Key Points
Rs12041331 is the first functional CpG-SNP related to platelet function whose regulatory mechanism depends on DNA methylation.
Rs12041331 marks allele-specific methylation at the CpG island encompassing the first untranslated exon during megakaryopoiesis.
Abstract
Genetic variation in the PEAR1 locus is linked to platelet reactivity and cardiovascular disease. The major G allele of rs12041331, an intronic cytosine guanine dinucleotide-single-nucleotide polymorphism (CpG-SNP), is associated with higher PEAR1 expression in platelets and endothelial cells than the minor A allele. The molecular mechanism underlying this difference remains elusive. We have characterized the histone modification profiles of the intronic region surrounding rs12041331 and identified H3K4Me1 enhancer-specific enrichment for the region that covers the CpG-SNP. Interestingly, methylation studies revealed that the CpG site is fully methylated in leukocytes of GG carriers. Nuclear protein extracts from megakaryocytes, endothelial cells, vs control HEK-293 cells show a 3-fold higher affinity for the methylated G allele compared with nonmethylated G or A alleles in a gel electrophoretic mobility shift assay. To understand the positive relationship between methylation and gene expression, we studied DNA methylation at 4 different loci of PEAR1 during in vitro megakaryopoiesis. During differentiation, the CpG-SNP remained fully methylated, while we observed rapid methylation increases at the CpG-island overlapping the first 5′-untranslated region exon, paralleling the increased PEAR1 expression. In the same region, A-allele carriers of rs12041331 showed significantly lower DNA methylation at CGI1 compared with GG homozygote. This CpG-island contains binding sites for the methylation-sensitive transcription factor CTCF, whose binding is known to play a role in enhancer activation and/or repression. In conclusion, we report the molecular characterization of the first platelet function-related CpG-SNP, a genetic predisposition that reinforces PEAR1 enhancer activity through allele-specific DNA methylation.
Introduction
PEAR1 encodes the Platelet-endothelial aggregation receptor 1 (PEAR1), a transmembrane tyrosine kinase receptor predominantly expressed in platelets and endothelial cells.1 In platelets, PEAR1 is involved in cell activation through its engagement in sustaining activation of the platelet integrin, αIIbβ3,2 and platelet FcεR1α has been identified as one of its activating ligands.3 PEAR1 is progressively more actively transcribed toward the later stages of megakaryocyte (MK) specification,4 following a similar pattern as that observed for GATA1.5,6 PEAR1 knock-down in CD34+ hematopoietic stem cells (HSCs) enhances megakaryopoietic progenitor proliferation,4 while in endothelial cells, it is implicated in the regulation of neo-angiogenesis.7 Both processes occur in a PI3K/PTEN-dependent manner.
Several epidemiological studies have identified PEAR1 as 1 potential candidate gene to explain platelet function variability in both healthy and pathological conditions.8-19
The noncoding PEAR1 single-nucleotide polymorphism (SNP) rs12041331 A allele was associated with lower PEAR1 expression9 and platelet response to agonists in different reports,8,9,13,14,17,19 whereas among patients with coronary artery disease taking antiplatelet agents, rs12041331 A-allele carriers surprisingly appeared to experience more adverse cardiovascular outcomes and had higher death rates than GG homozygotes.14 Rs12041331 seems to play a role in driving endothelial cell function too.20 Rs12041331 is a polymorphism that removes a cytosine guanine dinucleotide (CpG) site in the DNA sequence and is defined as a “CpG-SNP.” CpG sites in mammals influence gene regulation, dependent on their methylation status. DNA methylation has been extensively described in the control of cell specification, cancer, and other human diseases21 and cooperates with histone modifications in rendering the chromatin more or less accessible for transcription factors, thereby influencing gene expression.22 Despite reported associations of CpG-SNPs with multiple diseases,23-29 the role of CpG-SNPs in regulating platelet phenotypes was never investigated.
Two studies have already shown that PEAR1 rs12041331 G/A substitution leads to lower platelet PEAR1 expression9 and reduces endothelial cell migration in carriers of the A allele.20 In this study, we have addressed the transcriptional importance of CpG-SNP rs12041331 in the context of the PEAR1 gene and have identified, for the first time, allele-specific methylation as a regulatory mechanism capable of modulating gene expression at more than 1 level. We found that this type of epigenetic regulation contributes to the fine tuning of PEAR1 expression and provides a molecular explanation for population variability in PEAR1-dependent platelet function.
Methods
All human samples involved in the study were obtained from healthy volunteers after giving written informed consent. The study was approved by the Medical Ethical Committee of the University of Leuven (Reference number: S56976).
Chromatin immunoprecipitation (ChIP) assay
ChIP experiments have been performed using the megakaryocytic CHRF cell line. The detailed protocol, adapted from Pistoni et al.,30 is reported in supplemental Methods, available on the Blood Web site. We designed 10 amplicons P1-P10 (characteristics and genomic location are reported in supplemental Table 1) The ChIP-quantitative polymerase chain reaction (qPCR) data relative to histones are presented as % input/tot H3: 2^(−[CT Histone modification – CT 10% input])/2^(−[CT Total H3 – CT 10% input]).
Electrophoretic Mobility Shift Assays (EMSAs)
Nuclear proteins were extracted from CHRF-288-11, MEG01, EA.hy926, and HEK-293 cells, and their total protein concentration was determined by Bradford analysis. Single-stranded 33-bp 5′ biotin-labeled oligonucleotides were synthesized by Integrated DNA Technology (Integrated DNA Technologies, Leuven, Belgium) and then annealed to generate double-stranded probes. Three-probe combinations were generated containing either the A or the G allele with or without methylation at its preceding C, at the CpG-SNP position. Specific details on probes are reported in supplemental Table 2.
Luciferase gene-reporter assay
The PGL3 promoter vector and assay reagents were bought from Promega Corporation (Fitchburg, WI). A 223-bp DNA fragment containing the CpG-SNP was amplified (chr1:156899830-156900052, GRCh38/hg38 Assembly) by PCR with DNA from 2 subjects homozygous for the rs1204331 A/G polymorphism (G/G and A/A, respectively) and inserted in a PGL3 promoter vector (further details on protocol are reported in the supplemental Methods). The PGL3 plasmids and GFP control vector were cotransfected into HEK-293 cells using Polyplus transfection kit (Polyplus, Illkirch, France). Enhancer activity was expressed by relative light units of luciferase normalized to the luciferase measured in empty pGL3-promoter vector. One-way analysis of variance (ANOVA) was used to determine the significance of differences in luciferase expression among PGL3 promoter, with a significance level defined at P < .05.
Human CD34+ stem cell differentiation in vitro
Human CD34+ HSCs were separated by magnetic cell sorting from buffy coats isolated from healthy donor peripheral blood (Milteny Biotech, Auburn, CA) using a protocol modified from Kauskot et al.4 and described in the supplemental Methods.
The cultured CD34+ cells were harvested on days 0, 7, and 14, washed in Dulbecco phosphate-buffered saline before the cell pellets were either snap-frozen in liquid nitrogen and stored at −80°C for further DNA and RNA extraction or processed to detect CD34, CD41, and PEAR1 expression by flow cytometry using antigen-presenting cell–labeled CD34, fluorescein isothiocyanate–labeled CD41 (Becton Dickinson), and anti-hPEAR1 PE (clone: FAB45278; R&D Systems). PEAR1 and GAPDH expression analysis by quantitative reverse transcription (RT)-PCR was performed as described in the supplemental Methods.
DNA methylation analysis
CpG-SNP methylation was assessed by bisulfite-sequencing or bisulfite cloning and sequencing, using primers reported in supplemental Table 3. PEAR1 (CGI1, CGI2, and intron 1 region) methylation was evaluated using the Sequenom EpiTYPER MassARRAY platform as described.31,32 Details for the protocol used are described in the supplemental Methods. Statistical analysis was performed using one-way ANOVA with Friedman test, with a significance level of P < .05 to study the progression of methylation during MK differentiation and an unpaired t test, with a significance level of P < .05 to investigate differences between G and A allele carriers.
Results
Transcriptional activity control by epigenetic modification of rs12041331
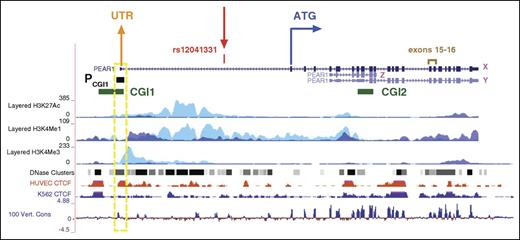
PEAR1 is encoded by 23 exons located on chromosome 1q23.1 (Figure 1) with 2 CpG islands covering (1) the first untranslated exon region (CGI1) and (2) the region comprising exons 8-9 (CGI2), both with a potential regulatory role in the expression of PEAR1. Several transcripts arise from the PEAR1 promoter but only 2 of them are translated into a full receptor (Figure 1). The G-to-A polymorphic variant rs12041331 (CpG-SNP) is located within a noncoding region of about 10 kb that links the first untranslated exon to the transcription start site of PEAR1, located downstream of the CpG-SNP (Figure 1). We examined the histone modification profile of this region obtained from ChIP-seq data in megakaryoblastic K562 and human umbilical vein endothelial cells (HUVECs) available in ENCODE. In general, gene activity is linked to high levels of H3K4Me3 and H3K27Ac.33-37 Enhancer regions are typically enriched for H3K4Me1 marking, while poised enhancers harbor H3K4me1 and H3K27Me3, and active enhancers are comarked by H3K4Me1 and H3K27Ac.38-40 From this first analysis, the PEAR1 intronic region where the CpG-SNP is located shows enrichment for histone modifications H3K4Me1 and H3K27Ac, with particular higher deposition in HUVECs (light blue peaks in Figure 1), and a DNase Hypersensitivity I-dependent open chromatin conformation (Figure 1).41
PEAR1 gene structure (chr1:156893731-156916434, GRCh38/hg38 Assembly). Exons are indicated by full blue boxes. The ATG codon is indicated at exon 1. The major PEAR1 transcript (ENST00000292357), translated into the full PEAR1 receptor, is indicated as “X” (in magenta). Two CpG islands are located in the gene (CGI1 and CGI2, depicted as green boxes on the left and right side, respectively; Location in chromosome 1: 156892430‐156893919 and 156907978‐156908857, GRCh38/hg38 Assembly). Rs12041331 (chr1:156899922, GRCh38/hg38 Assembly) is indicated in red. CpG islands are defined based on the formula described by Gardiner-Garden. H3K4Me1, H3K4Me3, and H3K27Ac profiles in HUVEC and K562 cell lines are displayed as colored overlayed histograms (light blue for HUVEC and purple for K562) in “auto-scale to data view” mode that takes the highest signal in the selected region as the 100% of the intensity and displays all other signals accordingly (data produced by the Bernstein Laboratory at the Broad Institute and the University of California, Santa Cruz and part of the ENCODE database). DNase Hypersensitivity 1 regions are displayed as gray to black boxes (from less to more open chromatin conformation). CTCF ChIP-seq data from HUVECs and K562 cell lines are depicted as colored histograms in full visualization and are normalized for immunoglobulin G signals (data produced by ENCODE). PEAR1 conservation across vertebrates is displayed as blue histograms at the bottom of the figure using the Vertebrate Multiz Alignment and Conservation (100 species) University of California, Santa Cruz track. “PCGI1” (also depicted as black box) refers to the region we analyzed for DNA methylation estimates in CD34+ HSCs into MK precursors and is better described in Figure 5. A yellow box (broken line) highlights the PCGI1 region that colocalizes with CTCF binding in HUVECs and shows conservation across different vertebrates. Exons 15 and 16 are indicated in brown and indicate the region used for RT-qPCR quantification of PEAR1 expression in the present study.
PEAR1 gene structure (chr1:156893731-156916434, GRCh38/hg38 Assembly). Exons are indicated by full blue boxes. The ATG codon is indicated at exon 1. The major PEAR1 transcript (ENST00000292357), translated into the full PEAR1 receptor, is indicated as “X” (in magenta). Two CpG islands are located in the gene (CGI1 and CGI2, depicted as green boxes on the left and right side, respectively; Location in chromosome 1: 156892430‐156893919 and 156907978‐156908857, GRCh38/hg38 Assembly). Rs12041331 (chr1:156899922, GRCh38/hg38 Assembly) is indicated in red. CpG islands are defined based on the formula described by Gardiner-Garden. H3K4Me1, H3K4Me3, and H3K27Ac profiles in HUVEC and K562 cell lines are displayed as colored overlayed histograms (light blue for HUVEC and purple for K562) in “auto-scale to data view” mode that takes the highest signal in the selected region as the 100% of the intensity and displays all other signals accordingly (data produced by the Bernstein Laboratory at the Broad Institute and the University of California, Santa Cruz and part of the ENCODE database). DNase Hypersensitivity 1 regions are displayed as gray to black boxes (from less to more open chromatin conformation). CTCF ChIP-seq data from HUVECs and K562 cell lines are depicted as colored histograms in full visualization and are normalized for immunoglobulin G signals (data produced by ENCODE). PEAR1 conservation across vertebrates is displayed as blue histograms at the bottom of the figure using the Vertebrate Multiz Alignment and Conservation (100 species) University of California, Santa Cruz track. “PCGI1” (also depicted as black box) refers to the region we analyzed for DNA methylation estimates in CD34+ HSCs into MK precursors and is better described in Figure 5. A yellow box (broken line) highlights the PCGI1 region that colocalizes with CTCF binding in HUVECs and shows conservation across different vertebrates. Exons 15 and 16 are indicated in brown and indicate the region used for RT-qPCR quantification of PEAR1 expression in the present study.
PEAR1 is mostly expressed in MKs, platelets, and endothelial cells.1 To be able to integrate and validate ENCODE data relative to histone modifications within the CpG-SNP region, we used 4 different hematopoietic cell lines (erythroblastoid K562, myeloid MEG01, and megakaryocytic DAMI and CHRF) to identify the 1 with the highest expression of PEAR1. Via quantitative RT-PCR, we observed that the CHRF cells express 8 times higher PEAR1 levels compared with K562, whereas MEG01 and DAMI show an intermediate PEAR1 expression (supplemental Figure 1). CHRF cells were, therefore, used to perform a series of ChIP experiments. We have designed 10 different amplicons to cover the full PEAR1 intron region, including the specific CpG-SNP position (amplicons P5-6) (Figure 2A). A glyceraldehyde-3-phosphate dehydrogenase (GAPDH) promoter-specific amplicon was used as positive or negative control to evaluate enrichment of 4 different histone modifications: H3K27Ac, H3K4Me3, H3K27Me3, and H3K4Me1. H3K27Ac and H3K4Me3 were strongly enriched at the GAPDH promoter amplicon, whereas a mild enrichment was observed for PEAR1 amplicons P2 and P3, indicating that the PEAR1 intron 1 region in CHRF cells does not contain promoter activity (Figure 2B). On the contrary, H3K4Me1, a known histone modification often associated with enhancers, was also enriched in amplicons P1-P6, including the CpG-SNP region (P5 and P6) over the GAPDH promoter signal for the same histone, whereas it was detectable in amplicons P7 to P10 (just upstream to the ATG starting codon) in a comparable way as the GAPDH promoter amplicon (Figure 2B). To further characterize the enhancer features of the PEAR1 intron 1 region, we analyzed the H3K27Me3 deposition in the same CHRF samples. H3K27Me3 in combination with H3K4Me1 defines closed or poised enhancers.42 We found that its enrichment was not significantly different from the signal for the GAPDH promoter region, thus suggesting that the PEAR1 intron 1, including the CpG-SNP region, contains a primed enhancer with potential binding sites for transcription factors that can regulate PEAR1 expression in the megakaryocytic specific CHRF cell line. The CpG site generated by the major G allele of PEAR1 CpG-SNP is a unique event in a region of around 200 bp downstream in the same locus, representing a relevant functional change that could influence gene regulation. We have genotyped leukocyte DNA from a total of 90 healthy individuals for the CpG-SNP: 60 European Caucasians and 30 east or south Asians. We identified 44 G/G (73.3), 16 G/A (26.7) in the Caucasian group and found 12 G/G (40.0), 14 G/A (46.7), and 4 A/A (13.3) individuals in the Asian group. Allelic frequency in both groups was in line with the Minor Allele Frequency of rs12041331 in Caucasian European (MAF = 0.9) and in east and south Asians (MAF = 0.46 and 0.37, respectively). We next investigated the methylation status of the CpG-SNP in 5 G/G, 3 G/A, and 2 A/A individuals via bisulfite-cloning and sequencing. This analysis revealed a fully methylated status of the C preceding the G in all G allele carriers (supplemental Figure 2). Full methylation for the C preceding the G was also observed in the different cell lines. This evidence indicated that the G allele is always characterized by a fully methylated C.
Histone modifications profiling of PEAR1 intron 1 region via ChIP. (A) Graphical representation of PEAR1 intron 1 region. Primers were randomly selected to cover the full intronic region, including rs12041331 (amplicons P5 and P6 in correspondence of rs12041331 indicated above the gene line) and are depicted as black boxes. (B) Levels of CHRF PEAR1 intron 1 (P1-P10) and GAPDH amplicons immunoprecipitated using H3K27me3, H3K4me3, H3K27ac, and H3K4me1 antibodies were measured following PCR amplification of DNA fragments. Signals from immunoglobulin G control antibody used for each ChIP were subtracted from each of the histone modification immunoprecipitation signals and the fold enrichment ratios of ChIP enriched vs total input DNA were represented (n = 4 for each histone, average ± standard error of the mean).
Histone modifications profiling of PEAR1 intron 1 region via ChIP. (A) Graphical representation of PEAR1 intron 1 region. Primers were randomly selected to cover the full intronic region, including rs12041331 (amplicons P5 and P6 in correspondence of rs12041331 indicated above the gene line) and are depicted as black boxes. (B) Levels of CHRF PEAR1 intron 1 (P1-P10) and GAPDH amplicons immunoprecipitated using H3K27me3, H3K4me3, H3K27ac, and H3K4me1 antibodies were measured following PCR amplification of DNA fragments. Signals from immunoglobulin G control antibody used for each ChIP were subtracted from each of the histone modification immunoprecipitation signals and the fold enrichment ratios of ChIP enriched vs total input DNA were represented (n = 4 for each histone, average ± standard error of the mean).
To evaluate the potential transcriptional activity of the G vs the A allele, we have used a luciferase gene reporter assay to investigate if a given DNA sequence is able to produce gene transcription as an enhancer region. For this purpose, we used the pGL3-promoter vector in which a sequence of about 200 bp flanking the CpG-SNP has been inserted. In agreement with previously reported findings,9 the G allele was associated with considerably higher luciferase transcription than the A allele (supplemental Figure 3), demonstrating a sequence-dependent role of this CpG-SNP in driving differential gene expression. Even the vectors used for the luciferase-gene reporter assay, after they had been purified from competent bacteria, showed a constantly complete methylation status for the G allele, directly linking this specific genetic-epigenetic combination to the higher transcriptional activity in the reporter assay.
To investigate the influence of methylation on the nuclear protein binding properties for G allele, we performed a classical EMSA using 5′-biotinylated probes corresponding to location chr1:156899906-156899938 (GRCh38/hg38 Assembly) and including the CpG-SNP. More specifically, we used 3 different double-stranded probes containing the G allele either with or without methylation at the C preceding the G (referred to as Gmeth and G, respectively) or the A allele and incubated these with nuclear extracts from CHRF, MEG01, EA.hy926, but also HEK-293 cells. In all conditions, the Gmeth resulted in significantly increased recruitment of nuclear proteins compared with the G allele, non-methylated at the preceding C, or the A allele (Figure 3), with different DNA-protein complexes formed for the various genotypes analyzed (Figure 3). Additional controls with scrambled 5′-biotinylated probes confirmed that increased nuclear protein binding only occurred when methylation was indeed present at the CpG-SNP position and not at another cytosine in the scrambled probe (not shown).
Transcriptional activity of rs12041331 and enhancing role of methylation at the G allele. EMSA analysis to characterize the methylated G allele vs the G allele and A allele binding properties to nuclear proteins. EMSA were performed using 5′biotinylated probes corresponding to location chr1:156869698-156869731 (GRCh38/hg38 Assembly) and including rs12041331. (A,C) Biotinylated double-stranded probes containing the G allele with (“Gmeth”) or without (“G”) methylation, or the A allele (“A”), were incubated in the presence of a 10× amount of cold probe (lanes 1-3), or alone with a fixed amount of nuclear extract from CHRF (A) or MEG01, EA.hy926, or HEK-293 cells (C). Lanes 7-9 show unbound biotinylated probes in all cases. Methylation significantly enhanced binding of the probe to nuclear extract proteins as shown by the increased normalized intensity of the signal in lane 4 vs that in lane 5 and 6 (B), quantified for 3 replicates of CHRF cells (A, mean ± standard deviation) and for the other 3 cell lines investigated (C), expressed as lane Integrated Density. (D) PROMO-based putative binding sites search for CpG-SNP region (25 bp). The G allele (top panel) produced specific binding prediction for c-Jun (indicated as number 7) and ATF3 (indicated as number 8), whereas for the A allele (bottom panel), YY1 (indicated as number 3) is predicted to bind the SNP site.
Transcriptional activity of rs12041331 and enhancing role of methylation at the G allele. EMSA analysis to characterize the methylated G allele vs the G allele and A allele binding properties to nuclear proteins. EMSA were performed using 5′biotinylated probes corresponding to location chr1:156869698-156869731 (GRCh38/hg38 Assembly) and including rs12041331. (A,C) Biotinylated double-stranded probes containing the G allele with (“Gmeth”) or without (“G”) methylation, or the A allele (“A”), were incubated in the presence of a 10× amount of cold probe (lanes 1-3), or alone with a fixed amount of nuclear extract from CHRF (A) or MEG01, EA.hy926, or HEK-293 cells (C). Lanes 7-9 show unbound biotinylated probes in all cases. Methylation significantly enhanced binding of the probe to nuclear extract proteins as shown by the increased normalized intensity of the signal in lane 4 vs that in lane 5 and 6 (B), quantified for 3 replicates of CHRF cells (A, mean ± standard deviation) and for the other 3 cell lines investigated (C), expressed as lane Integrated Density. (D) PROMO-based putative binding sites search for CpG-SNP region (25 bp). The G allele (top panel) produced specific binding prediction for c-Jun (indicated as number 7) and ATF3 (indicated as number 8), whereas for the A allele (bottom panel), YY1 (indicated as number 3) is predicted to bind the SNP site.
The formation of different DNA-protein complexes between the Gmeth vs A probes are suggestive for genotype-specific and methylation-dependent nuclear protein binding. To speculate whether the CpG-SNP disrupts possible transcription factor binding sites (TFBS), we have investigated the putative CpG-SNP region (25 bp) using the PROMO software with the presence of the G vs A allele.43,44 PROMO uses TRANSFAC databases to search for putative TFBS. PROMO analysis generated a prediction of at least 10 different interacting transcription factors for each of the alleles, but with 2 predicted to bind exclusively to the G allele (ie, ATF3 and c-Jun), whereas YY1 only binds only to the A allele (Figure 3). However, this software does not account for the impact of methylation status for the G allele on nuclear protein binding.
Role of CpG-SNP and DNA methylation in primary CD34+ HSC proliferation and differentiation to MKs
Our group previously reported a significant increase of PEAR1 expression in CD34+ HSC during their differentiation into MK.4
To elucidate whether increase in PEAR1 expression during MK differentiation would also be associated with the CpG-SNP, we isolated CD34+ cells from peripheral blood of 11 healthy donors (8 G/G, 2 A/G, and 1 A/A) and induced in vitro MK differentiation for 14 days. Cells were harvested at day 0, day 7 (proliferation phase), and day 14 (maturation phase). During this timeframe, MK differentiation (CD34 and CD41a) and PEAR1 expression were monitored via fluorescence-activated cell sorter (FACS) analysis (Figure 4A left panel). The analysis shows that CD41a expression is intermediate at day 7, whereas PEAR1 expression already reached maximal levels at this time point. Analysis of the median fluorescence intensity at this time point for all individuals revealed further and progressive differentiation for CD41a from day 7 to 14, whereas PEAR1 remained unchanged during MK maturation (Figure 4A right panel).
Influence of Rs12041331 on MK progenitor proliferation and PEAR1 expression during MK differentiation. (A) Left panel: histogram for CD41a and PEAR1 during differentiation of CD34+ cells from day 0 to day 14, as indicated. Right panel: CD34, CD41a, and PEAR1 expression measured by FACS analysis and expressed as mean fluorescence intensity during differentiation of CD34+ cells into MK precursors harvested at day 0 (D0), day 7 (D7), and day 14 (D14). (B) Left panel: Number of CFU-MK, defined as small (3-20 cells), medium (20-80 cells), and large (>80 cells) colonies, counted in a blinded fashion at day 8 after differentiation in GG (n = 3) vs AG+AA (n = 3) individuals. *P < .05, multiple unpaired Student t test with post-Holm-Sidak for multiple comparisons. Right panel: GG (n = 2) vs AG+AA (n = 3) PEAR1/CD41a ratios measured on CD41a-positive events at day 7 of differentiation. SP1, SP2, SP3 are CD41a-positive subpopulations defined by marker positivity and size (Forward Scatter, FSC) as defined in supplemental Figure 4. Comp-FITC-A, comparative mean fluorescene intensity for FITC-labeled antibody against CD41a; Comp-PE-A, comparative mean fluorescence intensity for PE-labeled antibody against PEAR1.
Influence of Rs12041331 on MK progenitor proliferation and PEAR1 expression during MK differentiation. (A) Left panel: histogram for CD41a and PEAR1 during differentiation of CD34+ cells from day 0 to day 14, as indicated. Right panel: CD34, CD41a, and PEAR1 expression measured by FACS analysis and expressed as mean fluorescence intensity during differentiation of CD34+ cells into MK precursors harvested at day 0 (D0), day 7 (D7), and day 14 (D14). (B) Left panel: Number of CFU-MK, defined as small (3-20 cells), medium (20-80 cells), and large (>80 cells) colonies, counted in a blinded fashion at day 8 after differentiation in GG (n = 3) vs AG+AA (n = 3) individuals. *P < .05, multiple unpaired Student t test with post-Holm-Sidak for multiple comparisons. Right panel: GG (n = 2) vs AG+AA (n = 3) PEAR1/CD41a ratios measured on CD41a-positive events at day 7 of differentiation. SP1, SP2, SP3 are CD41a-positive subpopulations defined by marker positivity and size (Forward Scatter, FSC) as defined in supplemental Figure 4. Comp-FITC-A, comparative mean fluorescene intensity for FITC-labeled antibody against CD41a; Comp-PE-A, comparative mean fluorescence intensity for PE-labeled antibody against PEAR1.
To evaluate a possible influence of the CpG-SNP on MK progenitors during the first 7 differentiation days, we performed a clonogenic culture assay of CD34+ HSC isolated from 3 G/G, 2 A/G, and 1 A/A carriers (all Asians). Total colony-forming unit (CFU)-MK colonies were counted in a blinded manner at day 8 after isolation, and although the total number of colonies was not different according to the genotype, we observed significant differences in the size of single MK colonies. The A allele carriers had significantly larger CFU-MK colonies compared with G/G carriers that mainly have small MK colonies (Figure 4B left panel). We did not obtain enough cells to study MK ploidy, but genetic defects in MK maturation, ploidy, and proplatelet formation (eg, GATA1,45,46 DIAPH147 ) typically have enlarged and dense MK colonies. In addition, we have shown before that human PEAR1 knock-down MK progenitors proliferate more than progenitors that can upregulate PEAR1 during differentiation.4
Quantitative RT-PCR (n = 4) also confirmed that PEAR1 expression increases during differentiation, as reported before4 (data not shown). To investigate whether the PEAR1 expression would be different in cells committed toward MK maturation, we investigated the median fluorescence intensity of PEAR1 in CD41a-positive cells (selection shown in supplemental Figure 4). We were not able to identify significant differences in the CD41a-normalized PEAR1 expression between G/G and A allele carriers (Figure 4B right panel) at day 7 or day 14 of MK progenitor differentiation.
During HSC differentiation to MKs, the C preceding the G variant in the G carriers was constantly fully methylated. To identify whether other methylation-sensitive loci would play a role in PEAR1 expression, we selected 3 other regions within the PEAR1 locus (Figure 5A) and performed methylation estimate measurements using the Sequenom EpiTYPER. This methylation analysis involves both CpG islands (“PCGI1” and “PCGI2” in Figure 5A) and the P3 intronic region where the strongest enrichment of histone H3K27Me3 and H3K4Me1 was observed in previous ChIP experiments (indicated as “Pintron1” in Figure 5A) for a total of 67 CpG sites. Significantly increasing methylation was only observed during MK progenitor differentiation for the CGI1 region, which colocalizes with the first untranslated exon of PEAR1 but also is located close to the predicted enhancer region (Figure 2). We evaluated total methylation as an average of 32 CpG sites and observed that during MK differentiation it significantly increased from 3% ± 1% at day 0 to 14% ± 4% at day 14 (one-way ANOVA with Friedman test, P = .0011) (Figure 5B). Specific individual CpG-site analysis showed that CpG7-8 and CpG19-22 significantly increased methylation from day 0 to day 7 and further mildly increased from day 7 to day 14 (Figure 5C top panel). Pintron1 and PCGI2 remained highly methylated throughout HSC to MK differentiation (Figure 5B).
Increased PEAR1 expression during megakaryopoiesis parallels DNA methylation enrichment at CGI1. (A) Schematic representation of PEAR1 regions investigated in the methylation study indicated as black vertical boxes (“PCGI1” in CpG-island 1 [CGI1], “Pintron1” in intron 1 of PEAR1, “rs12041331”, “PCGI2” in CpG-island 2 [CGI2]). (B) Sequenom methylation estimates (% average) for PCGI1, Pintron1, and PCGI2 (bar representation, average ± standard deviation, n = 8). (C) Single CpG (dot line representation) during differentiation of CD34+ cells into MK precursors harvested at day 0 (D0), day 7 (D7), and day 14 (D14), and (D) of GG (n = 5) vs AG+AA (n = 3) methylation at day 14. *P < .05 one-way ANOVA.
Increased PEAR1 expression during megakaryopoiesis parallels DNA methylation enrichment at CGI1. (A) Schematic representation of PEAR1 regions investigated in the methylation study indicated as black vertical boxes (“PCGI1” in CpG-island 1 [CGI1], “Pintron1” in intron 1 of PEAR1, “rs12041331”, “PCGI2” in CpG-island 2 [CGI2]). (B) Sequenom methylation estimates (% average) for PCGI1, Pintron1, and PCGI2 (bar representation, average ± standard deviation, n = 8). (C) Single CpG (dot line representation) during differentiation of CD34+ cells into MK precursors harvested at day 0 (D0), day 7 (D7), and day 14 (D14), and (D) of GG (n = 5) vs AG+AA (n = 3) methylation at day 14. *P < .05 one-way ANOVA.
Although overall PEAR1 methylation was not different at PIntron and PCGI2 between G/G and A allele carriers, a significantly higher methylation for the CpG sites 7-8, 11-13, 19-22, and 31-32 in the PCGI1 region was observed for G/G carriers at the end of differentiation day 14 (unpaired Student t test, P < .05) (Figure 5D).
Discussion
PEAR1 is highly expressed in platelets and MKs and participates in both platelet activation and MK progenitor proliferation.2,4 The minor A allele of rs12041331 was found to be associated with lower PEAR1 expression in platelets,9 and associations with interindividual variability in the responses to antiplatelet drugs have been shown. A recent genome-wide study also associated rs12566888 (the only SNP in high linkage disequilibrium with rs12041331 in the all PEAR1 gene region) with platelet aggregation. Despite this evidence, in a study involving 2 independent populations, although the minor A allele was still linked to lower platelet function on aspirin, it was also found to be a risk factor for cardiovascular events.14 All the evidence produced together with this apparently divergent result fostered our present investigations focusing on the molecular mechanisms by which this variant would modulate PEAR1 expression. We hypothesized that such a mechanism of fine-tuning expression, able to interfere with residual platelet activation variability, as shown in the different epidemiological studies, could be mediated by a combination of genetic predisposition and epigenetic mechanisms.
PEAR1 variants associated with platelet function and individual variability are located both in coding and in noncoding regions of the PEAR1 gene, and 4 (rs2768759, rs12041331, rs11264579, and rs3737224) of 8 are CpG-SNPs. Three of these CpG-SNPs are positioned either upstream the CGI1 (rs2768759) or in intron 1 of the PEAR1 gene (rs12041331 and rs11264579). The only non CpG-SNP present in the same region (rs12566888) is in high linkage disequilibrium with rs12041331.
Association studies between CpG-SNPs and certain phenotypes are increasing,23-29 but the molecular characterization of their mechanism of action still remains a challenge. Most of the CpG-SNPs functionally characterized are located in the 5′-untranslated region or promoter regions,48-51 while PEAR1 rs12041331 is located in a noncoding intronic region, yet with specific enhancer features. Independently of their genomic location, CpG-SNPs collectively appear to act by impeding or facilitating the binding of specific factors that define their functional role in gene expression. In the case of rs12041331, the latter is suggested by the enhanced DNA-protein binding for the major G allele, which we observed in EMSA coupled to DNA methylation. Our study is in line with previous reports on the impact of rs12041331 on platelet9 and endothelial cell20 function, and it adds epigenetic and allele-specific methylation as new regulatory mechanism for PEAR1 regulation.
Three different transcription factors are predicted to bind to the G or the A allele: ATF3 and c-Jun and YY1, respectively. Even if the prediction of these factors does not take in to account the influence of DNA methylation, it is interesting to know that both YY1 and c-Jun have been found to be highly enriched in enhancer regions in a number of cell lines including HUVECs.52 In addition to that, a recent study defines the binding profile of TFs to the DNA Methyl Transferases (DNMTs) as the enzymes responsible for the maintenance or the de novo deposition of DNA methylation.53 ATF3, c-Jun, and YY1 have all been found to interact with DNMT3L, the enzyme that participates in de novo DNA methylation by interacting with DNMT3A and DNMT3B. This interaction could be both activating and inhibiting the action of DNMT3L, representing a possible mechanism by which the G-methylated allele contributes to the CGI1 de novo methylation during MK differentiation (Figure 6).
Epigenetic-dependent regulatory mechanism of rs12041331 in PEAR1 expression. The major G allele of the CpG-SNP introduces a CpG site in the enhancer region that binds with higher affinity to several nuclear proteins than the A allele. At the same time, the G-methylated allele marks higher methylation at CGI1, the CpG island located at the untranslated region of PEAR1 that contains several binding sites for CTCF, important for chromosome looping and architecture. When higher methylation is present, CTCF binds with lower affinity to the CGI1 region, liberating the enhancer activity of intron 1, where the CpG-SNP is also located. On the contrary, lower methylation at CGI1 is associated with the minor A allele of the CpG-SNP, reinforcing CTCF binding and partially inhibiting gene expression. This model advances a positive effect of methylation on gene expression and is in accordance with the association between increasing methylation and PEAR1 expression during early MK progenitor development.
Epigenetic-dependent regulatory mechanism of rs12041331 in PEAR1 expression. The major G allele of the CpG-SNP introduces a CpG site in the enhancer region that binds with higher affinity to several nuclear proteins than the A allele. At the same time, the G-methylated allele marks higher methylation at CGI1, the CpG island located at the untranslated region of PEAR1 that contains several binding sites for CTCF, important for chromosome looping and architecture. When higher methylation is present, CTCF binds with lower affinity to the CGI1 region, liberating the enhancer activity of intron 1, where the CpG-SNP is also located. On the contrary, lower methylation at CGI1 is associated with the minor A allele of the CpG-SNP, reinforcing CTCF binding and partially inhibiting gene expression. This model advances a positive effect of methylation on gene expression and is in accordance with the association between increasing methylation and PEAR1 expression during early MK progenitor development.
Methylation at rs12041331 is associated with a higher PEAR1 expression in platelets9 and differential function in both platelets9-11,14,16-18 and endothelial cells.20 Although an inverse correlation between DNA methylation and gene expression is very well known, the opposite is not yet well understood, although reported in some large-scale studies54 and highly dependent on the genomic context.55-57 To better understand this unusual relation between DNA methylation and gene expression, we have studied PEAR1 DNA methylation, during early and late megakaryopoiesis. We found that parallel progressive elevation of PEAR1 CGI1 methylation and PEAR1 expression occurs in early differentiation. At the same time, we also observed that rs12041331 was constitutively methylated throughout differentiation. CpG-SNP allele-specific methylation occurs at the same time also at the CGI1 region of the gene, where A allele carriers present with significantly lower methylated CpG sites. Taken together with the different methylation at CGI1, rs12041331 contributes to drive and fine-tune PEAR1 expression in platelets. This interpretation is in line with the reported platelet PEAR1 protein differences between individuals with the 2 genotypes9 as well as with the luciferase gene-reporter assay, which measured the isolated effect of the methylated G allele for rs12041331. Although we could not detect significant differences in MK PEAR1 expression at the end of differentiation, we hypothesize that its expression might be more important for proliferation at the very early stages of differentiation, where we observed differences in the size of the CFU-MK between A allele carriers and GG homozygotes (Figure 4B left panel). In fact, PEAR1 reaches maximal expression levels already at day 7 of differentiation, suggesting that a still earlier time point might better reveal differences in PEAR1 expression between the GG and A allele carriers. We could not include such analysis in our present study, because of the limited sample availability (comprising a single AA carrier only) and of the necessity to already determine membrane protein expression studies on these samples (FACS), CFU-MK colony assays, and DNA methylation studies. We did analyze PEAR1 expression as a function of the Forward Scatter, an index of cellular size, and hence, indirectly of MK maturation, during which process the higher ploidy results in larger cell masses.
The CpG sites of the CGI1 regions that undergo the most significant methylation changes identified in the MK progenitor-specific analysis reside within predicted CTCF binding sites. CTCF is an important player regulating complex processes, including transcription, imprinting, long-range chromatin interactions, and subnuclear localization in health and disease. Its binding has been shown to be highly influenced by DNA methylation both in vitro and in vivo and, in combination with other regulatory mechanisms, drives spatial and temporal-dependent gene expression. ENCODE CTCF annotations for HUVECs that express high levels of PEAR1 show binding of CTCF at the CGI1 region, and moreover, specifically located in that region investigated by us in the methylation study (Figure 1). At the same region, specific active enhancer-interacting promoter histone marks52 are also present (H3K27Ac and H3K4Me3), whereas H3K4Me1, typically defining enhancers (such as the rs12031441 region), is not comparably enriched. This region is highly conserved across vertebrates (Figure 1 last track), suggesting its possible relevance during evolution. CTCF binding may play an important role in mediating interactions between the CGI1 region and the gene transcription start site, located approximately 10 kb downstream. CTCF binding depends on higher CpG content58 and DNA methylation restricting enhancer-mediated transcriptional inductions or working as a chromatin barrier separating active and repressive domains at gene promoters or enhancers.59 CTCF binding at PEAR1 CGI1 would then be prevented by elevated methylation, allowing the downstream locus to act as an active enhancer in the PEAR1 transcription in G-carriers. This model is in line with the higher G allele–dependent expression and methylation observed. At the same time, PEAR1 CGI1 could be the enhancer of more distant promoters along chromosome 1, influencing transcription of other genes.60
In conclusion, we have mechanistically characterized the role of the CpG-SNP rs12041331 as genetic predisposition that reinforces PEAR1 enhancer activity through allele-specific methylation. Future studies are needed to investigate whether this mechanism of gene regulation is highly specific for PEAR1 exclusively or also applies to more platelet function- or megakaryopoiesis-related genes.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Ploi Yibmantasiri and Antonio Passarella for graphical assistance.
This work was supported by the “Fonds voor Wetenschappelijk Onderzoek (FWO) Vlaanderen” grant G0A6514N and by the “Programmafinanciering KU Leuven (PF/10/014).” B.I. and M.P. are both FWO postdoctoral fellows (12M2715N and 1288714N, respectively); P.V. holds a FWO senior clinical investigator grant (1801414N).
Authorship
Contribution: B.I. designed and performed experiments, analyzed data, and wrote the manuscript; M.P. performed experiments and advised on ChIP research methodology; K.C. and P.A. performed experiments; D.L. advised on Sequenom methodology and reviewed the manuscript; C.V. and P.V. reviewed the manuscript; K.F. and M.F.H. designed experiments and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Marc F. Hoylaerts, Center for Molecular and Vascular Biology, Department of Cardiovascular Sciences, University of Leuven, Herestraat 49, 3000 Leuven, Belgium; e-mail: marc.hoylaerts@med.kuleuven.be.




![Figure 5. Increased PEAR1 expression during megakaryopoiesis parallels DNA methylation enrichment at CGI1. (A) Schematic representation of PEAR1 regions investigated in the methylation study indicated as black vertical boxes (“PCGI1” in CpG-island 1 [CGI1], “Pintron1” in intron 1 of PEAR1, “rs12041331”, “PCGI2” in CpG-island 2 [CGI2]). (B) Sequenom methylation estimates (% average) for PCGI1, Pintron1, and PCGI2 (bar representation, average ± standard deviation, n = 8). (C) Single CpG (dot line representation) during differentiation of CD34+ cells into MK precursors harvested at day 0 (D0), day 7 (D7), and day 14 (D14), and (D) of GG (n = 5) vs AG+AA (n = 3) methylation at day 14. *P < .05 one-way ANOVA.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/128/7/10.1182_blood-2015-11-682153/6/m_1003f5.jpeg?Expires=1764965645&Signature=xl-zjKgv--SW4r9IXEYBoc~ZggW~XTGWZk64FiWjYfM97Pq85jYsT4PVn0fQCixeQY3SX5k80SaUydAggH69kDOldMIzQ8TAPcCQr8Ctk0be~KP2nA2q0Gsn8AAUxBTDu4xl6tH08g6x~6KvOOytxOXrSD8Dvy~ZidLd1MLFE0uR7aIbWwcvAEPYzio5EyMRTKjBZYK10VY4IukFZ2F8MwzALbxrvhAv1hZqFicJN6IBpUJ9akaKjVVNQzvvU8pXUj9goFvVpABkXknDQ6mz0V915ooWkBhtDJJjnXEccmLBhGaX2LIzxHR5ZsfPFLnOMOUpOixT0aaH72q~0ZJ1XA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
