Key Points
Output from individual rhesus macaque hematopoietic stem and progenitor cells is stable for years, with little evidence of clonal succession.
Individual clones may display stable myeloid or lymphoid bias for many years.
Abstract
Autologous transplantation of hematopoietic stem and progenitor cells lentivirally labeled with unique oligonucleotide barcodes flanked by sequencing primer targets enables quantitative assessment of the self-renewal and differentiation patterns of these cells in a myeloablative rhesus macaque model. Compared with other approaches to clonal tracking, this approach is highly quantitative and reproducible. We documented stable multipotent long-term hematopoietic clonal output of monocytes, granulocytes, B cells, and T cells from a polyclonal pool of hematopoietic stem and progenitor cells in 4 macaques observed for up to 49 months posttransplantation. A broad range of clonal behaviors characterized by contribution level and biases toward certain cell types were extremely stable over time. Correlations between granulocyte and monocyte clonalities were greatest, followed by correlations between these cell types and B cells. We also detected quantitative expansion of T cell–biased clones consistent with an adaptive immune response. In contrast to recent data from a nonquantitative murine model, there was little evidence for clonal succession after initial hematopoietic reconstitution. These findings have important implications for human hematopoiesis, given the similarities between macaque and human physiologies.
Introduction
The pathways by which functional blood-cell heterogeneity is developed and maintained are important to understanding leukemogenesis and hematopoietic responses to stress, aging, or marrow toxic drugs and to improving the efficacy and safety of hematopoietic stem-cell (HSC) transplantation and gene therapies. Developmental hierarchies connecting self-renewing long-term repopulating HSCs to terminally differentiated daughter cells have been mapped over the past 3 decades based on murine transplantation and both murine and human in vitro assays.1,2 Associating hematopoietic potential and lifespan with cell-surface protein expression through limit dilution in vitro differentiations, human-murine xenografts, or murine autologous transplants has enabled construction of a proposed hematopoietic tree with self-renewing HSCs giving rise to a variety of transient and cell type–restricted progenitors.3-7 Although these assays provide important information regarding what rare cell populations can do under extreme replicative stress, the extrapolation of conclusions to steady-state human hematopoiesis or non–dose-limited transplantation may not be straightforward.8,9 In particular, the generation of consistently myeloid- or lymphoid-biased daughter-cell populations in serial transplantation of single stem cells indicates that surface protein expression is not yet sufficient for delineation of HSC behavior, and unknown, possibly epigenetic, factors have an impact on HSC and progenitor-cell (HSPC) output.10,11 Significant differences between humans and small rodents in terms of HSPC phenotype, lifelong hematopoietic demand, cytokine utilization, and marrow niche characteristics also limit extrapolation of steady-state or posttransplantation human HSPC behavior from in vitro, xenograft, and murine transplantation models.2,12-14
As an alternative approach, we and others have made use of clonal labeling strategies, which enable detection of the progeny of individual, labeled HSPCs in diverse hematopoietic cell types over time in a clinically relevant, non–limit dilution setting.15,16 These experiments, which have their origin in proviral integration site analysis via Southern blot after retroviral transduction of HSPC in mice, enable both identification of proportional biases in HSPC output from various HSPC classes and inference of the rates at which cellular output from individual progenitors appears, expands, and exhausts.17 Although low HSPC survival rates and likely perturbation of HSPC behavior after transduction with oncogenic murine retroviral vectors have limited the applicability of older results,18 both initial and subsequent murine studies using modern transduction and labeling methods have generally matched limit-dilution results, with initial engraftment from non–self-renewing progenitors being followed by more stable long-term engraftment from multipotent HSPCs. HSPC tracking via vector insertion site (VIS) retrieval also now exists for humans, both from xenograft models19 and from patients enrolled in pioneering gene therapy trials, with caveats for clonal skewing and a high risk for development of leukemia in older trials.20 VIS retrieval from patients enrolled in more recent trials utilizing less genotoxic lentiviral vectors has shown persistence of diverse clonal repertoires, but VIS retrieval is semiquantitative at best; underlying disease state and prior treatment of these patients may affect HSPC behavior, and repeated sampling of blood and marrow is limited by clinical and ethical restrictions.21-24
We used the rhesus macaque autologous transplantation model to interrogate in vivo HSPC clonal behavior, given the close phylogenetic similarity and shared HPSC characteristics with humans.25,26 High-throughput sequencing of lentiviral vectors containing high-diversity genetic barcodes from macaques receiving HSPC transplants probabilistically guaranteed to contain a unique clone-labeling barcode sequence flanked by polymerase chain reaction primer sites avoids amplification across the variable provirus-host genomic boundary and results in a more reliable correspondence between the normalized number of barcode sequencing reads generated and the abundance of cells to which they are associated, thereby enabling accurate quantitative assessment of HSPC outputs and biases.15,16,27 We previously reported novel findings regarding lineage relationships during initial hematopoietic reconstitution after transplantation in this model.15 In the current study, we analyzed clonal behavior for up to 49 months posttransplantation. We observed the gradual stabilization of hematopoietic output from individual clones, characterized by the emergence and increasing prevalence of cells generated from stable multipotent progenitors by 12 months posttransplantation. We found no evidence for a significant degree of clonal succession after stabilization, at least for the clones constituting the vast majority of hematopoiesis that can be reliably sampled and detected using our methodologies. The diverse and stable biases of these long-lived progenitors, including T cell–biased, myeloid-biased, B cell– and myeloid-biased, and multipotent HPSCs, indicate that the range of transplanted CD34+ HSPC fates posttransplantation is extremely broad. The information provided by our approach for the first time conclusively demonstrates stability of clonal output over time, even in the context of a highly complex and diverse population of clones in a large animal or human setting.
Methods
Autologous rhesus macaque model
All procedures were approved by the National Heart, Lung and Blood Institute Animal Care and Use Committee. Barcode library preparation and CD34+ cell collection, transduction, and transplantation for animals were as previously described15 and are summarized in Figure 1, along with information on 2 additional animals included in the current study. CD34+ HSPCs were transduced with green fluorescent protein–labeled lentiviral vector libraries carrying high-diversity oligonucleotide barcodes and reinfused into the autologous animal after pretransplantation conditioning with 1000 rads total-body irradiation.28 We previously verified that the majority of transduced HSPCs in these animals contained only a single barcode and that the barcoded vector libraries used had sufficient diversity to ensure that each individual barcode marked only a single engrafting HSPC and its progeny.15
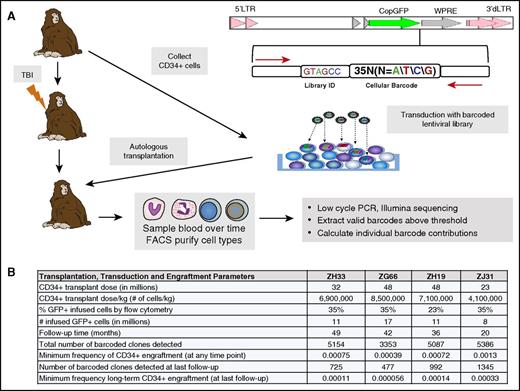
Rhesus macaque autologous transplantation and hematopoietic barcoding. (A) Experimental summary. The replication-incompetent HIV-derived lentiviral barcoding vector used is diagrammed at the top right. The barcode consists of a 6–base pair library identification (ID) followed by a 35–base pair high-diversity cellular barcode. This vector was used to transduce rhesus macaque CD34+ cells, and these cells were reinfused after myeloablative total-body irradiation (TBI; 1000 rads) of the autologous recipient. Purified blood cells from various lineages underwent low-cycle polymerase chain reaction (PCR) amplification utilizing primers bracketing the barcode (red arrows in diagram), followed by Illumina sequencing and data processing, as described in supplemental Data. (B) Transplantation and engraftment parameters. The table summarizes CD34+ cell collection, transduction, and transplantation parameters, as well as total clone numbers and clone frequencies for each animal, after applying the threshold of a clone contributing at least 0.05% to at least 1 cell type at a minimum of at least 1 time point. GFP, green fluorescent protein; FACS, fluorescence-activated cell sorting; LTR, long terminal repeat; WPRE, woodchuck hepatitis posttranscriptional regulatory element.
Rhesus macaque autologous transplantation and hematopoietic barcoding. (A) Experimental summary. The replication-incompetent HIV-derived lentiviral barcoding vector used is diagrammed at the top right. The barcode consists of a 6–base pair library identification (ID) followed by a 35–base pair high-diversity cellular barcode. This vector was used to transduce rhesus macaque CD34+ cells, and these cells were reinfused after myeloablative total-body irradiation (TBI; 1000 rads) of the autologous recipient. Purified blood cells from various lineages underwent low-cycle polymerase chain reaction (PCR) amplification utilizing primers bracketing the barcode (red arrows in diagram), followed by Illumina sequencing and data processing, as described in supplemental Data. (B) Transplantation and engraftment parameters. The table summarizes CD34+ cell collection, transduction, and transplantation parameters, as well as total clone numbers and clone frequencies for each animal, after applying the threshold of a clone contributing at least 0.05% to at least 1 cell type at a minimum of at least 1 time point. GFP, green fluorescent protein; FACS, fluorescence-activated cell sorting; LTR, long terminal repeat; WPRE, woodchuck hepatitis posttranscriptional regulatory element.
Cell purification and barcode retrieval
Rhesus macaque peripheral blood cells were isolated via density gradient separation and sorted after staining with CD3, CD20, CD14, and CD33 antibodies to detect T, B, monocyte, and granulocyte cells, respectively, as listed in supplemental Table 1 (available on the Blood Web site; BD FACSAriaII, BD Biosciences, San Jose, CA) and as described.15 DNA was extracted with the DNeasy Blood and Tissue Kit (Qiagen, Germantown, MD); 200 to 500 ng of sample DNA underwent low-cycle polymerase chain reaction with primers bracketing the barcode and Illumina HiSeq2000 sequencing as described.15
Data processing and analysis
An explanation of the methodology used for the processing and analysis of sequencing files is included in the supplemental data, with links to raw data and custom Python and R code. In addition, we used R packages pheatmap, DiversitySampler, RColorBrewer, nnet, foreach, stringr, biclust, and scales. Validation of sequence output processing and application of a conservative barcode detection threshold are given in supplemental Figures 1 and 2.
Results
Marking level and clonal diversity
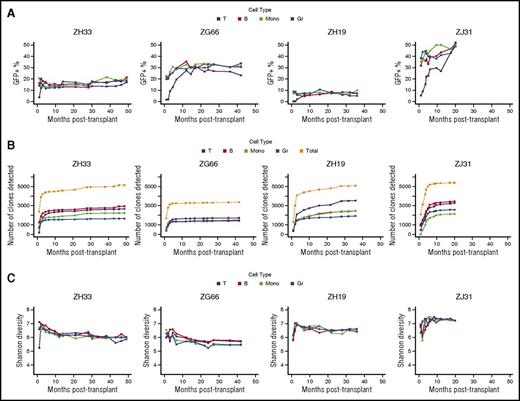
We quantitatively assessed individual CD34+ HSPC contributions to T, B, monocyte, and granulocyte cells in 4 rhesus macaques at time points up to 49 months after lentivirally-barcoded autologous CD34+ transplantation (Figure 1A). All animals received doses of transduced CD34+ cells within a relatively narrow range (11 to 17 million), with similar overall transduction efficiencies as assessed by green fluorescent protein positivity at the end of transduction (Figure 1B). Myeloid- and B-cell marking levels were stable as soon as 1 month post-transplantation, although barcoded T-cell production lagged behind that of the other cell types and only reached equilibrium at 5 to 17 months (Figure 2A). Animal-to-animal variability in marking level likely reflects variance in recovery of endogenous hematopoiesis after high-dose, but not completely myeloablative, irradiation. We detected stable and consistent clone numbers over time, with the number of total HSPC-contributing clones plateauing by 4 to 6 months in all animals (Figure 2B). Although the large number of clones narrowly exceeding the applied sampling/sequencing error threshold of 0.05% of marked hematopoietic production indicates that this approach likely excludes real, but low, contributing clones that cannot be reliably detected (Figure 2B; supplemental Figure 2E), any real excluded clones are very low contributors and represent a relatively low fraction of total hematopoiesis. The application of this threshold affects clone number and sets a lower limit for the true number of repopulating clones. Other research groups utilizing barcoding and a variety of analytic approaches have also documented and discussed the technical issues inherent in analyzing very low-frequency barcodes.27,29 However, the relationship between clonal abundance and retrieved barcode read number for clones above such thresholds has been shown to be linear and reproducible by our group and others.15,27,29
Longitudinal vector marking and overall clonal diversity. (A) Marking levels. The percentage of peripheral blood cells positive for the barcode/green fluorescent protein (GFP) vector is shown for each hematopoietic cell lineage over time for animals ZH33, ZG66, ZH19, and ZJ31. T cells (T), black; B cells (B), red; monocytes (Mono), green; granulocytes (Gr), dark blue. (B) Cumulative detected clone numbers. The cumulative number of clones over time contributing above the threshold at a minimum of 1 time point is shown for individual lineages and overall. The rapid increase in number of detected clones after engraftment corresponds to initial posttransplantation hematopoietic reconstitution with ≥1 waves of transient clones and emergence of long-term repopulating clones. The flat areas subsequent on the curves indicate broad clonal persistence and lack of emergence of new clones after initial posttransplantation reconstitution with long-term repopulating clones. These plateaus imply that capture of clones is substantially complete after several early time points. The cumulative numbers of clones detected within each individual cell type are color coded. The overall numbers of cumulatively detected clones in all cell types are plotted in gray. (C) Overall clonal diversity. Shannon entropy as a measure of diversity depends on both the number of detected clones and the distribution of their sizes. Given a number of detected clones, higher diversity corresponds to a more even distribution of sizes. Here we show that diversity is high, similar among animals, constant among cell types, and stable after initial reconstitution.
Longitudinal vector marking and overall clonal diversity. (A) Marking levels. The percentage of peripheral blood cells positive for the barcode/green fluorescent protein (GFP) vector is shown for each hematopoietic cell lineage over time for animals ZH33, ZG66, ZH19, and ZJ31. T cells (T), black; B cells (B), red; monocytes (Mono), green; granulocytes (Gr), dark blue. (B) Cumulative detected clone numbers. The cumulative number of clones over time contributing above the threshold at a minimum of 1 time point is shown for individual lineages and overall. The rapid increase in number of detected clones after engraftment corresponds to initial posttransplantation hematopoietic reconstitution with ≥1 waves of transient clones and emergence of long-term repopulating clones. The flat areas subsequent on the curves indicate broad clonal persistence and lack of emergence of new clones after initial posttransplantation reconstitution with long-term repopulating clones. These plateaus imply that capture of clones is substantially complete after several early time points. The cumulative numbers of clones detected within each individual cell type are color coded. The overall numbers of cumulatively detected clones in all cell types are plotted in gray. (C) Overall clonal diversity. Shannon entropy as a measure of diversity depends on both the number of detected clones and the distribution of their sizes. Given a number of detected clones, higher diversity corresponds to a more even distribution of sizes. Here we show that diversity is high, similar among animals, constant among cell types, and stable after initial reconstitution.
Similar clone numbers found in all animals, even those such as ZH19 and ZG66, which have large differences in marking level, may reflect both similar starting cell doses and similar transduction efficiencies, as well as the fact that clones in ZH19, which contribute to barcode-marked hematopoiesis at the rate of more abundant clones in ZG66, are actually smaller in absolute terms, and clones with absolute abundance that would be below the limit of detection in ZG66 may be above the threshold in ZH19. To incorporate information on both the number of clones and the distribution of their sizes in a manner that was less affected than overall clone number with respect to application of a threshold (supplemental Figure 2B-C), we calculated the Shannon diversity of recovered barcodes in each sample (Figure 2C). ZH33, ZG66, and ZH19 all had similar degrees of diversity and cumulative retrieved clone numbers (supplemental Figure 3). The relatively higher diversity of ZJ31 was not reflected by a larger postthreshold clone number but rather in its larger number of relatively high-contributing clones (supplemental Figure 2A), underscoring the presence of a quantitative notion of clonal diversity in stem-cell behavior.
Hematopoietic clonality
The clonality of a sample—the quantitative contribution of various HSPCs to that sample—can be assessed using a variety of statistical methods. As shown in our previous report, low Pearson correlations between cell-type clonalities immediately after transplantation indicate that hematopoiesis during this period is primarily supported by cell type–restricted clones.15 Over time, correlations between cell types increased because of the proliferation of multipotent clones. The highest correlations were between monocyte and granulocyte clonalities, and these stabilized within 3 months after transplantation in all animals (Figure 3A; supplemental Figure 4B). Although correlations of myeloid clonalities with B cells were only slightly lower than intramyeloid correlations, this gap was stable and reproducible in multiple animals, suggesting that these cell types are ontogenically less similar. Correlations between B-cell/myeloid (granulocyte and monocyte) clonalities and T-cell clonalities lagged behind intra–B-cell/myeloid correlations temporally and stabilized concurrently with T-cell reconstitution from barcoded cells. T-cell clonality was least correlated with other cell types and declined over time in ZH33.
Similarities between clonal contributions to hematopoietic cell types. (A) Pearson correlation measures similarity between the sets of clones from which specific cell populations are descended. Each line gives the Pearson correlations over time between samples from 2 cell types, with each line consisting of 2 colors corresponding to the cell types being compared (T cells [T], black; B cells [B], magenta; monocytes [Mono], green; granulocytes [Gr], light blue). Mono-Gr clonalities are most similar, followed by Gr-B or Mono-B and Gr/B-T or Mono/B-T. These similarities generally stabilize, with greatest volatility in correlation comparisons involving T cells. (B) Heat map showing the natural log fractional abundances of the highest contributing clones in ZH33 over time, defined as the set of all barcodes present as a top 10 highest contributing barcode in ≥1 of the samples shown. Each row corresponds to 1 barcode. The barcodes are organized by unsupervised hierarchical clustering using the Euclidean distance between barcodes’ log fractional abundances, with relative contribution shown as a magenta-to-light blue gradient, representing high contribution to no contribution, respectively. Clones are clustered along the y-axis to place similar clones next to each other. *Indicates the top 10 clones in a given sample; thus, there are 10 in each column and ≥1 in each row. Note transient clones contribute to each lineage at 1 month and then disappear, replaced by multipotent clones beginning at months 2 to 3, with contributions persisting long term.
Similarities between clonal contributions to hematopoietic cell types. (A) Pearson correlation measures similarity between the sets of clones from which specific cell populations are descended. Each line gives the Pearson correlations over time between samples from 2 cell types, with each line consisting of 2 colors corresponding to the cell types being compared (T cells [T], black; B cells [B], magenta; monocytes [Mono], green; granulocytes [Gr], light blue). Mono-Gr clonalities are most similar, followed by Gr-B or Mono-B and Gr/B-T or Mono/B-T. These similarities generally stabilize, with greatest volatility in correlation comparisons involving T cells. (B) Heat map showing the natural log fractional abundances of the highest contributing clones in ZH33 over time, defined as the set of all barcodes present as a top 10 highest contributing barcode in ≥1 of the samples shown. Each row corresponds to 1 barcode. The barcodes are organized by unsupervised hierarchical clustering using the Euclidean distance between barcodes’ log fractional abundances, with relative contribution shown as a magenta-to-light blue gradient, representing high contribution to no contribution, respectively. Clones are clustered along the y-axis to place similar clones next to each other. *Indicates the top 10 clones in a given sample; thus, there are 10 in each column and ≥1 in each row. Note transient clones contribute to each lineage at 1 month and then disappear, replaced by multipotent clones beginning at months 2 to 3, with contributions persisting long term.
Although hematopoietic output is supported by hundreds or thousands of clones at any one time, Pearson correlation is sensitive to a handful of high-contributing biased clones. Although multipotent clones, defined as those contributing to all cell lineages, were only 27%, 43%, 33%, and 36% of detected clones at the longest follow-up, they contributed 79%, 95%, 72%, and 67% to granulocyte marking at this time point in ZH33, ZG66, ZH19, and ZJ31, respectively. These large clones are biologically important because they contribute a disproportionate amount to hematopoiesis (supplemental Figures 3 and 5). Furthermore, their size reduces susceptibility to sequencing or sampling errors or low-level contamination between sorted cell types. To visualize the clonal behaviors that lead to the observed correlations, we tracked the hematopoietic contributions of the 10 largest clones in T cells, B cells, monocytes, and granulocytes and ordered them to group clones with similar behavior. Within the range of observed behaviors, certain categories of long-term HSPC clonal outputs were evident. Waves of short-lived cell type–restricted clones predominated immediately after transplantation, before being replaced by long-lived multipotent clones at 2 to 4 months (Figure 3B). We observed similar polyclonal stable multipotent output from these high-contributing clones long term in the other 3 animals (supplemental Figures 4 and 6). The differences in the number of early transient repopulating generations of clones between animals, with only 1 transient group of clones detected in ZH33 versus 2 waves in the other animals, suggest that additional transitory generations of early repopulating clones may be contributing between the early time points that we sampled. In ZH33 and, to a lesser extent, in other animals, we observed expansions of contributions from oligoclonal sets of highly T cell–biased clones at time points later than 9 months, consistent with effector memory clonal T-cell expansions in response to environmental cues such as viruses (Figure 3B; supplemental Figure 4). The same patterns of clone types were apparent when increasing the numbers of assayed top clones to 100 per sample, suggesting that clonal behavior is similar at a range of clonal sizes (supplemental Figure 6). The minimum frequency of long-term engrafting clones can be calculated from the clone number at longest follow-up and ranged from .0056% to .014% of initial transduced CD34+ cells, biologically plausible numbers (Figure 1B) and within ranges estimated in recent human gene therapy clinical trials utilizing insertion-site retrieval.30
Clonal stability
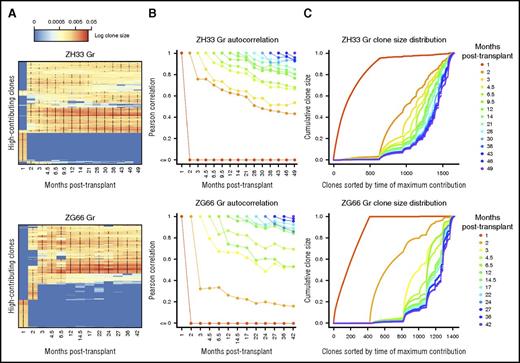
Given the short half-life of hours to days for granulocytes,31,32 we focused on this cell type to assess the kinetics of ongoing hematopoiesis in animals with the longest follow-up. Longitudinal granulopoietic clonal stability reflects continuous output from HSPCs, in contrast to longer-lived and tissue-migrating B cells and monocytes and self-renewing mature T cells. There was a clear distinction between early repopulating clones that contributed for 1 to 2 months and then disappeared and stable long-term clones, which appeared as early as 2 months and persisted for up to 49 months (Figure 4A). Once these long-term clones appeared, their contributions were extremely stable over time at a population level (Figure 4B), with minimal evidence for granulopoietic clonal succession characterized by either disappearance of previously contributing or appearance of previously quiescent clones. Granulocyte clonality did evolve in a slow and predictable manner; sample clonalities were most correlated with the previous and subsequent time points, and correlations between granulocyte clonalities declined at a stable rate over time because of some minor transience in clonal contributions (Figure 4B). However, visualization of the entire clonal repertoire shows that these variations were minor when considered in the context of quantitative hematopoietic output (Figure 4C). We also observed clonal stability when considering only if barcodes were detected and ignoring their quantitative contribution (supplemental Figure 7). The relatively early appearance of long-term dominant clones suggests that the replication of HSPCs in this crucial early time period leads to clonal persistence.
Stability of granulopoiesis over time. (A) Heat map showing the natural log fractional abundances of the highest contributing granulocyte (Gr) barcodes (clones) in ZH33 and ZG66 over time, defined as the set of all barcodes present as a top 100 highest contributing barcode in ≥1 of the samples shown. Each row corresponds to 1 barcode. The barcodes are organized by unsupervised hierarchical clustering using the Euclidean distance between barcodes’ log fractional abundances, with relative contribution shown as a red-to-blue gradient, representing high contribution to no contribution, respectively. Clones are clustered along the y-axis to place similar clones next to each other. *Indicates the top 100 clones in a given sample; thus, there are 100 in each column and ≥1 in each row. (B) The Pearson correlations between Gr clonalities at a certain time point post-transplantation and all subsequent Gr clonalities are plotted as individual lines for ZH33 and ZG66. Each time point is shown as a different color line, so, for example, the correlation of the 1-month sample with the 2-month sample is shown by the position of the red line at the 2-month time point on the y-axis. (C) The total clonal repertoires of barcoded granulopoiesis are displayed for ZH33 and ZG66 as cumulative distribution curves (supplemental Data provide more information on the analytic methodology used). Each position on the x-axis is an individual clone, and each line is the cumulative distribution of clonal contributions at the specified time point. Note that although these clones appear as lines, they are actually discrete sets of points (clones). The height of the line at an index on the x-axis is the sum of the clonal contributions of the clones with an index less than or equal to the index in question. Lower indices indicate earlier clones, and the clone ordering on the x-axis is determined by time of maximum contribution, showing emergence of new clones over time. This ordering is shared among time points, enabling comparison of the behavior of individual clones across time. Because contribution is assessed fractionally, the y-axis is from 0 to 1.
Stability of granulopoiesis over time. (A) Heat map showing the natural log fractional abundances of the highest contributing granulocyte (Gr) barcodes (clones) in ZH33 and ZG66 over time, defined as the set of all barcodes present as a top 100 highest contributing barcode in ≥1 of the samples shown. Each row corresponds to 1 barcode. The barcodes are organized by unsupervised hierarchical clustering using the Euclidean distance between barcodes’ log fractional abundances, with relative contribution shown as a red-to-blue gradient, representing high contribution to no contribution, respectively. Clones are clustered along the y-axis to place similar clones next to each other. *Indicates the top 100 clones in a given sample; thus, there are 100 in each column and ≥1 in each row. (B) The Pearson correlations between Gr clonalities at a certain time point post-transplantation and all subsequent Gr clonalities are plotted as individual lines for ZH33 and ZG66. Each time point is shown as a different color line, so, for example, the correlation of the 1-month sample with the 2-month sample is shown by the position of the red line at the 2-month time point on the y-axis. (C) The total clonal repertoires of barcoded granulopoiesis are displayed for ZH33 and ZG66 as cumulative distribution curves (supplemental Data provide more information on the analytic methodology used). Each position on the x-axis is an individual clone, and each line is the cumulative distribution of clonal contributions at the specified time point. Note that although these clones appear as lines, they are actually discrete sets of points (clones). The height of the line at an index on the x-axis is the sum of the clonal contributions of the clones with an index less than or equal to the index in question. Lower indices indicate earlier clones, and the clone ordering on the x-axis is determined by time of maximum contribution, showing emergence of new clones over time. This ordering is shared among time points, enabling comparison of the behavior of individual clones across time. Because contribution is assessed fractionally, the y-axis is from 0 to 1.
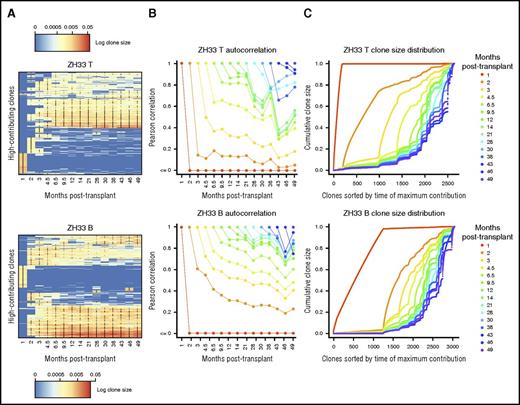
Despite the dissimilarity of mature B- and T-cell life histories compared with that of mature granulocytes, long-term clonality of these cell types was also very stable. Clones that contributed to B and T cells after the transient initial posttransplantation period tended to keep doing so over the entire observation period. Lack of clonal exhaustion or clonal expansion in circulating B cell clones after the first several months posttransplantation intimates ongoing production of circulating B cells from the marrow, similar to the circumstances driving stability in granulocytes. Similar to granulocytes, B-cell clonalities evolved slowly and steadily over time but were to a great extent shared between all long-term time points (Figure 5A). In contrast, circulating T-cell clonalities could vary rapidly, perhaps as part of an antigen-specific response (Figure 5B). Antigen-experienced, clonally-expanded memory B cells and plasma cells may reside instead primarily outside the circulation. The lower correlation of the most recent T-cell samples with past clonalities in ZH33 was due to emergence of several large T cell–producing clones, which expanded rapidly at these later time points (Figure 5C). Identifying these emergent clones in the top clone heatmap (Figure 3B) shows that they are T cell biased. Given the overall long-term similarity of T-cell clonality with that of other cell types, these clones seem more likely to be part of an adaptive immune response rather than altered production from the marrow.
Stability of lymphopoiesis over time. (A) Heat map showing the natural log fractional abundances of the highest contributing T-cell (T; top) and B-cell (B; bottom) barcodes (clones) in ZH33 over time, defined as the set of all barcodes present as a top 100 highest contributing barcode in ≥1 of the samples shown. Each row corresponds to 1 barcode. The barcodes are organized by unsupervised hierarchical clustering using the Euclidean distance between barcodes’ log fractional abundances, with relative contribution shown as a red-to-blue gradient, representing high contribution to no contribution, respectively. Clones are clustered along the y-axis to place similar clones next to each other. *Indicates the top 100 clones in a given sample; thus, there are 100 in each column and ≥1 in each row. (B) Pearson correlations between T-cell (top) and B-cell (bottom) clonalities in ZH33 at a certain time point posttransplantation and all subsequent T-cell or B-cell clonalities are plotted as individual lines. Each time point is shown as a different color line, so, for example, the correlation of the 1-month sample with the 2-month sample is shown by the position of the red line at the 2-month time point on the y-axis. These correlations depict the stability of T-cell or B-cell clonality over time. (C) The total clonal repertoires of barcoded T-cell (top) and B-cell (bottom) populations are displayed as cumulative distribution curves over time for ZH33. Each position on the x-axis is an individual clone, and each line is the cumulative distribution of clonal contributions at the specified time point. That is, the height of the line at an index on the x-axis is the sum of the clonal contributions of the clones with an index less than or equal to the index in question, at the time point being plotted. Lower indices indicate earlier clones, and the clone ordering on the x-axis is determined by time of maximum contribution. The ordering is shared among time points, enabling comparison of the behavior of individual clones across time. Thus, this figure shows emergence of new clones over time. Because contribution is assessed fractionally, the y-axis is from 0 to 1.
Stability of lymphopoiesis over time. (A) Heat map showing the natural log fractional abundances of the highest contributing T-cell (T; top) and B-cell (B; bottom) barcodes (clones) in ZH33 over time, defined as the set of all barcodes present as a top 100 highest contributing barcode in ≥1 of the samples shown. Each row corresponds to 1 barcode. The barcodes are organized by unsupervised hierarchical clustering using the Euclidean distance between barcodes’ log fractional abundances, with relative contribution shown as a red-to-blue gradient, representing high contribution to no contribution, respectively. Clones are clustered along the y-axis to place similar clones next to each other. *Indicates the top 100 clones in a given sample; thus, there are 100 in each column and ≥1 in each row. (B) Pearson correlations between T-cell (top) and B-cell (bottom) clonalities in ZH33 at a certain time point posttransplantation and all subsequent T-cell or B-cell clonalities are plotted as individual lines. Each time point is shown as a different color line, so, for example, the correlation of the 1-month sample with the 2-month sample is shown by the position of the red line at the 2-month time point on the y-axis. These correlations depict the stability of T-cell or B-cell clonality over time. (C) The total clonal repertoires of barcoded T-cell (top) and B-cell (bottom) populations are displayed as cumulative distribution curves over time for ZH33. Each position on the x-axis is an individual clone, and each line is the cumulative distribution of clonal contributions at the specified time point. That is, the height of the line at an index on the x-axis is the sum of the clonal contributions of the clones with an index less than or equal to the index in question, at the time point being plotted. Lower indices indicate earlier clones, and the clone ordering on the x-axis is determined by time of maximum contribution. The ordering is shared among time points, enabling comparison of the behavior of individual clones across time. Thus, this figure shows emergence of new clones over time. Because contribution is assessed fractionally, the y-axis is from 0 to 1.
Clonal bias
The relative contributions of individual HSPCs to differentiated cell types may imply the proximity of these cell types in a hematopoietic hierarchy. When a clone contributes only to some cell types, but not others, as in the case of expanding T-cell clones, it is straightforward to hypothesize the presence of a cell type–restricted progenitor or mature cell with expansion potential. However, most long-term clones that we observed are not cell type restricted in an absolute sense, but rather have some quantitative level of bias, which may change over time. We consider a clone to be unbiased between 2 samples if we detect it at similar frequency in both samples. Large unbiased clones suggest that HSPC proliferation to multiple differentiated cell types is due to production of proliferative multipotent progenitors rather than proliferative cell type–specific progenitors. That the largest clones are the least biased further supports this theory; proliferative potential in self-renewing progenitors restricted to one cell type would lead to bias, unless it were exactly balanced by that of progenitors restricted to another.
Bias distributions generally reflect the correlations shown in Figure 2. The high correlation between monocyte and granulocyte clonalities results from hundreds or thousands of clones that produce monocytes and granulocytes at a fixed ratio, which is shared between clones and stable over time at an individual clonal level (Figure 6A). This behavior emerges after 1 to 2 months. The tight range of biases between these 2 myeloid cell types indicates that proliferative potential is being retained in progenitor cells contributing to both myeloid cell types. There is minimal evidence for 1 myeloid branch of a given clone extinguishing while the other survives (Figure 3B). The largest clones are also the least biased, and their unbiasedness varies little over time. We observed similar behavior when considering bias between B cells and granulocytes (Figure 6B). That B cell– and granulocyte-producing clones rarely contributed to only one of these cell types indicates that a multipotent progenitor with self-renewal capacity was jointly contributing to these 2 cell types long term. Although completely B cell– or myeloid-restricted clones are rare or nonexistent, the range of biases displayed by high-contributing clones is generally broader for the comparison of B cells and granulocytes than for that of monocytes and granulocytes. No matter the bias value, these B-cell and granulocyte biases were stable over time for individual clones, even at levels other than 1:1 (Figure 6B), plausibly because of inheritable epigenetic programming of multipotent progenitors to favor a differentiated product or stochasticity of the differentiation processes. Again, variances were smallest for the largest clones. Individual clonal bias was greater comparing T-cell and B-cell contributions (Figure 6C). Clone-specific bias tracking allowed us to see that certain clones contributed almost exclusively to T cells over several time points, and the biases of individual clones could change markedly. These data suggest that ongoing B-cell production occurs jointly with T-cell production, but that highly biased T-cell clones can then arise via clone-specific expansion, which begins at some specific time point.
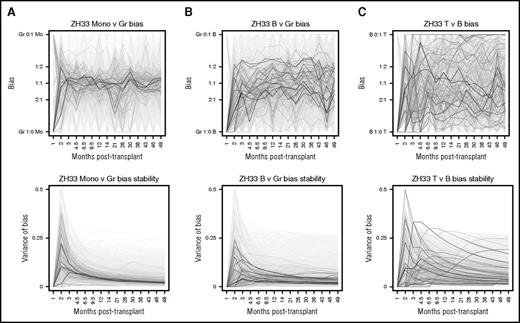
Clonal biases and variance of biases. (A) The ratio of the fractional contributions of each clone to monocytes (Mono) versus granulocytes (Gr) at each time point is mapped over time for ZH33 (top). All clones are shown; however, larger clones have darker lines and are plotted in the foreground. Smaller clones are shown with lighter lines in the background. Small (light) clones are more likely to appear highly biased because of sampling constraints. Note that clones are overall unbiased in monocyte versus granulocyte production, and individual clone bias is generally stable over time. The variance of these individual clonal biases up to the specified time point for Mono/Gr are shown at the bottom, demonstrating a marked stabilization of bias over time. The precise definition of bias is given in the supplemental data. (B) The same analyses as in (A) are shown for B cells versus granulocytes (B/Gr). Clones may be biased in production of B cells versus granulocytes, but individual clone bias is generally stable over time. Larger clones generally have smaller variance. (C) The same analyses as in (A) are shown for T cells versus B cells. Clones may be highly and stably biased toward T-cell (and away from B-cell) production and are less stable over time (vertical movement in bottom graph). Larger clones do not necessarily have smaller variance in this bias comparison, in contrast to the comparisons shown in (A) and (B).
Clonal biases and variance of biases. (A) The ratio of the fractional contributions of each clone to monocytes (Mono) versus granulocytes (Gr) at each time point is mapped over time for ZH33 (top). All clones are shown; however, larger clones have darker lines and are plotted in the foreground. Smaller clones are shown with lighter lines in the background. Small (light) clones are more likely to appear highly biased because of sampling constraints. Note that clones are overall unbiased in monocyte versus granulocyte production, and individual clone bias is generally stable over time. The variance of these individual clonal biases up to the specified time point for Mono/Gr are shown at the bottom, demonstrating a marked stabilization of bias over time. The precise definition of bias is given in the supplemental data. (B) The same analyses as in (A) are shown for B cells versus granulocytes (B/Gr). Clones may be biased in production of B cells versus granulocytes, but individual clone bias is generally stable over time. Larger clones generally have smaller variance. (C) The same analyses as in (A) are shown for T cells versus B cells. Clones may be highly and stably biased toward T-cell (and away from B-cell) production and are less stable over time (vertical movement in bottom graph). Larger clones do not necessarily have smaller variance in this bias comparison, in contrast to the comparisons shown in (A) and (B).
Discussion
HSPC lifespan, ontogenic hierarchies, and proliferative potential are incompletely understood, particularly in humans, with a lack of longitudinal, quantitative data on contributions of individual HSPCs to all cell types in the setting of normal hematopoiesis.2,33 In this report, we used clonal barcoding to map output from thousands of HPSCs for up to 49 months after autologous transplantation in rhesus macaques, a large animal model with relevance to human hematopoiesis,26 extending our observations in this model immediately after engraftment15 and now focusing on questions regarding clonal stability and bias. We observed short-lived cell type–restricted clones immediately after engraftment, supplanted by highly stable long-term multipotent clones in all assayed cell types that persisted for at least 49 months. The overall stability that we observed after initial reconstitution suggests that even biased clonal contribution levels were fixed early after transplantation, resulting from degree of initial proliferation of each transduced HPSC. Even in the wide spectrum of clonal behaviors that we observed, we did not detect B or T cell–restricted or highly biased multilymphoid progenitors, indicating that this hypothetical cell type was not contained in our transplant dose at a detectable level and was not preferentially generated by long-term HSCs, although we cannot rule out the possibility that they were generated by long-term HSCs as part of a balanced set of contributions.
Long-term clonal stability is concordant with prior large animal transplantation studies and human gene therapy trials utilizing lentiviral vectors and insertion-site retrieval, where individual clones were retrieved on multiple occasions over time from myeloid blood cells after disappearance of short-term engrafting clones.21,22,34-36 However, these studies used vector integration site retrieval to track clones, which requires large amounts of DNA because of inefficiency, is only semiquantitative, and can be inconsistent even when run on biological replicates.37 The average capture rate for individual clones in a human gene therapy clinical trial at individual time points was at best estimated to be approximately 50% and is generally much lower.30 Furthermore, the mere detection of a clone at 2 time points does not indicate whether or not it has increased or decreased in size. Earlier studies utilizing retroviral vectors in murine models or human-murine xenografts observed many fewer clones and relied on less sensitive or quantitative approaches, but also overall suggested clonal stability from at least some multipotent cells after disappearance of short-term progenitors.18,38,39 Our study focused only on clones large enough to be reliably detected by our methodology, accounting for >80% of marked hematopoiesis in 3 of the 4 animals. It is possible that real clones falling outside our analyses could manifest different characteristics, including clonal succession; however, this seems unlikely, because we saw minimal differences in behavior between the largest and smallest clones that fell above our analytic thresholds.
The long-term retention of a particular hematopoietic output pattern for individual HSPC clones supports the presence of maintained epigenetic control of fate decisions within self-renewing HSPC populations and is direct quantitative evidence that all self-renewing or long-lived HSPCs are not equivalent.10 For example, numerous clones exhibit consistent quantitative bias to myeloid cell types over a long period of time. However, it is hard to determine whether that result is subsequent to stochastic production of a prolific myeloid progenitor by an unbiased long-term HSC or a myeloid-biased long-term HSC, in which the production of myeloid progenitors is intrinsic to the long-term HSC.11 Stability may also depend on the number of cells delivered; limiting HSPC doses can result in wide swings in contributions based on impact of stochastic factors.40 The high correlation between contributions to granulocytes and monocytes is especially interesting, because a variable number of circulating mature monocytes and granulocytes over time suggests that although the ratio of production is stable between clones, it may not be fixed, and that a developmental switch controlling monocyte output is shared between myeloid progenitors.
Our results contrast with findings of granulopoietic clonal transience or periodicity in a recent report tracking HSPC clonal behavior in mice not receiving transplants, via transient activation of a Sleeping Beauty (SB) transposase, resulting in semirandom insertion of the SB transposons in HSPCs and allowing nonquantitative retrieval of SB insertion-site clonal tags.41 This study reported that hematopoiesis was supported by output from waves of cell type–restricted clones, most detectable at only 1 time point, in mice observed for up to 1 year. The difference between these results and ours may stem from 2 factors. First, the SB method avoids myeloablative impact on marrow niches, within which normal hematopoietic development occurs, and the replicative stress of reconstitution, which may alter normal ontogeny.42 In particular, the SB study showed that clonal transience was somewhat reduced by myeloablation. However, it is difficult to explain such marked differences in HSPC behavior long term based on myeloablation years previously. In humans, multiple acquired somatic mutations in HSPCs from humans with paroxysmal nocturnal hemoglobinuria in the PIG-A gene have also been reported to show stability in myeloid cells over time in a nontransplantation, nonablative setting.43 Second, the SB method is binary, with clones scored as present or absent, whereas our method is quantitative. Inefficient label retrieval or sparse sampling results in erroneous nondetection of small clones, which are, despite their relative nonabundance, counted as equivalent to high-contributing clones by binarizing methods. Therefore, nonquantitative analyses have a bias toward concluding that clonal cycling or succession has occurred, and they preclude the observation that large clones are the most stable. We nevertheless observed some polyclonal stability even when disregarding the quantitative information in our data (supplemental Figure 7), but such a binarization would somewhat weaken our conclusions. In a rhesus transplantation model, Kim et al36 modified VIS retrieval to be more quantitative on samples and analyzed samples collected up to 10 years post-transplantation and described clonal contributions that fluctuated between detectable and nondetectable levels. We have not observed this type of pattern to date in our barcoding model, with shorter follow-up, which, however, extends beyond the time point Kim et al noted clonal fluctuations.44
Consistent with recent literature, our data challenge a firm delineation between self-renewing stem versus progenitor cells, as in traditional conceptions of hematopoietic hierarchies. Additionally, observation of highly biased erythroid and megakaryocytic lineages in murine models and human cells studied in vitro calls for revision of classical hematopoietic lineage trees.45,46 Many of these data are coming together to suggest a continuous notion of the differentiation state, based on transcriptomic, proteomic, or epigenetic factors.47,48 Connecting single-cell phenotypes to ontologically informative individual barcodes in our rhesus model will create a more definitive mapping of these complex relationships. Progress in systems biology, genomics, and statistics suggests a myriad of questions that can be answered with clonal labeling strategies. We have recently used these rhesus macaque data sets to derive stochastic compartmental models providing probabilistic descriptions of how hematopoietic cells divide and differentiate.44 As sequencing costs fall, it may be possible to use deep sequencing of benign sporadic genomic mutations to extend methods that track clonal contribution by malignant mutations to normal humans. This approach has already uncovered surprising degrees of clonal hematopoiesis in the elderly.49
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the National Institutes of Health (NIH), National Heart, Lung and Blood Institute (NHLBI) DNA Sequencing and Genomics Core, the NHLBI Flow Cytometry Core, Stephanie Sellers, and Keyvan Keyvanfar for technical assistance, and the staff of the NHLBI Primate Facility for excellent animal care.
This research was supported by the intramural research program of the NIH, NHLBI and the following grants: NIH, NHLBI K99-HL11304 (R.L.) and California Institute for Regenerative Medicine TG2-01159 (R.L.).
Authorship
Contribution: S.J.K., C.W., and C.E.D. conceived the study; S.J.K. and C.E.D. wrote the paper; S.J.K., D.A.E., C.W., and B.L. performed experiments; R.E.D. was responsible for all animal care; R.L. provided reagents and analyses; and S.J.K., J.X., and D.A.E. conceived and performed quantitative and statistical analyses and prepared figures.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Cynthia E. Dunbar, Molecular Hematopoiesis Section, Hematology Branch, National Heart, Lung and Blood Institute, National Institutes of Health, Bethesda, MD 20892; e-mail: dunbarc@nhlbi.nih.gov.

![Figure 3. Similarities between clonal contributions to hematopoietic cell types. (A) Pearson correlation measures similarity between the sets of clones from which specific cell populations are descended. Each line gives the Pearson correlations over time between samples from 2 cell types, with each line consisting of 2 colors corresponding to the cell types being compared (T cells [T], black; B cells [B], magenta; monocytes [Mono], green; granulocytes [Gr], light blue). Mono-Gr clonalities are most similar, followed by Gr-B or Mono-B and Gr/B-T or Mono/B-T. These similarities generally stabilize, with greatest volatility in correlation comparisons involving T cells. (B) Heat map showing the natural log fractional abundances of the highest contributing clones in ZH33 over time, defined as the set of all barcodes present as a top 10 highest contributing barcode in ≥1 of the samples shown. Each row corresponds to 1 barcode. The barcodes are organized by unsupervised hierarchical clustering using the Euclidean distance between barcodes’ log fractional abundances, with relative contribution shown as a magenta-to-light blue gradient, representing high contribution to no contribution, respectively. Clones are clustered along the y-axis to place similar clones next to each other. *Indicates the top 10 clones in a given sample; thus, there are 10 in each column and ≥1 in each row. Note transient clones contribute to each lineage at 1 month and then disappear, replaced by multipotent clones beginning at months 2 to 3, with contributions persisting long term.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/129/11/10.1182_blood-2016-07-728691/4/m_blood728691f3.jpeg?Expires=1767716233&Signature=uqleOJgEzpcJH3YiiuDMt7IiYNFc8IhH5K6qlugSjFw8ZYJc-mySAUU6kvgDP542XmtZCqHlT2g~zrYq-ZaH21UBS~toogCpKO9oNR20JMmIWojxJsKN8imyEeWvl0RdA8TcCMJPmoIQGGYZeE0~GuAt4gwh-JStBhNRBpGDjvTy9U0rfDoGsLrTk4maofjj23qvYp~AGlR52ncBp4QE8S98Vkrefep1beG8IYZiXMHvkAS64qeC1JeCNRFK6DCdbsdQum6Tat68mSx0tiLtvTgiksfHR~5YsKf1nDSqer3Ix4~TFtC2hUrvzPhDb8l9xXZcghwFsoN5oobjrB1r3g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal