Key Points
Marker chromosomes are frequently found in AML, particularly among aneuploid adverse-risk karyotypes and confer a poor prognosis.
About one-third of marker and ring chromosome karyotypes arise from chromothripsis.
Abstract
Metaphase karyotyping is an established diagnostic standard in acute myeloid leukemia (AML) for risk stratification. One of the cytogenetic findings in AML is structurally highly abnormal marker chromosomes. In this study, we have assessed frequency, cytogenetic characteristics, prognostic impact, and underlying biological origin of marker chromosomes. Given their inherent gross structural chromosomal damage, we speculated that they may arise from chromothripsis, a recently described phenomenon of chromosome fragmentation in a single catastrophic event. In 2 large consecutive prospective, randomized, multicenter, intensive chemotherapy trials (AML96, AML2003) from the Study Alliance Leukemia, marker chromosomes were detectable in 165/1026 (16.1%) of aberrant non–core-binding-factor (CBF) karyotype patients. Adverse-risk karyotypes displayed a higher frequency of marker chromosomes (26.5% in adverse-risk, 40.3% in complex aberrant, and 41.2% in abnormality(17p) karyotypes, P < .0001 each). Marker chromosomes were associated with a poorer prognosis compared with other non-CBF aberrant karyotypes and led to lower remission rates (complete remission + complete remission with incomplete recovery), inferior event-free survival as well as overall survival in both trials. In multivariate analysis, marker chromosomes independently predicted poor prognosis in the AML96 trial ≤60 years. As detected by array comparative genomic hybridization, about one-third of marker chromosomes (18/49) had arisen from chromothripsis, whereas this phenomenon was virtually undetectable in a control group of marker chromosome-negative complex aberrant karyotypes (1/34). The chromothripsis-positive cases were characterized by a particularly high degree of karyotype complexity, TP53 mutations, and dismal prognosis. In conclusion, marker chromosomes are indicative of chromothripsis and associated with poor prognosis per se and not merely by association with other adverse cytogenetic features.
Introduction
Cytogenetic testing is routinely performed in newly diagnosed acute myeloid leukemia (AML) patients for risk stratification and treatment recommendation. Elaborate risk classifications based on karyotyping results are provided by both the European Leukemia Net (ELN)1 and the Medical Research Council (MRC).2,3 Complex aberrant karyotypes, which harbor at least 3 (ELN) or 4 (MRC) chromosomal aberrations, are known to confer a poor prognosis.3-8 Monosomal karyotypes, which are defined by at least 2 autosomal monosomies or 1 autosomal monosomy in combination with a structural aberration have been shown to be particularly unfavorable.9-11 Recently, clonal heterogeneity at the cytogenetic level has been identified as an additional risk factor.12-14 Also, abnormalities of chromosome 17p13, which lead to TP53 inactivation, are associated with a particularly dismal prognosis.15-18 In addition, the prognostic impact of individual chromosomal aberrations has been thoroughly elucidated.3 In contrast, the role of marker chromosomes has remained elusive so far. In cytogenetic nomenclature, marker chromosomes designate chromosomes that are rearranged to a level that prevents its allocation to 1 of the known 23 chromosomes.19 Marker chromosomes thus reflect gross structural chromosomal damage.
It was the aim of this study to assess the frequency, cytogenetic characteristics, and prognostic impact of marker chromosome karyotypes as well as the underlying biological mechanisms of marker chromosome formation. For that, 2 large clinical trials from the German Study Alliance Leukemia (SAL) were analyzed.
Methods
Patients and treatment
This analysis included all patients enrolled into the 2 large consecutive, prospective, randomized, multicenter AML96 and AML2003 trials of SAL.20,21 Patients had non-M3 AML and were previously untreated. Both trials permitted the inclusion of patients with prior myelodysplastic syndrome (MDS) and therapy-related AML. All adult patients fit for intensive chemotherapy were eligible for the AML96 trial, whereas age was restricted to 16 to 60 years in the AML2003 trial.21,22 Both studies have been previously described in detail.20-22 Approval was obtained from the respective ethics committees of the participating centers. Patients gave written informed consent in accordance with the Declaration of Helsinki. Both trials had been registered at www.clinicaltrials.gov (AML96, NCT00180115; AML2003, NCT00180102).
Karyotype analysis
Conventional karyotyping of metaphases was performed according to routine protocols. For this project, all karyotypes from the AML96 and AML2003 trials have been retrospectively reviewed and screened for marker chromosomes. As a control, all other aberrant karyotypes were selected. In total, 1653 (88.8%) of 1862 patients in the AML96 trial and 986 (83.6%) of 1179 patients in the AML2003 trial were evaluable, among them 792 (47.9%) and 482 (48.9%) with aberrant karyotypes, and 123 (7.4%) and 44 (4.5%) with marker chromosomes, respectively. Constitutional karyotypes were not counted as aberrant for this analysis. As described by the International System for Human Cytogenetic Nomenclature guidelines, chromosomal gains and structural abnormalities had to be detected in at least 2 metaphases and chromosomal losses in at least 3 metaphases to be acknowledged as clonal.19 Complex aberrant karyotypes were determined based on the MRC adverse-risk criteria requiring at least 4 chromosomal aberrations.3 Monosomal karyotypes were classified according to Breems et al.9 The criteria for cytogenetic clonal heterogeneity have previously been described.12 Deletions of the long arm of chromosome 5 and loss of the whole chromosome 5 were subsumed as abnormality (abnl)(5q). Abnl(7q) was defined analogously. Abnl(17p) was defined as previously described.15,17,18 For confirmation of abnl(17p), interphase fluorescence in situ hybridization (FISH) analysis using a 17p13 probe (Abbott, Wiesbaden, Germany) was performed.
Chromothripsis detection by array comparative genomic hybridization
Array comparative genomic hybridization (array-CGH) was used for the detection of chromothripsis. Chromothripsis was defined according to the criteria by Rausch et al, which require at least 10 changes in segmental copy number (CN) involving 2 or 3 distinct CN states on a single chromosome.23 In detail, for each sample, 50 ng of DNA was hybridized to an Affymetrix CytoScan HD Oligo/single-nucleotide polymorphism array according to the manufacturer's instructions. Arrays were scanned with the Affymetrix GeneChip Scanner 3000 7G, and CN analysis was done with Affymetrix Chromosome Analysis Suite software version 2.1.0.16 (r6634) and Annotation Net Affx Build 33. Interpretation was based on human reference sequence GRCh37/hg19, February 2009. The complete data set was visually analyzed. All patients with marker or ring chromosomes and double minute karyotypes identified in the AML96 and AML2003 trials and available DNA samples from first diagnosis were analyzed, provided the blast purity in the sample was ≥50%. As a control group, all samples from patients with complex aberrant karyotypes without marker, ring, or derivative chromosomes and double minutes were used. Patients with an incomplete karyotype formula were excluded from the control group. The threshold of a blast purity ≥50% was equally applied.
Array data have been deposited in the Gene Expression Omnibus, accession number GSE93886.
Multicolor FISH of chromothripsis-positive patients
Multiplex fluorescence in situ hybridization (M-FISH) was performed as described.24 Briefly, 7 pools of flow-sorted human whole chromosome painting probes were amplified and directly labeled using 7 different fluorochromes (DEAC, FITC, Cy3, Cy3.5, Cy5, Cy5.5, and Cy7) using degenerative oligonucleotide primed polymerase chain reaction (PCR). Metaphase chromosomes immobilized on glass slides were denatured in 70% formamide/2× SSC pH 7.0 at 72°C for 2 minutes followed by dehydration in a degraded ethanol series. A hybridization mixture containing combinatorially labeled painting probes, an excess of unlabeled cot1 DNA, 50% formamide, 2× SSC, and 15% dextran sulfate was denatured for 7 minutes at 75°C, preannealed at 37°C for 20 minutes, and hybridized at 37°C to the denaturated metaphase preparations. After 48 hours, the slides were washed in 2× SSC at room temperature 3 times 5 minutes followed by 2 washes in 0.2× SSC/0.2% Tween-20 at 56°C for 7 minutes each. Metaphase spreads were counterstained with 4′,6-diamidino-2-phenylindole and covered with antifade solution. Metaphase spreads were recorded using a DM RXA epifluorescence microscope (numerical aperture 1, objective lens 63×, magnification ×63; Leica Microsystems, Bensheim, Germany) equipped with a Sensys CCD camera (Photometrics, Tucson, AZ). Camera and microscope were controlled by the Leica Q-FISH software, and images were processed on the basis of the Leica MCK software and presented as multicolor karyograms (Leica Microsystems Imaging Solutions, Cambridge, UK). For barcode FISH, multicolor fluorochrome banding probe kits for chromosomes 2, 3, 7, 8, 9, and 17 (XCyte; MetaSystems, Neulussheim, Germany) were hybridized according to the manufacturer's instructions. Multicolor fluorochrome banding probe kits comprise overlapping partial chromosome paints, which resemble a false-color banding pattern, when visualized in the M-FISH software, allowing information on intrachromosomal rearrangements.
Molecular analyses
FLT3-ITD and NPM1 mutational testing.
TP53 mutational testing.
For library preparation, the multiplex PCR-based Ion Torrent AmpliSeq technology (Thermo Fisher Scientific, Waltham, MA), together with the custom-designed Lung Cancer Panel v2 (LCPv2; Thermo Fisher Scientific), was used as described previously.27,28 Amplicon library preparation was performed with the Ion AmpliSeq Library Kit v2.0. For mutation analysis, the LCPv2 panel was employed. The LCPv2 panel has 2 primer pools with 234 amplicons and covers 42 genes. For amplification, ∼10 ng of DNA, determined by quantitative polymerase chain reaction (qPCR) assay, was used. Briefly, the DNA was mixed with the primer pool and the AmpliSeq HiFi Master Mix in a 20-µL reaction volume and transferred to a PCR cycler (Biometra, Göttingen, Germany). After the end of the PCR reaction, amplicons were partially digested using FuPa reagent, followed by the ligation of barcoded sequencing adapters (Ion Xpress Barcode Adapters; Thermo Fisher Scientific). The final library was purified using AMPure XP magnetic beads (Beckman Coulter, Krefeld, Germany) and quantified using qPCR (Ion Library Quantitation Kit, Thermo Fisher Scientific) on a StepOne Plus qPCR machine (Thermo Fisher Scientific). The individual libraries were diluted to a final concentration of 100 pM. Eight to 10 libraries were pooled and processed to library amplification on Ion Spheres using Ion PGM Hi-Q OT2 200 Kit. Unenriched libraries were quality controlled using Ion Sphere quality control measurement on a QuBit instrument. After library enrichment (Ion OneTouch ES, Thermo Fisher Scientific), the library was processed for sequencing using the Ion Torrent Ion PGM Hi-Q sequencing chemistry, and the barcoded libraries were loaded onto a 318v2 chip. For data analysis, raw sequencing data were processed using the implemented Torrent Suite Software (version 5.0.2) and aligned against the human genome (version hg19) using the Torrent Mapping Alignment Program algorithm. For DNA mutation analysis, the aligned reads were processed using the built-in Variant Caller plug-in (version 5.0.2.1). Variant annotation was performed using a custom-built variant annotation pipeline in the CLC Genomics Workbench (version 8.0.2). For visualization of sequencing and fusion reads, the Integrative Genomic Browser (http://www.broadinstitute.org/igv/) was used.
Statistical analysis
Associations between marker chromosome karyotypes and cytogenetic groups, clinical characteristics, and response rates were analyzed by univariate statistics. Wilcoxon rank sum tests were used to compare the marker chromosome-positive and -negative groups for continuous (clinical or cytogenetic) values. Fisher’s exact tests were implemented to compare the distribution of categorical factors between these 2 groups. For the analysis of marker chromosome karyotypes with cytogenetic and clinical characteristics, P values were adjusted for multiple testing using the method of Holm.29 (Adjusted) P values ≤.05 were considered to be significant.
In univariate survival analyses, the prognostic value of marker chromosome karyotypes was analyzed for the outcomes event-free survival (EFS) and overall survival (OS), which were measured from the start of therapy until event (death from any cause, therapy failure, or relapse) and from the start of therapy until death, respectively. EFS was not censored at the time of allogeneic stem cell transplantation. Kaplan-Meier survival curve estimates and corresponding log-rank tests were computed based on the data sets of AML96, AML96 patients ≤60 years, AML96 patients >60 years, AML96 patients with adverse-risk cytogenetics, AML2003, and AML2003 patients with adverse-risk cytogenetics, respectively.
Multivariate survival analysis was done using Cox proportional hazards models, including variables describing marker chromosome karyotypes, age, prior MDS, therapy-related AML, and adverse-risk cytogenetics according to MRC criteria. P values ≤.05 were considered to be significant.
The statistical analysis was performed using R version 3.1.1 and 3.3.1 (www.r-project.org).
Results
Frequency of marker chromosome karyotypes and distribution among cytogenetic groups
Overall, marker chromosomes were detectable in 165/1026 (16.1%) aberrant non–core-binding-factor (CBF) karyotypes. Their frequency largely depended on the cytogenetic risk group. Adverse-risk karyotypes as defined by MRC criteria harbored marker chromosomes in 148/558 (26.5%) cases, whereas their frequency was lower among intermediate-risk aberrant karyotypes (17/468, 3.6%) and particularly low for CBF leukemias with a mere 2/248 (0.8%) cases. Therefore, this cytogenetic subgroup was excluded from further analysis.
Within the cytogenetic adverse-risk group, marker chromosomes were particularly frequent in patients with complex aberrant (40.3%), monosomal (50.2%), abnl(5q) (43.4%), abnl(7q) (32.9%), and abnl(17p) (41.2%) karyotypes (Table 1). Likewise, the marker chromosome frequency was also high in patients with cytogenetic clonal heterogeneity (33.6%). On the other hand, adverse-risk defining recurrent translocations and inversions displayed fewer marker chromosome karyotypes: 9.4% in abnl(3q), 4.3% in t(6;9)(p23;q34), and 0% in t(v;11)(v;q23). Among all aberrant non-CBF karyotypes, marker chromosomes were overrepresented in the derivate (der) chromosome-positive group (24.7% vs 14.0%, P = .006). In contrast, marker chromosomes were rare in NPM1 (3.8%) or FLT3-ITD (3.1%) mutated aberrant non-CBF karyotype AML.
Frequencies of marker chromosomes by cytogenetic entities
| . | No. of patients . | Marker positivity (%) . | P value* . |
|---|---|---|---|
| All | 1026 | 165/1026 (16.1) | |
| Cytogenetics | |||
| Adverse risk (MRC) | 558 | 148/558 (26.5) | <.001 |
| Complex aberrant (≥4) | 352 | 142/352 (40.3) | <.001 |
| Monosomal | 249 | 125/249 (50.2) | <.001 |
| Abnl(17p) | 131 | 54/131 (41.2) | <.001 |
| Abnl(5q) | 226 | 98/226 (43.4) | <.001 |
| Abnl(7q) | 243 | 80/243 (32.9) | <.001 |
| Abnl(3q) | 53 | 5/53 (9.4) | .93 |
| t(6;9)(p23;q34) | 23 | 1/23 (4.3) | .82 |
| t(9;11)(p22;q23) | 47 | 2/47 (4.3) | .21 |
| t(11;19)(q23;p13) | 12 | 0/12 (0) | .93 |
| t(v;11)(v;q23) | 47 | 0/47 (0) | .04 |
| Subclone formation | 342 | 115/342 (33.6) | <.001 |
| Derivate chromosome | 198 | 49/198 (24.7) | .006 |
| Molecular genetics | |||
| NPM1 mutation | 104 | 4/104 (3.8) | .003 |
| FLT3-ITD mutation | 128 | 4/128 (3.1) | <.001 |
| . | No. of patients . | Marker positivity (%) . | P value* . |
|---|---|---|---|
| All | 1026 | 165/1026 (16.1) | |
| Cytogenetics | |||
| Adverse risk (MRC) | 558 | 148/558 (26.5) | <.001 |
| Complex aberrant (≥4) | 352 | 142/352 (40.3) | <.001 |
| Monosomal | 249 | 125/249 (50.2) | <.001 |
| Abnl(17p) | 131 | 54/131 (41.2) | <.001 |
| Abnl(5q) | 226 | 98/226 (43.4) | <.001 |
| Abnl(7q) | 243 | 80/243 (32.9) | <.001 |
| Abnl(3q) | 53 | 5/53 (9.4) | .93 |
| t(6;9)(p23;q34) | 23 | 1/23 (4.3) | .82 |
| t(9;11)(p22;q23) | 47 | 2/47 (4.3) | .21 |
| t(11;19)(q23;p13) | 12 | 0/12 (0) | .93 |
| t(v;11)(v;q23) | 47 | 0/47 (0) | .04 |
| Subclone formation | 342 | 115/342 (33.6) | <.001 |
| Derivate chromosome | 198 | 49/198 (24.7) | .006 |
| Molecular genetics | |||
| NPM1 mutation | 104 | 4/104 (3.8) | .003 |
| FLT3-ITD mutation | 128 | 4/128 (3.1) | <.001 |
Adjusted for multiple testing.
Association of marker chromosome karyotypes with clinical parameters
Karyotypes with marker chromosomes were more frequent among elderly patients (Table 2). We observed a trend for an antecedent MDS, although statistical significance was not reached. Markedly, karyotypes with marker chromosomes were significantly associated with lower bone marrow blasts and lower leukocyte and platelet counts, findings that are suggestive of an antecedent MDS.
Clinical characteristics of patients with marker chromosomes
| . | No marker (n = 861) . | Marker (n = 165) . | P value* . |
|---|---|---|---|
| Age, y (median, range) | 55 (17-83) | 58 (15-77) | .002 |
| Sex, % (male/female) | 53.0/47.0 | 53.9/46.1 | .93 |
| FAB code | .006 | ||
| FAB M0 (%) | 67/807 (8.3) | 11/147 (7.5) | |
| FAB M1 (%) | 166/ 807 (20.6) | 23/147 (15.6) | |
| FAB M2 (%) | 238/807 (29.5) | 57/147 (38.8) | |
| FAB M4/M5 (%) | 275/807 (34.1) | 32/147 (21.8) | |
| FAB M6 (%) | 32/807 (4.0) | 14/147 (9.5) | |
| FAB M7 (%) | 11/ 807 (1.4) | 1/147 (0.7) | |
| RAEB (%) | 18/807 (2.2) | 9/147 (6.1) | |
| Prior MDS (%) | 166/849 (19.6) | 38/163 (23.3) | .93 |
| Therapy-related AML (%) | 63/849 (7.4) | 18/163 (11.0) | .82 |
| BM blasts, % (median, range) | 61 (6-99) | 48 (5-95) | <.001 |
| Leukocytes, /nL (median, range) | 10.39 (0.2-465.9) | 3.95 (0.6-171.3) | <.001 |
| Platelets, /nL (median, range) | 55 (1-1043) | 40.5 (4-433) | .002 |
| LDH value, U/L (median, range) | 415 (27-7369) | 324 (97-5489) | .24 |
| . | No marker (n = 861) . | Marker (n = 165) . | P value* . |
|---|---|---|---|
| Age, y (median, range) | 55 (17-83) | 58 (15-77) | .002 |
| Sex, % (male/female) | 53.0/47.0 | 53.9/46.1 | .93 |
| FAB code | .006 | ||
| FAB M0 (%) | 67/807 (8.3) | 11/147 (7.5) | |
| FAB M1 (%) | 166/ 807 (20.6) | 23/147 (15.6) | |
| FAB M2 (%) | 238/807 (29.5) | 57/147 (38.8) | |
| FAB M4/M5 (%) | 275/807 (34.1) | 32/147 (21.8) | |
| FAB M6 (%) | 32/807 (4.0) | 14/147 (9.5) | |
| FAB M7 (%) | 11/ 807 (1.4) | 1/147 (0.7) | |
| RAEB (%) | 18/807 (2.2) | 9/147 (6.1) | |
| Prior MDS (%) | 166/849 (19.6) | 38/163 (23.3) | .93 |
| Therapy-related AML (%) | 63/849 (7.4) | 18/163 (11.0) | .82 |
| BM blasts, % (median, range) | 61 (6-99) | 48 (5-95) | <.001 |
| Leukocytes, /nL (median, range) | 10.39 (0.2-465.9) | 3.95 (0.6-171.3) | <.001 |
| Platelets, /nL (median, range) | 55 (1-1043) | 40.5 (4-433) | .002 |
| LDH value, U/L (median, range) | 415 (27-7369) | 324 (97-5489) | .24 |
BM blasts, bone marrow blast percentage; FAB, French-American-British; LDH, lactate dehydrogenase.
Adjusted for multiple testing.
Impact of marker chromosomes on remission achievement, EFS, and OS
To analyze their prognostic impact, karyotypes with marker chromosomes were compared with all other aberrant non-CBF karyotypes. In younger patients, marker chromosomes conferred a poorer response to chemotherapy. The combined rate of complete remission (CR) plus complete remission with incomplete recovery of counts (CRi) was significantly lower for marker chromosome-positive patients ≤60 years in both the AML96 (36.0% vs 55.8%, P = .01) and the AML2003 trials (14.3% vs 44.1%; P < .001), respectively (supplemental Table 1, available on the Blood Web site). In contrast, remission rates were similarly poor for patients >60 years with and without marker chromosomes in the AML96 study (26.8% vs 22.3%; P = .43).
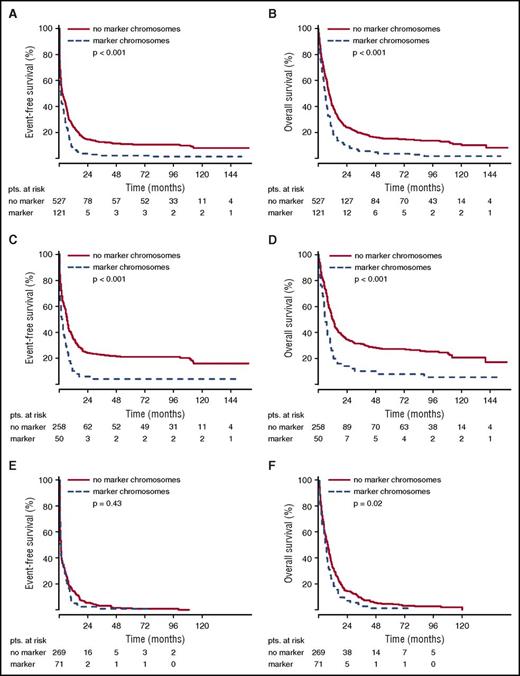
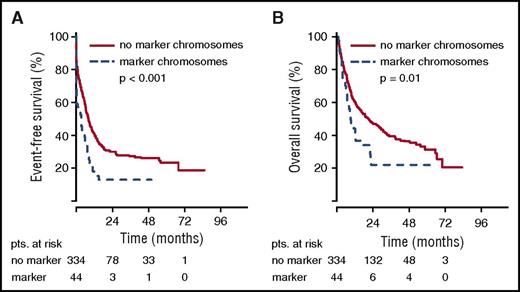
The poor remission rate of younger patients with marker chromosome karyotypes translated into inferior EFS. For the total group of patients in the AML96 trial, the detection of marker chromosomes led to a shortened EFS of 1.15 vs 2.66 months (P < .001) (Figure 1). Here, the negative prognostic effect of marker chromosomes was most pronounced in the age group ≤60 years with an EFS of 2.24 for the marker chromosome–positive group vs 6.54 months for the control group (P < .001). For patients >60 years, the EFS was equally poor with 0.82 vs 0.92 months (P = .43). Similarly, in the AML2003 study, a shortened EFS of 3.45 vs 8.03 months was observed for patients with marker chromosomes (P < .001) (Figure 2). The adverse effect of marker chromosomes on EFS could be consistently detected throughout all randomization strata in both AML96 and AML2003 (data not shown).
EFS and OS for non-CBF aberrant karyotypes in the AML96 trial depending on marker chromosome detection. EFS (A,C,E) and OS curves (B,D,F) are provided for the total AML96 patient group (A, B), the age subgroup ≤60 years (C,D), and the age subgroup >60 years (E,F).
EFS and OS for non-CBF aberrant karyotypes in the AML96 trial depending on marker chromosome detection. EFS (A,C,E) and OS curves (B,D,F) are provided for the total AML96 patient group (A, B), the age subgroup ≤60 years (C,D), and the age subgroup >60 years (E,F).
EFS and OS for non-CBF aberrant karyotypes in the AML2003 trial depending on marker chromosome detection. EFS (A) and OS curves (B) are provided for the total AML2003 patient group. pts., patients.
EFS and OS for non-CBF aberrant karyotypes in the AML2003 trial depending on marker chromosome detection. EFS (A) and OS curves (B) are provided for the total AML2003 patient group. pts., patients.
OS analysis yielded the same effects. For the total group of patients in the AML96 trial, the median OS was inferior for the marker chromosome–positive group (5.65 vs 9.17 months, P < .001), with the largest difference in the subgroup ≤60 years (5.72 vs 11.87 months, P < .001) (Figure 1). However, for the age group >60 years, a significant OS difference was observed as well (5.65 vs 7.33 months, P = .02). Again, the poor OS of marker chromosome–positive patients was confirmed in the AML2003 trial with median OS reduced to 8.68 as compared with 20.78 months for the control group without marker chromosomes (P = .01) (Figure 2).
Prognostic impact of marker chromosomes in the cytogenetic adverse-risk group
Most marker chromosome karyotypes fall into the adverse-risk category. We therefore analyzed whether the detection of marker chromosomes is a discriminating factor within this adverse-risk category as defined by the MRC. Subgroup analysis indeed showed that marker chromosomes added prognostic information in the adverse-risk group of the AML96 trial (median EFS: 0.96 vs 1.68 months, P = .02; median OS: 5.38 vs 7.20 months, P < .001) (supplemental Figure 1). Statistical significance was not reached in the AML2003 trial (median EFS: 3.45 vs 5.53 months, P = .19; median OS: 8.02 vs 9.63 months, P = .24).
Prognostic impact of marker chromosomes in multivariate analysis
We assessed whether marker chromosomes constitute an independent risk factor for EFS and OS by multivariate Cox regression analysis. As covariables, age, prior MDS, therapy-related AML, and adverse-risk cytogenetics according to MRC criteria were chosen. Given the broad overlap between marker chromosomes and adverse-risk karyotypes, inclusion of this parameter as covariable was particularly important.
In the AML96 trial, marker chromosomes emerged as an independent prognostic variable for OS (P = .02) but not EFS (P = .28) when all patients with non-CBF AML and aberrant karyotypes were considered (Table 3). In the age group ≤60 years, statistical significance was reached for both OS and EFS (P = .01 and P = .008, respectively). Age and adverse-risk karyotype emerged as additional significant variables, whereas prior MDS and therapy-related AML were not statistically significant.
Multivariate analysis of impact of risk factors on EFS and OS
| . | . | EFS . | OS . | ||||
|---|---|---|---|---|---|---|---|
| Study . | Risk factor . | HR . | 95% CI . | P value . | HR . | 95% CI . | P value . |
| AML96 | Age | 1.33 | (1.24-1.42) | <.001 | 1.33 | (1.24-1.42) | <.001 |
| Prior MDS | 1.05 | (0.87-1.28) | .60 | 0.94 | (0.77-1.15) | .57 | |
| Therapy-related AML | 0.95 | (0.67-1.33) | .75 | 1.17 | (0.83-1.64) | .37 | |
| Adverse risk (MRC) | 1.69 | (1.41-2.04) | <.001 | 1.82 | (1.52-2.17) | <.001 | |
| Marker chromosome | 1.13 | (0.91-1.40) | .28 | 1.28 | (1.03-1.58) | .02 | |
| AML96 ≤60 y | Age | 1.18 | (1.05-1.32) | .005 | 1.24 | (1.10-1.41) | .001 |
| Prior MDS | 1.10 | (0.79-1.55) | .57 | 0.99 | (0.70-1.40) | .95 | |
| Therapy-related AML | 1.11 | (0.72-1.72) | .64 | 1.30 | (0.84-2.02) | .24 | |
| Adverse risk (MRC) | 1.89 | (1.45-2.50) | <.001 | 2.17 | (1.67-2.86) | <.001 | |
| Marker chromosome | 1.56 | (1.12-2.16) | .008 | 1.54 | (1.11-2.14) | .01 | |
| AML96 >60 y | Age | 1.19 | (0.93-1.52) | .17 | 1.43 | (1.13-1.82) | .003 |
| Prior MDS | 1.04 | (0.82-1.33) | .73 | 0.94 | (0.74-1.21) | .65 | |
| Therapy-related AML | 0.85 | (0.49-1.46) | .55 | 1.07 | (0.62-1.85) | .81 | |
| Adverse risk (MRC) | 1.56 | (1.23-2.00) | <.001 | 1.59 | (1.23-2.04) | <.001 | |
| Marker chromosome | 0.93 | (0.70-1.23) | .61 | 1.19 | (0.89-1.57) | .24 | |
| AML2003 | Age | 1.08 | (0.96-1.20) | .21 | 1.18 | (1.04-1.34) | .01 |
| Prior MDS | 1.02 | (0.73-1.43) | .92 | 0.89 | (0.61-1.29) | .53 | |
| Therapy-related AML | 1.41 | (0.97-2.05) | .08 | 1.94 | (1.31-2.88) | .001 | |
| Adverse risk (MRC) | 1.67 | (1.30-2.13) | <.001 | 1.82 | (1.39-2.38) | <.001 | |
| Marker chromosome | 1.36 | (0.95-1.97) | .10 | 1.16 | (0.79-1.72) | .45 | |
| . | . | EFS . | OS . | ||||
|---|---|---|---|---|---|---|---|
| Study . | Risk factor . | HR . | 95% CI . | P value . | HR . | 95% CI . | P value . |
| AML96 | Age | 1.33 | (1.24-1.42) | <.001 | 1.33 | (1.24-1.42) | <.001 |
| Prior MDS | 1.05 | (0.87-1.28) | .60 | 0.94 | (0.77-1.15) | .57 | |
| Therapy-related AML | 0.95 | (0.67-1.33) | .75 | 1.17 | (0.83-1.64) | .37 | |
| Adverse risk (MRC) | 1.69 | (1.41-2.04) | <.001 | 1.82 | (1.52-2.17) | <.001 | |
| Marker chromosome | 1.13 | (0.91-1.40) | .28 | 1.28 | (1.03-1.58) | .02 | |
| AML96 ≤60 y | Age | 1.18 | (1.05-1.32) | .005 | 1.24 | (1.10-1.41) | .001 |
| Prior MDS | 1.10 | (0.79-1.55) | .57 | 0.99 | (0.70-1.40) | .95 | |
| Therapy-related AML | 1.11 | (0.72-1.72) | .64 | 1.30 | (0.84-2.02) | .24 | |
| Adverse risk (MRC) | 1.89 | (1.45-2.50) | <.001 | 2.17 | (1.67-2.86) | <.001 | |
| Marker chromosome | 1.56 | (1.12-2.16) | .008 | 1.54 | (1.11-2.14) | .01 | |
| AML96 >60 y | Age | 1.19 | (0.93-1.52) | .17 | 1.43 | (1.13-1.82) | .003 |
| Prior MDS | 1.04 | (0.82-1.33) | .73 | 0.94 | (0.74-1.21) | .65 | |
| Therapy-related AML | 0.85 | (0.49-1.46) | .55 | 1.07 | (0.62-1.85) | .81 | |
| Adverse risk (MRC) | 1.56 | (1.23-2.00) | <.001 | 1.59 | (1.23-2.04) | <.001 | |
| Marker chromosome | 0.93 | (0.70-1.23) | .61 | 1.19 | (0.89-1.57) | .24 | |
| AML2003 | Age | 1.08 | (0.96-1.20) | .21 | 1.18 | (1.04-1.34) | .01 |
| Prior MDS | 1.02 | (0.73-1.43) | .92 | 0.89 | (0.61-1.29) | .53 | |
| Therapy-related AML | 1.41 | (0.97-2.05) | .08 | 1.94 | (1.31-2.88) | .001 | |
| Adverse risk (MRC) | 1.67 | (1.30-2.13) | <.001 | 1.82 | (1.39-2.38) | <.001 | |
| Marker chromosome | 1.36 | (0.95-1.97) | .10 | 1.16 | (0.79-1.72) | .45 | |
Statistically significant effects are set in bold. P values of the Wald tests for the model coefficients HRs and 95% CIs are given for the respective clinical trials. The Cox models for all patients were stratified by study.
CI, confidence interval; HR, hazard ratio; t-AML, therapy-related acute myeloid leukemia.
In the AML2003 trial, adverse-risk karyotype was a statistically significant variable for both EFS and OS. For OS, additionally, age and therapy-related AML reached statistical significance (Table 3), whereas marker chromosomes did not constitute an independent prognostic marker for EFS or OS, respectively (P = .10 and P = .45). The higher allogeneic transplantation rate in the AML2003 trial of 64.6% among aberrant non-CBF karyotypes as compared with only 23.0% in the AML96 trial could not be shown to account for this discrepancy between the 2 trials in a multivariate analysis with allogeneic transplantation as a time-dependent variable (data not shown).
When multivariate analysis was limited to the adverse-risk subgroup, marker chromosomes were independently statistically significant only in the AML96 trial regarding OS (supplemental Table 2).
Frequency of chromothripsis as detected by array-CGH in marker chromosome karyotypes
Among patients with marker chromosome-positive karyotypes, chromothripsis was detected by array-CGH in as many as 18/49 patients (36.7%), including 3 patients with an array-CGH pattern typical of chromothripsis, although formal diagnostic criteria were marginally missed with 8 or 9 CN changes, respectively (supplemental Table 3). On the contrary, only a single patient (1/34, 2.9%) of the complex aberrant control group without marker chromosomes tested positive for chromothripsis. The association of marker chromosome karyotypes and chromothripsis detection by array-CGH was statistically highly significant (P < .001).
Chromothripsis in marker chromosome karyotypes typically involved a single chromosome (n = 11), with 2 or 3 chromosomes affected in 5 and 2 patients, respectively. There was no predilection for a particular chromosome (supplemental Figure 2). In 12/18 (66.7%) cases, at least 1 of the chromosomes identified as chromothriptic was reported as monosomic in the karyotype formula. Thus, chromothriptic chromosomes seem unrecognizable by metaphase karyotyping and therefore fall into the marker chromosome category, obviously leading to the annotation of a putative chromosomal loss of the affected chromosome. Among the 18 chromothripsis-positive samples, a hypodiploid karyotype was found in 11, a pseudodiploid karyotype in 4, a hyperdiploid karyotype in 2, and a near-tetraploid karyotype in 1 patient, respectively.
When comparing the chromothripsis-positive and -negative subgroups among the marker chromosome-positive patients, clinical characteristics were evenly distributed. As for the cytogenetic categories, chromothripsis-positive patients displayed a particularly high degree of karyotype complexity (Table 4): a complex aberrant karyotype according to MRC criteria (≥4 aberrations) was detected in 100% vs 64.5% (P = .04), a monosomal karyotype in 88.9% vs 45.2% (P = .04), and subclone formation in 88.9% vs 51.6% (P = .13). Markedly, the chromothripsis-positive subgroup also displayed a higher frequency of abnl(17p) with 50.0% vs 16.1%, although statistical significance was lost following adjustment for multiple testing (P = .20).
Clinical characteristics depending on detection of marker chromosomes, chromothripsis, and ring chromosome groups
| . | Group 1 marker chr. pos. chromothripsis pos. (n = 18)* . | Group 2 marker chr. pos. chromothripsis neg. (n = 31) . | P value† group 1 vs 2 . | Group 3 ring chr. pos. (n = 7)* . | Group 4 control group (complex aberrant w/o marker, ring, dm, der) (n = 34) . |
|---|---|---|---|---|---|
| Clinical parameters | |||||
| Age, y (median, range) | 58 (35-76) | 58 (18-77) | 1.0 | 68 (49-78) | 47 (18-75) |
| Gender (male/female) | 8/10 | 17/14 | 1.0 | 4/3 | 18/16 |
| FAB (0/1/2/4,5/6) | 1/5/4/5/3 | 3/6/11/10/1 | 1.0 | 0/0/4/2/1 | 0/14/3/15/0 |
| Prior MDS (no./N) | 2/17 (11.8%) | 5/31 (16.1%) | 1.0 | 0/6 (0%) | 2/34 (5.9%) |
| Leukocytes, /nL (median, range) | 8.5 (1.6-171.3) | 19.4 (0.6-159.8) | 1.0 | 3.8 (2.5-148.6) | 26.5 (0.4-212.0) |
| LDH, U/mL (median, range) | 716 (182-4160) | 518 (165-5489) | 1.0 | 207 (118-849) | 480 (27-3438) |
| Cytogenetic parameters | |||||
| Complex ab. (≥4) (no./N) | 18/18 (100%) | 20/31 (64.5%) | .04 | 5/7 (71.4%) | 34/34 (100%) |
| Monosomal (no./N) | 16/18 (88.9%) | 14/31 (45.2%) | .04 | 5/7 (71.4%) | 9/34 (26.5%) |
| Subclone formation (no./N) | 16/18 (88.9%) | 16/31 (51.6%) | .13 | 4/7 (57.1%) | 25/34 (73.5%) |
| Abnl(5q) (no./N) | 11/17 (64.7%) | 12/31 (38.7%) | 1.0 | 5/7 (71.4%) | 6/34 (17.6%) |
| Abnl(7q) (no./N) | 8/17 (47.1%) | 10/31 (32.3%) | 1.0 | 1/7 (14.3%) | 5/34 (14.7%) |
| Abn(17q) (no./N) | 9/18 (50.0%) | 5/31 (16.1%) | .20 | 2/7 (28.6%) | 5/34 (14.7%) |
| Number markers (median, range) | 2 (1-11) | 1 (1-7) | .01 | 0 (0-11) | 0 (0-0) |
| Derivate chromosome (no./N) | 8/18 (44.4%) | 5/31 (16.1%) | .41 | 2/7 (28.6%) | 0/34 (0%) |
| Remission status | |||||
| CR+CRi | 2/16 (12.5%) | 10/31 (32.3%) | .18 | 2/7 (28.6%) | 10/34 (29.4%) |
| . | Group 1 marker chr. pos. chromothripsis pos. (n = 18)* . | Group 2 marker chr. pos. chromothripsis neg. (n = 31) . | P value† group 1 vs 2 . | Group 3 ring chr. pos. (n = 7)* . | Group 4 control group (complex aberrant w/o marker, ring, dm, der) (n = 34) . |
|---|---|---|---|---|---|
| Clinical parameters | |||||
| Age, y (median, range) | 58 (35-76) | 58 (18-77) | 1.0 | 68 (49-78) | 47 (18-75) |
| Gender (male/female) | 8/10 | 17/14 | 1.0 | 4/3 | 18/16 |
| FAB (0/1/2/4,5/6) | 1/5/4/5/3 | 3/6/11/10/1 | 1.0 | 0/0/4/2/1 | 0/14/3/15/0 |
| Prior MDS (no./N) | 2/17 (11.8%) | 5/31 (16.1%) | 1.0 | 0/6 (0%) | 2/34 (5.9%) |
| Leukocytes, /nL (median, range) | 8.5 (1.6-171.3) | 19.4 (0.6-159.8) | 1.0 | 3.8 (2.5-148.6) | 26.5 (0.4-212.0) |
| LDH, U/mL (median, range) | 716 (182-4160) | 518 (165-5489) | 1.0 | 207 (118-849) | 480 (27-3438) |
| Cytogenetic parameters | |||||
| Complex ab. (≥4) (no./N) | 18/18 (100%) | 20/31 (64.5%) | .04 | 5/7 (71.4%) | 34/34 (100%) |
| Monosomal (no./N) | 16/18 (88.9%) | 14/31 (45.2%) | .04 | 5/7 (71.4%) | 9/34 (26.5%) |
| Subclone formation (no./N) | 16/18 (88.9%) | 16/31 (51.6%) | .13 | 4/7 (57.1%) | 25/34 (73.5%) |
| Abnl(5q) (no./N) | 11/17 (64.7%) | 12/31 (38.7%) | 1.0 | 5/7 (71.4%) | 6/34 (17.6%) |
| Abnl(7q) (no./N) | 8/17 (47.1%) | 10/31 (32.3%) | 1.0 | 1/7 (14.3%) | 5/34 (14.7%) |
| Abn(17q) (no./N) | 9/18 (50.0%) | 5/31 (16.1%) | .20 | 2/7 (28.6%) | 5/34 (14.7%) |
| Number markers (median, range) | 2 (1-11) | 1 (1-7) | .01 | 0 (0-11) | 0 (0-0) |
| Derivate chromosome (no./N) | 8/18 (44.4%) | 5/31 (16.1%) | .41 | 2/7 (28.6%) | 0/34 (0%) |
| Remission status | |||||
| CR+CRi | 2/16 (12.5%) | 10/31 (32.3%) | .18 | 2/7 (28.6%) | 10/34 (29.4%) |
ab., aberrant; chr., chromosome; dm, double minutes; neg., negative; pos., positive; w/o, without.
The single patient with both marker and ring chromosomes is listed in both categories.
Adjusted for multiple testing.
Chromothripsis-positive patients had a particularly dismal prognosis as compared with the chromothripsis-negative group with a combined CR+CRi rate of 2/16 vs 10/31 (P = .18). The chromothripsis-positive subgroup also displayed inferior EFS and OS, although statistical significance was not reached for either endpoint, likely due to the already poor prognosis of the marker chromosome-positive/chromothripsis-negative comparator arm (supplemental Figure 3).
Association of chromothripsis with TP53 mutations
We performed TP53 sequencing in 40 patients, including all chromothripsis-positive patients from the marker chromosome group and the single chromothripsis-positive patient from the control group. The overall frequency of TP53 mutations was 20/40 (50%). The TP53 mutation frequency in chromothripsis-positive patients was 14/19 (73.7%) as compared with only 6/21 (28.6%) in chromothripsis-negative group (P = .01) (supplemental Table 4). Likewise, TP53 mutations were much more abundant among abnl(17p) patients with 13/15 (86.7%) positive patients as compared with 7/25 (28%) in abnl(17p)-negative patients (P < .001). However, there were also 5 chromothripsis-positive cases in both abnl(17p)-negative and TP53 unmutated cases, suggesting that chromothripsis can also occur independently of TP53 aberrations.
Ring chromosomes, double minutes, and derivative chromosomes as products of chromothripsis
Ring chromosomes were detectable in 12/1026 (1.2%) of aberrant non-CBF karyotypes. Seven of these samples, along with another 4 from the registry, could be assessed by array-CGH. The frequency of chromothripsis was as high as 5/7 (71.4%) in the study group and 8/11 (72.7%) in the overall group (supplemental Table 3). Another cytogenetic phenomenon suggestive of chromothripsis is double minutes, which were detectable in 4/1026 of aberrant non-CBF karyotypes (0.4%), with 1 of 2 samples analyzed positive for chromothripsis by array-CGH. Although karyotypes with derivative chromosomes were not specifically considered, 13/49 marker chromosome karyotypes analyzed by array-CGH also included a derivative chromosome, among them 8 chromothripsis-positive samples. In 4/8 of these cases, at least 1 of the chromosomes identified as chromothriptic was annotated as derivative.
M-FISH and barcode FISH
For 3 patients, we performed M-FISH to further characterize their unclassified chromosome rearrangements. In addition, the chromothriptic chromosomes of the respective metaphases were analyzed by barcode FISH.
Patient 1.
Conventional cytogenetic analysis revealed in 19 of 20 analyzed metaphase spreads a male karyotype with an additional ring chromosome of unknown origin. M-FISH analysis identified the origin of the ring chromosome as material from chromosome 8: 47,XY,+r(8)[19]/46,XY[1] (supplemental Figure 4A). Chromothripsis was found by array-CGH in the long arm of chromosome 8 (supplemental Figure 4B) corresponding to the dispersed barcode pattern of chromosome 8q material (supplemental Figure 4C).
Patient 2.
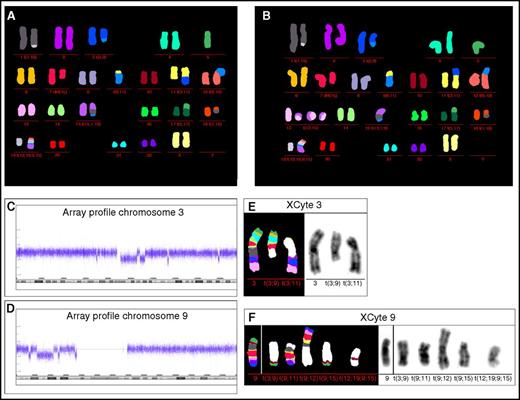
Karyotyping showed the following complex aberrant female karyotype: 44,XX,−1,−2,−3,del(5)(q?)(FISH),del(7)(q22)(FISH),−9,−9,−11,−11,−12,−15,−17,−18,−19,+mar1,+mar2,+mar3, +mar4,+mar5,+mar6,+mar7,+mar8,+mar9,+mar10. In 5 metaphases, a ring chromosome was observed. M-FISH analysis was able to characterize the unclassified marker and ring chromosomes: 39∼45,XX,der(1)t(1;19)[10],der(3)t(3;9)[10],−5[10],del(7q)[10],−9[10],der(9) t(9;11)[10],der(11)t(3;11)[8],der(12)t(9;12)[10],r(13;15)[2],+r(13;15)[2],+r(13;15)[1],der(15)t(15;1;9)[9],der(17)t(5;17)[10],der(18)t(1;18)[10],der(19)t(12;19;9;15)[10][cp10]/46,XX[4] (Figure 3A-B). Chromothripsis was identified for chromosomes 3 and 9 (Figure 3C-D). XCyte 3 and XCyte 9 bar coding revealed chromosome 3 and 9 specific banding pattern on the translocation chromosomes (Figure 3E-F).
M-FISH, barcode FISH, and array profile patient 2. (A-B) M-FISH identified the additional marker and ring chromosomes. Both metaphases show the same aberrations, but in panel B, the ring chromosomes were identified as composed of material from chromosomes 13 and 15. Array profile revealed chromothripsis of the long arm of chromosome 3 and from the short arm of chromosome 9 (C-D). In this patient, the ring chromosome material 13 and 15 showed no chromothriptic pattern in array analysis; however, due to the low number of metaphases showing ring chromosomes (2 of 14), array-CGH may not be able to detect a chromothriptic event. (E) XCyte bar coding of chromosome 3 showed the banding pattern of the normal chromosome 3 and additionally the p-arm banding pattern on the derivative translocation chromosome t(3;9) and long arm banding pattern on the derivative translocation chromosome t(3;11). The centromeric banding pattern of chromosome 3 is not detectable according to the chromothriptic array profile of chromosome 3. (F) XCyte 9 barcode. A normal control hybridization of XCyte barcode 9 is presented on the left side. In the patient, the XCyte barcode is distributed on 5 translocation chromosomes. Chromosomal material involved in chromothriptic region 9p could be localized on the translocation chromosomes t(3;9), t(9;15), and the complex rearranged translocation chromosome t(12;19;9;15). Note, unstained translocation partners are indicated in white.
M-FISH, barcode FISH, and array profile patient 2. (A-B) M-FISH identified the additional marker and ring chromosomes. Both metaphases show the same aberrations, but in panel B, the ring chromosomes were identified as composed of material from chromosomes 13 and 15. Array profile revealed chromothripsis of the long arm of chromosome 3 and from the short arm of chromosome 9 (C-D). In this patient, the ring chromosome material 13 and 15 showed no chromothriptic pattern in array analysis; however, due to the low number of metaphases showing ring chromosomes (2 of 14), array-CGH may not be able to detect a chromothriptic event. (E) XCyte bar coding of chromosome 3 showed the banding pattern of the normal chromosome 3 and additionally the p-arm banding pattern on the derivative translocation chromosome t(3;9) and long arm banding pattern on the derivative translocation chromosome t(3;11). The centromeric banding pattern of chromosome 3 is not detectable according to the chromothriptic array profile of chromosome 3. (F) XCyte 9 barcode. A normal control hybridization of XCyte barcode 9 is presented on the left side. In the patient, the XCyte barcode is distributed on 5 translocation chromosomes. Chromosomal material involved in chromothriptic region 9p could be localized on the translocation chromosomes t(3;9), t(9;15), and the complex rearranged translocation chromosome t(12;19;9;15). Note, unstained translocation partners are indicated in white.
Patient 3.
Chromosome banding analysis of only 5 metaphase spreads identified a complex aberrant male karyotype with 4 varying karyograms: 43,XY,add(2)(p?21),del(5),−7,−21,−21[1]/44,XY,add(2)(p?21),del(5)(q31),−7,−17,−21,+mar[2]/44,XY,add(2),del(5),−7,−21[1]/44,XY,add(2),del(5),−7,−17[1]. M-FISH analysis of 15 metaphase spreads identified 4 different subclones and was able to characterize the complex rearranged chromosomes: 43∼44,XY,der(2p),der(5)t(5;12),−7,der(12)t(7;12),der(16)t(16;21),der(17)t(4;17),der(21)t(17;21),−21[6]/43∼44,der(2)t(2;4),der(5)t(5;12),−7,der(12)t(7;12),der(16)t(16;21), del(17),der(21)t(17;21)[4]/44,XY,der(2)t(2;4),der(5)t(5;12),−7,der(12)t(7;12),der(15)t(1;15),der(16)t(16;21),del(17),der(21)t(17;21)[3]/42∼43,XY,der(5)t(5;12),−7,der(12)t(7;12), der(16)t(16;21),der(17)t(17;21),−21,−21[2] (supplemental Figure 5A-D). Array-CGH identified chromothripsis on chromosome 2p, chromosome 7, and the long arm of chromosome 17 (supplemental Figure 5E-G). XCyte barcode analysis localized the banding pattern of the respective chromosomes 2, 7, as well as 17 in the normal and translocation chromosomes, and furthermore, for some regions of these chromosomes, an aberrant or dispersed banding pattern was recognizable (supplemental Figure 5E-G).
Discussion
This study establishes marker chromosomes as a frequent phenomenon in AML. Marker chromosomes were particularly frequent in adverse-risk karyotypes. Interestingly, marker chromosome frequencies largely differed within the adverse-risk category. In karyotypes with recurrent translocations like inv(3)(q21q26.2) or variant MLL translocations, which define adverse-risk per se, marker chromosome frequency was low. In contrast, complex aberrant and monosomal karyotypes displayed a high frequency of marker chromosomes with 40.3% and 50.2%, respectively. Thus, markers occur together with other manifestations of aneuploidy and appear to be an indicator of chromosomal instability as well as karyotype complexity.
Obviously, marker chromosomes overlap with other unfavorable cytogenetic features, including complex aberrant,4 monosomal,9 abnl(5q),30 abnl(7q),31 and abnl(17p)15-18 karyotypes. However, the poor prognosis of marker chromosomes does not purely reflect their association with these adverse-risk categories as, at least in the AML96 trial, marker chromosomes proved to be an independent risk factor even when adverse-risk cytogenetics3 were included as a covariable in a multivariate model. Thus, marker chromosomes add prognostic information by themselves and do not merely reflect the poor prognosis of established adverse-risk criteria.
Unlike the AML96 study, marker chromosomes did not entail an independent adverse prognosis by multivariate analysis in the AML2003 trial. One possible explanation for this discrepancy is that the AML96 trial was larger than the AML2003 trial and also included elderly patients, which translated into a higher absolute number of patients with aberrant karyotypes and marker chromosomes.12,21
It lies in the very nature of marker chromosomes to be heavily rearranged and to reflect gross chromosomal damage. Therefore, they are candidates to have arisen from chromothripsis. This recently discovered phenomenon denotes a single catastrophic event in which a chromosome arm or whole chromosome is shattered into numerous fragments.32-35 After an error-prone reparation process, in which the chromosomal segments are pieced back together again, the chromosome reemerges heavily rearranged. In tune with this concept, 2 AML patients with chromothripsis displayed marker chromosomes by metaphase karyotyping in a case report by Mackinnon and Campbell.36
In our study, we unequivocally detected chromothripsis in about one-third of marker chromosome karyotypes, whereas this phenomenon was virtually absent in the control group composed of marker chromosome-negative complex aberrant karyotypes. We conclude that in many cases marker chromosomes reflect heavily rearranged chromosomes following chromothripsis. Chromothripsis negativity likely reflects the fact that marker chromosomes are heterogeneous, ranging from heavily rearranged chromothriptic chromosomes to chromosomes with lesser structural aberrations, which nevertheless impaired proper identification.37,38 In any case, marker chromosome-positive karyotypes should raise the suspicion of chromothripsis, which helps to select candidates for follow-up analyses by array-CGH, multicolor FISH, or sequencing approaches.
In our analysis, chromothripsis-positive marker chromosome karyotypes displayed a particularly high level of karyotype complexity: All 18 chromothripsis-positive samples met the MRC criteria of karyotype complexity, which require a minimum of 4 aberrations. A previous study suggested that chromothripsis is most abundant in TP53-mutated AML cases,23 but this association was not observed by others.39 We found strongly elevated frequencies of abnl(17p) and TP53 mutations in chromothripsis-positive marker karyotypes, thus confirming the association between chromothripsis and TP53 mutations previously published by Rausch et al.23 However, chromothripsis is not exclusively reserved to abnl(17p) and TP53 mutated cases.
Although chromothripsis has not been thoroughly studied in AML yet, it seems to be associated with poor prognosis in several other malignancies.23,35,40-43 In line with these observations, the prognosis of the marker chromosome/chromothripsis double-positive subgroup in this study was dismal. Thus, chromothripsis likely contributes to the poor prognostic impact of marker chromosomes in general in AML.
Ring chromosomes have been described to arise from chromothriptic fragmentation in liposarcoma, where ring formation appears to stabilize the fragmented chromosome.44 Our study now shows that AML ring chromosomes, which are rare in this disorder, typically evolve from chromothripsis as well. Previous studies have established that double minutes arise as well from chromothripsis and the subsequent repair of chromosomal fragments, which cannot be stitched back together again to full chromosomes.23,32,36,45,46 A recent study in AML has shown that double-minute karyotypes in AML are associated with complex aberrant karyotypes, frequent abnl(17p)/TP53 mutations, micronuclei formation, myelodysplastic features, and dismal prognosis, which in combination are highly suggestive hallmarks of chromothripsis.47 As further proof, these AML samples also displayed an amplification of MYC as a feature of proliferation and genomic instability47 reminiscent of the prototypically chromothriptic small cell lung cancer line SCLC-21H, which displays a large number of double minutes as well.32 Our data also suggest that derivate chromosomes are possible by-products of chromothripsis as previously suggested.36,48,49
In conclusion, this is the first study showing that marker chromosomes are a frequent finding predominantly in adverse-risk AML and independently associated with an adverse prognosis. We confirm chromothripsis as a genetic phenomenon in AML and show for the first time that it is associated with marker chromosome formation.
The data reported in this article have been deposited in the Gene Expression Omnibus database (accession number GSE93886).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank all physicians and patients of the Study Alliance Leukemia study group and all geneticists contributing metaphase karyotyping results. Furthermore, the authors thank Michelle Ebentheuer, Evelyn Fey, Alexandra Koeppel, and Kristin Schmitt for excellent technical assistance in CN analysis as well as Brigitte Schoell for excellent technical assistance in performing M-FISH and bar coding FISH experiments.
This work was supported by the Wilhelm-Sander Foundation for cancer research (A.K., A.J., T.B.). M.K.-K. was supported by the Rahel Goitein-Straus Program of the Medical Faculty Heidelberg.
Authorship
Contribution: T.B. and A.K. designed the study; M.G., F.S., B.M., and A.J. undertook cytogenetic analysis; M.G., M.K.-K., K.H., and A.J. performed array-CGH analysis; V.E., M. Kirchner, and A.S. undertook TP53 testing; C.T. undertook NPM1 and FLT3-ITD testing; T.B., F.S., M.B., G.E., A.D.H., and A.K. included patients as investigators; T.B., M.G., F.S., B.M., C.E.H., M. Kramer, C.T., M.B., G.E., A.D.H., A.J., and A.K. collected data; C.K. and A.B. performed statistical analysis; T.B., M.G., F.S., C.K., A.B., A.J., and A.K. interpreted data; and T.B., M.G., C.K., A.S., and A.K. wrote the manuscript. All authors revised and approved the manuscript.
Conflict-of-interest disclosure: C.T. indicates ownership of AgenDix. The remaining authors declare no competing financial interests.
A complete list of the members of the Study Alliance Leukemia investigators appears in “Appendix.”
Correspondence: Alwin Krämer, Clinical Cooperation Unit Molecular Hematology/Oncology, German Cancer Research Center, Department of Internal Medicine V, University of Heidelberg, Im Neuenheimer Feld 280, 69120 Heidelberg, Germany; e-mail: a.kraemer@dkfz.de.
Appendix
Study Alliance Leukemia investigators: Aachen: T. Brümmendorf, S. Koschmieder; Altenburg: A. Schulz-Abelius; Bamberg: R. Seggewiß-Bernhardt; Bayreuth: A. Kiani; Berlin: B. Dörken, A. Pezzutto, C. Baldus; Berlin Vivantes: C. Scholz; Bielefeld: M. Görner; Bochum: D. Behringer; Bremen: R. Trappe; Brno: J. Mayer; Chemnitz: M. Hänel; Dresden: G. Ehninger, M. Bornhäuser, C. Röllig, C. Thiede, U. Platzbecker; Düren: M. Flaßhove; Erlangen: A. Mackensen, S. Krause; Essen: U. Dührsen, R. Noppeney; Frankfurt (Oder): M. Kiehl; Frankfurt (Main): H. Serve, C. Brandts, G. Bug, B. Steffen; Fulda: H.G. Höffkes; Halle: C. Müller-Tidow; Hamburg St. Georg: N. Schmitz, A. Elmaagacli; Heidelberg: A.D. Ho, A. Krämer; Hildesheim: U. Kaiser; Jena: A. Hochhaus; Kaiserslautern: H. Link; Koblenz: R. Naumann; Leipzig St. Georg: L. Mantovani Löffler; Marburg: A. Neubauer; Münster: W. Berdel; Nürnberg: M. Wilhelm, K. Schäfer-Eckart; Offenburg: A. Jakob; Regensburg: W. Herr, A. Reichle; Riesa: J. Schubert; Rotenburg: F. Heits; Schwäbisch-Hall: T. Geer; Sindelfingen: M. Ritter; Stuttgart: E. Aulitzky, M. Kaufmann; Trier: R. Mahlberg; Wiesbaden: N. Frickhofen; Winnenden: M. Schaich; Würzburg: H. Einsele, V. Kunzmann.
References
Author notes
T.B. and M.G. contributed equally to this study.
A.J. and A.K. contributed equally to this study.