Percentages are shown in all quadrants. (B) Graph is generated using the slope of the line of best fit from the data in panel A. (C) Collated ligand uptake efficiency data are shown for CTLA-4 and LRBA mutations.
Discussion
CTLA-4 deficiency is a rare autosomal-dominant disorder identified in patients with CVID with a range of autoimmune complications.5,6 In contrast, LRBA deficiency is a recessive disorder where biallelic mutations result in aberrant trafficking of proteins including CTLA-4,7 resulting in an earlier onset but phenotypically similar disease.9,27 In order to understand the impact of different CTLA-4 and LRBA mutations, we have probed a number of aspects of CTLA-4 biology. These include the level of detectable protein expression within T-cell subsets and the assessment of protein trafficking coupled to the ability to interact with natural ligands. Together, these approaches can be used to estimate the functional capacity of CTLA-4, without the need for specialized reagents or complex assays.
Using these approaches, we identified characteristic features relating to both CTLA-4 and LRBA deficiency. The most robust estimate of CTLA-4 deficiency resulted from comparison of total CTLA-4 levels in mTregs with CTLA-4 levels in CD4+ nTcons in the same individual. Because nTcons express little or no CTLA-4, this provides a reliable internal control with which to compare Treg expression of CTLA-4. This reveals differences in level of expression in healthy mTregs, which on average are ∼10-fold those of nTcons. In CTLA-4 haploinsufficient patients, this difference is reduced to fivefold or less and in LRBA patients approximately threefold. In general, LRBA deficiency resulted in lower levels of CTLA-4 compared with CTLA-4 heterozygous mutations, which may contribute to its generally earlier disease onset. Although this approach to CTLA-4 staining is generally adequate, the extent of reduced CTLA-4 staining can be mutation dependent. Ultimately, not all mutations in CTLA-4 will affect antibody staining and therefore be revealed by a simple staining approach. For example, a mutation in the CTLA-4 ligand-binding site, gave limited differences in CTLA-4 antibody staining when compared with control. Therefore, in cases where there is no obvious deficit in total CTLA-4 it is important to consider defects in CTLA-4 trafficking and ligand binding. Accordingly, the P137R mutation, which occurs within the well-described CTLA-4 ligand-binding site,28 displayed defects in soluble ligand uptake in our assays.
It is increasingly clear that a major aspect of CTLA-4 function relates to Treg biology and the ability of CTLA-4 to compete for CD28 ligands.16,29,30 The ability of CTLA-4 to physically capture its ligands via transendocytosis22 from antigen-presenting cells is predictive of CTLA-4 function on Tregs.11 Here, we used a simplified ligand uptake assay, which uses soluble CD80-Ig, to test the key features of CTLA-4 function, namely ligand binding and CTLA-4 trafficking. We have previously shown that uptake of antibodies and ligands by CTLA-4 at 37°C is a convenient measure of CTLA-4 trafficking.25 By gating on Foxp3+ cells, this provides an estimate of CTLA-4 function in Tregs. Although direct studies of CTLA-4–dependent Treg suppression are functionally relevant, in reality, accurate measurement is technically difficult, requiring large numbers of T cells to generate meaningful data.5 The popular surrogate of measuring Treg suppression using anti-CD3/anti-CD28 bead stimulation does not measure CTLA-4–dependent suppressive function in our view.11 Accordingly, the assays outlined here represent a compromise, allowing testing of the largely agreed requirements for CTLA-4 function, that is, level of expression, inducibility, trafficking, and ligand binding. Importantly, all of these assays can be carried out using standard flow cytometric approaches, using commercially available reagents and can therefore be readily adopted.
Some studies have suggested that increased Tcon cell proliferation or inability to control interleukin 2 production may result from CTLA-4 mutation or deficiency.6,31,32 We have been repeatedly unable to show any intrinsic effects of CTLA-4 deficiency on CD4 T-cell responses in the absence of Treg5 and are likewise unable to demonstrate an effect of anti-CTLA-4 blockade on proliferation of CD4 Tcon, suggesting they are not subject to intrinsic CTLA-4 regulation.11 We would urge caution in using CD4 T-cell proliferation as a measure of CTLA-4 defects because there is abundant literature showing that CTLA-4 has little intrinsic ability to directly affect these aspects of T-cell function.15,33 In contrast, the cell-extrinsic (regulatory) function of CTLA-4 is borne out by numerous studies.34-36
In the present study, we did not identify deficits in Treg numbers associated with CTLA-4 or LRBA deficiency and observed that brief stimulation was a useful tool for confirming Foxp3 expression. Studies by Sakaguchi and colleagues have shown that both Foxp3hi and Foxp3lo cells exist in human blood.37 Because Foxp3 expression is influenced by levels of CD25 expression, factors such as interleukin 2 consumption, CD4 lymphopenia, and immunosuppressive treatments may all affect Foxp3. Thus, although Foxp3 and CD25 expression may be decreased in CTLA-4 and LRBA deficiency, this may not indicate low Treg numbers per se. Indeed, it was reported in IPEX patients that numbers of natural Tregs were underestimated due to decreased CD25 staining.38 Charbonnier et al also recently reported decreased Treg frequencies associated with LRBA deficiency.39 Although we did not observe this in our study, we noted that short in vitro stimulation increased the percentage of Foxp3+ cells without inducing their proliferation. This effect was particularly obvious in LRBA patients. Brief stimulation may therefore help to reveal Tregs, which may otherwise have low expression of critical markers such as CD25 and Foxp3 resulting in underestimates. Our findings of increased Tregs are consistent with the fact that in mice, CTLA-4 deficiency clearly promotes expansion of the Treg compartment.26 Such an expansion might therefore be expected in LRBA deficiency where CTLA-4 levels are typically very low. Treg expansion is also seen in patients with CTLA-4 heterozygous mutations, however, this occurs only in some individuals and may therefore be mutation specific. It is also important to note that the induction of Foxp3 can occur in Tcons40 ; therefore, determining whether stimulated T cells expressing Foxp3 are natural Tregs is complex. It is likely that analysis of the methylation status of the Foxp3 locus will be informative in this situation.41 Nonetheless, from a functional perspective it is clear that expression of CTLA-4 itself is sufficient to confer suppressive activity in both Tcons as well as Tregs,42-44 indicating the Foxp3+ CTLA-4+ T cells we observe after stimulation are nonetheless likely to be suppressive.
One feature that appears to distinguish LRBA from CTLA-4 deficiency is the upregulation of CTLA-4 in response to stimulation. Upregulation of CTLA-4 was higher in patients with LRBA mutations, consistent with the fact that there is no defect in CTLA-4 itself and synthesis is likely to be normal. Thus, during acute stimulation, the ability to synthesize new CTLA-4 appears to outweigh any enhanced destruction due to the LRBA defect. This results in significant recovery of CTLA-4 from a very low baseline, providing a useful indicator of LRBA deficiency. In addition, the response of cells to BafA, which inhibits lysosomal degradation, gave a more significant enhancement of CTLA-4 staining in the case of LRBA mutations. This was clearer in some LRBA individuals than others and it will be interesting to determine whether the effect of BafA depends on particular LRBA mutations. Finally, functional efficacy of CTLA-4 proteins as measured by the slope of ligand uptake displayed very limited difference from controls, again showing that ligand capture is broadly unimpaired in LRBA deficiency. Taken together, the high fold increase in response to stimulation, enhanced response to BafA, and unimpaired ligand capture appear to be characteristics that distinguish LRBA from direct CTLA-4 mutations.
In summary, CTLA-4 expressed by Tregs acts as a major mechanism to control self-reactive T cells by regulating CD28 ligand availability. The approaches described here can be used to identify functional deficits in the CTLA-4 pathway.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the National Institute for Health Research (NIHR) Rare Diseases Translational Research Collaboration via the NIHR University College London Hospitals Biomedical Research Centre. T.Z.H. was funded by NIHR. B.R. and B.S. were funded by the Biotechnology and Biological Sciences Research Council (BBSRC). N.H. was funded by a Wellcome Trust Clinical PhD Studentship. W.Q. was supported by the NIHR and Great Ormond St. Biomedical Research Centre.
Authorship
Contribution: T.Z.H. developed methods, performed experiments, analyzed data, and helped write the manuscript; N.V. and J.W. performed experiments; B.S., A.K., D.J., N.H., and B.R. contributed to the experimental design, developed methods, and helped with data interpretation; A.W., W.Q., H.B., S.S., O.N., P.O., S.H., and P.D.A. provided clinical samples and contributed to data interpretation; H.S., L.S.K.W., and S.O.B. supervised the experiments, contributed to data analysis, and cowrote the paper; and D.M.S. conceived of the experiments, supervised the project, interpreted results, and wrote the paper.
Conflict-of-interest disclosure: H.S. is a consultant and shareholder in Cell Medica. The remaining authors declare no competing financial interests.
Correspondence: David M. Sansom, Department of Immunology, Institute of Immunity and Transplantation, University College London, Royal Free Hospital, Rowland Hill St, London NW3 2PF, United Kingdom; e-mail: d.sansom@ucl.ac.uk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal