Key Points
FBXO11 loss in mice enhances GC B-cell formation and leads to increased BCL6 expression.
FBXO11 inactivation, mimicking genetic alterations identified in human lymphomas, represents an alternative mechanism of BCL6 deregulation.
Abstract
The BCL6 proto-oncogene encodes a transcriptional repressor that is required for the germinal center (GC) reaction and is implicated in lymphomagenesis. BCL6 protein stability is regulated by F-box protein 11 (FBXO11)–mediated ubiquitination and degradation, which is impaired in ∼6% of diffuse large B-cell lymphomas that carry inactivating genetic alterations targeting the FBXO11 gene. In order to investigate the role of FBXO11 in vivo, we analyzed GC-specific FBXO11 knockout mice. FBXO11 reduction or loss led to an increased number of GC B cells, to an altered ratio of GC dark zone to light zone cells, and to higher levels of BCL6 protein in GC B cells. B-cell receptor–mediated degradation of BCL6 was reduced in the absence of FBXO11, suggesting that FBXO11 contributes to the physiologic downregulation of BCL6 at the end of the GC reaction. Finally, FBXO11 inactivation was associated with the development of lymphoproliferative disorders in mice.
Introduction
The germinal center (GC) reaction is an essential component of the T cell–dependent immune response leading to the generation of memory B cells and plasma cells with high affinity for antigens.1,2 During the GC reaction, B cells undergo immunoglobulin somatic hypermutation (SHM) and class switch recombination (CSR) in order to acquire the ability to produce high affinity antibodies of different isotype classes.1,2 The GC structure is characterized by 2 histologically and functionally distinct compartments: the dark zone (DZ), where GC B cells proliferate and undergo SHM, and the light zone (LZ), where cells are selected based on the affinity of their immunoglobulin receptors.2 Although essential for a successful immune response, the immunoglobulin loci remodeling mechanisms have also been shown to lead to genetic accidents contributing to lymphomagenesis.3 Among the GC-derived malignancies, diffuse large B-cell lymphoma (DLBCL) represents the most common type of non-Hodgkin lymphoma with a remarkably diverse clinical presentation and outcome, likely reflecting its pathogenetic and biologic heterogeneity.3,4
The initiation and maintenance of the GC reaction is dependent on BCL6, a transcriptional repressor belonging to the BTB/POZ/zinc finger family of transcription factors. Mice lacking BCL6 cannot form GCs nor produce high-affinity antibodies.5 In GC B cells, BCL6 contributes to the physiologic immunoglobulin remodeling processes by negatively modulating the DNA damage response and impairs premature B-cell activation and differentiation by repressing several signaling pathways and master regulators of the post-GC fate.6 BCL6 expression is restricted to the GC reaction and must be switched off in order to proceed in the differentiation process. The importance of BCL6 repression for a proper post-GC differentiation is emphasized by the fact that deregulated BCL6 expression is associated with malignant transformation. Multiple signaling pathways contribute to BCL6 downregulation at the end of the GC reaction including engagement of the B-cell receptor (BCR) and T cell–mediated CD40 signaling.7,8 Genetic alterations targeting the BCL6 locus are found in ∼30% of DLBCLs and include translocations that deregulate BCL6 expression through a promoter substitution mechanism and mutations that affect regulatory elements in the BCL6 gene.6 In addition, deregulation of BCL6 can occur via genetic alterations affecting BCL6 modulators including CREBBP and EP3009 and MEF2B.10 The oncogenic function of BCL6 deregulation was demonstrated in a mouse model engineered to recapitulate a DLBCL-associated translocation, which develops lymphomas analogous to the human DLBCL.11
It has been shown that F-box protein 11 (FBXO11) is involved in the ubiquitylation and subsequent degradation of BCL6.12 F-box proteins function as substrate-recognition subunits of SKP1–cullin-1–F-box protein (SCF) E3 ligase complexes, thus ensuring substrate-specific ubiquitylation and subsequent degradation of target proteins.13 F-box proteins include a protein-protein interaction domain, the F-box motif, that directly binds to SKP1 for recruitment into the SCF complex and various carboxy-terminal domains that are required for substrate specific recognition.14 FBXO11 was found to be inactivated by mutations or deletions in ∼6% of DLBCLs.12,15,16 Monoallelic genetic inactivation of FBXO11 in DLBCL has been shown to result in increased levels of BCL6, suggesting that it may contribute to the pathogenesis of DLBCL.12 Together with the fact that monoallelic FBXO11 mutations are also present in ovary, lung, and colon cancer,17 these observations suggest that FBXO11 may function as a haploinsufficient tumor suppressor. In the present study, we generated conditional FBXO11 knockout (KO) mice to investigate in vivo the role of FBXO11 in the GC reaction as well as in lymphomagenesis.
Materials and methods
Generation of FBXO11 conditional mice
The conditional FBXO11 allele was generated by insertion of 2 loxP sites into intronic regions ∼250 bp upstream and downstream of exon 4 by embryonic stem (ES) cell targeting. Correct homologous recombination was confirmed by Southern blotting, and the targeted mouse W9.5 ES cells were injected into blastocysts derived from C57BL/6 mice to generate chimeras. The neomycin-resistance marker, flanked by frt sites, was deleted by crossing the mice to Flp-transgenic mice. The FBXO11fl/+ mice were bred with Cre-transgenic mice, expressing Cre under the control of the Cγ1 promoter (F1 generation) to allow specific deletion of FBXO11 in GC-derived B cells only.18 Mice from the F1 generation were intercrossed to generate age-matched cohorts of FBXO11fl/fl-Cγ1cretg/+ (KO), FBXO11fl/+-Cγ1cretg/+ (heterozygous [HET]), and FBXO11+/+-Cγ1cretg/+ (wild-type [WT]) littermates.
Mouse immunization, immunofluorescence, and flow cytometry
Mice were housed in a dedicated pathogen-free environment. All experiments and procedures conformed to ethical principles and guidelines revised and approved by the Institutional Animal Care and Use Committee at Columbia University. Three-month-old mice were immunized by intraperitoneal injection of 109 sheep red blood cells (SRBCs) or 100 μg of NP (4-hydroxy-3-nitrophenyl-acetyl) conjugated to keyhole limpet hemocyanin to trigger T cell–dependent immune responses and were euthanized 10 days (or at the time points specified in “Results”) upon immunization. Spleens were isolated and divided into 2 fragments, which were processed for immunohistochemistry/immunofluorescence and flow cytometry, respectively. Splenic mononuclear cells were isolated by straining the tissue through 40-μm cell strainers in 1× phosphate-buffered saline + 0.5% bovine serum albumin followed by red blood cell lysis. Mononuclear cell suspensions were stained for 20 minutes on ice with antigen-specific fluorochrome-conjugated antibodies (supplemental Table 1, available on the Blood Web site) and analyzed with a BD LSR II (BD Biosciences); 50 000 to 500 000 events were collected per each sample and were analyzed with FlowJo Software (FlowJo LLC). For intracellular staining, the cells were fixed and permeabilized using the BD Cytofix/Cytoperm buffer (BD Biosciences) following the manufacturer’s instructions. For immunofluorescence, 3-μm-thick formalin-fixed, paraffin-embedded sections were stained as previously published.5 Heat-induced epitope retrieval was performed in citrate buffer (pH 6.0). Primary antibodies were incubated overnight at 4°C (supplemental Table 1). After repeated washes in 1× phosphate-buffered saline + 0.1% Tween20, tissue sections were incubated with isotype-specific fluorochrome-conjugated secondary antibodies (supplemental Table 1). Slides were then washed and mounted. Immunohistochemistry stainings were performed as previously described.11
Expression constructs
The FBXO11 short hairpin RNAs (shRNAs) were designed using the “sensor” algorithm19 : FBXO11-shRNA #2, 5′-TGCTGTTGACAGTGAGCGCCAGATAGTAATCCTACACTAATAGTGAAGCCACAGATGTATTAGTGTAGGATTACTATCTGTTGCCTACTGCCTCGGA-3′; FBXO11-shRNA #5, 5′-TGCTGTTGACAGTGAGCGCAAGGGACAAGGAGTAATAGAATAGTGAAGCCACAGATGTATTCTATTACTCCTTGTCCCTTTTGCCTACTGCCTCGGA-3′; FBXO11-shRNA #7, 5′-TGCTGTTGACAGTGAGCGATAGAGTTTATTAGACATGATATAGTGAAGCCACAGATGTATATCATGTCTAATAAACTCTACTGCCTACTGCCTCGGA-3′.
Cell lines
Daudi cells were obtained from the American Type Culture Collection and maintained at the recommended concentration in Iscove modified Dulbecco medium (Gibco), supplemented with 10% fetal calf serum, 100 U/mL penicillin, and 0.1 mg/mL streptomycin at 37°C in 5% CO2. BCR stimulation was performed using 10 µg/mL immunoglobulin M (IgM) (SouthernBiotech) antibodies. Daudi cell lines with inducible expression of the FBXO11-shRNAs were established by electroporation (960 μFD/250 V) and subsequently selected with hygromycin (600 μg/mL).
Immunoblot
Total protein extracts were prepared from frozen pellets of cultured cells or isolated mouse B cells. Whole cell lysates were prepared as previously described7 and analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis and immunoblotting using the antibodies listed in supplemental Table 1. ImageJ software was used for densitometry.
Quantitative reverse transcription PCR
RNA was isolated from purified GC B cells using the NucleoSpin RNA XS kit (Macherey-Nagel), and retrotranscription was performed starting from 500 ng of total RNA by SuperScript II Reverse Transcriptase kit (Thermo Fisher Scientific). Quantitative polymerase chain reaction (PCR) was performed using primers for BCL6 (forward: 5′-AGGCCTCCTTCCGCTACAAG; reverse: 5′-CAAATGTTACAGCGATAGGGTTTCT) and actin B (forward: 5′-ATGGAGGGGAATACAGCCC; reverse: 5′-TTCTTTGCAGCTCCTTCGTT), SYBRgreen Absolute (Thermo Fisher Scientific), and the Applied Biosystems 7300 Real-Time PCR System, as recommended by the manufacturer.
Statistics
One-way analysis of variance (ANOVA) paired with Tukey’s multiple-comparison test was performed with Prism (GraphPAD Software).
The binomial distribution P value was calculated as P = 1/2n, where n is the number of independent observations.
Results
Generation of mice conditionally deleting FBXO11 in GC B cells
The FBXO11 gene is conserved from nematodes to mammals and is located on mouse chromosomal region 17qE4, containing 23 exons and spanning 75-kb genomic DNA. In order to investigate the consequences of FBXO11 deletion in vivo, we generated a conditional allele by inserting 2 loxP sites into intronic regions ∼250 bp upstream and downstream of exon 4 and performed gene targeting in ES cell (Figure 1A). The deletion of exon 4 causes a translational reading frame shift resulting in the truncation of the FBXO11 protein. Correct homologous recombination was confirmed by Southern blotting (Figure 1B). The targeted mouse ES cells were injected into blastocysts to generate chimeras, from which germ-line transmission of the “floxed” allele of FBXO11 was achieved. The neomycin-resistance marker, used to select targeted ES cells, was subsequently deleted via flanking frt sites and expression of Flp recombinase by crossing the “floxed” FBXO11 mice with Flp-transgenic mice. Next, the FBXO11fl/+ mice were bred with Cre-transgenic mice, expressing Cre under the control of the Cγ1 promoter (F1 generation) to allow specific deletion of FBXO11 in GC-derived B cells only.18 Mice from the F1 generation were intercrossed to generate age-matched cohorts of FBXO11fl/fl-Cγ1cretg/+ (KO), FBXO11fl/+-Cγ1cretg/+ (HET), and FBXO11+/+-Cγ1cretg/+ (WT) littermates. Mice were born at the expected Mendelian frequencies. To confirm the correct deletion of FBXO11 in GC B cells, mice were challenged with SRBCs as T cell–dependent antigens. Immunofluorescence staining showed that FBXO11 protein was expressed, together with BCL6, in GC B cells of WT mice, as expected, whereas no expression of FBXO11 was detected in KO mice, confirming that targeting of exon 4 impairs FBXO11 protein expression (Figure 1C).
Conditional targeting of FBXO11 in mice. (A) Schematic representation of the FBXO11 targeting strategy. Indicated are restriction sites as well as expected fragment sizes detectable by Southern blot in the WT allele as well as in the targeted allele before and after Flp-mediated deletion of the neomycin-resistance cassette. CL, conditional allele; TV, targeting vector. (B) Southern blot analysis of BamHI digested genomic DNA displays the 15-kb WT band along with the 10-kb band after correct targeting in ES cells using a 5′ probe (left). Southern blot analysis of StuI-digested DNA displays the 20-kb WT band along with the 8.5-kb band after correct targeting in ES cells using a 3′ probe (right). (C) Double immunofluorescence staining of FBXO11 (green) and BCL6 (red) of a GC in FBXO11+/+-Cγ1cretg/+ (WT) and FBXO11fl/fl-Cγ1cretg/+ (KO) mice.
Conditional targeting of FBXO11 in mice. (A) Schematic representation of the FBXO11 targeting strategy. Indicated are restriction sites as well as expected fragment sizes detectable by Southern blot in the WT allele as well as in the targeted allele before and after Flp-mediated deletion of the neomycin-resistance cassette. CL, conditional allele; TV, targeting vector. (B) Southern blot analysis of BamHI digested genomic DNA displays the 15-kb WT band along with the 10-kb band after correct targeting in ES cells using a 5′ probe (left). Southern blot analysis of StuI-digested DNA displays the 20-kb WT band along with the 8.5-kb band after correct targeting in ES cells using a 3′ probe (right). (C) Double immunofluorescence staining of FBXO11 (green) and BCL6 (red) of a GC in FBXO11+/+-Cγ1cretg/+ (WT) and FBXO11fl/fl-Cγ1cretg/+ (KO) mice.
GC-specific deletion of FBXO11 leads to increased numbers of GC B cells and affects the DZ/LZ ratio
In order to investigate the effects of FBXO11 deletion on the formation of GC, mice were immunized with SRBCs and analyzed on day 10 after immunization, corresponding to the peak of the GC response. Following acute immunization, all mice displayed normal B-cell distributions in the non-GC compartments (supplemental Figure 1). GC B cells were analyzed in 16 to 18 animals/genotype by staining mononuclear cells obtained from spleen for lectin peanut agglutinin (PNA) and the B-cell marker B220 (Figure 2A). FBXO11 KO mice showed a significantly higher number of GC B cells as compared with the WT and the HET mice by cytofluorimetric analysis (Figure 2B). These results were confirmed by immunostaining of spleen sections with anti-PNA antibodies and analysis of the GC number and size. Compared with the WT, the KO mice displayed a significantly higher number of GCs and GC to spleen area ratio, whereas HET mice, although displaying a trend in the same direction, would not show a statistically significant increase in GC number and/or size (Figure 2C). Analogous results were obtained by using BCL6 as a marker of GC B cells (data not shown).
FBXO11 deletion leads to an increase in GC B cells and affects the DZ/LZ ratio. (A) Flow cytometry analysis of splenic mononuclear cells from age-matched FBXO11+/+-Cγ1cretg/+ (WT), FBXO11fl/+-Cγ1cretg/+ (HET), and FBXO11fl/fl-Cγ1cretg/+ (KO) for lectin PNA and the B-cell marker B220. Mice were immunized with SRBCs 10 days prior to the analysis. (B) Percentages of B220+/PNA+ splenic GC B cells. n = number of analyzed mice, average ± standard deviation [SD]; *P < .05 (1-way ANOVA). (C) Average GC size (left), numbers (middle) and GC/spleen area ratios obtained by staining with anti-PNA at least 3 independent spleen sections for each mouse. Pix, pixels. n = number of analyzed mice, average ± SD; *P < .05 (1-way ANOVA). (D) Representative flow cytometry analysis of splenic GC DZ and LZ cells as defined by CXCR4 and CD86 expression. (E) DZ/LZ cell ratios determined as in panel D in FBXO11 WT, HET, and KO mice. n = number of analyzed mice, average ± SD; *P < .05 (1-way ANOVA). (F) DZ/LZ cell ratios in Iμ-HA-BCL6 mice and WT littermates. n = number of analyzed mice, average ± SD; *P < .05 (1-way ANOVA).
FBXO11 deletion leads to an increase in GC B cells and affects the DZ/LZ ratio. (A) Flow cytometry analysis of splenic mononuclear cells from age-matched FBXO11+/+-Cγ1cretg/+ (WT), FBXO11fl/+-Cγ1cretg/+ (HET), and FBXO11fl/fl-Cγ1cretg/+ (KO) for lectin PNA and the B-cell marker B220. Mice were immunized with SRBCs 10 days prior to the analysis. (B) Percentages of B220+/PNA+ splenic GC B cells. n = number of analyzed mice, average ± standard deviation [SD]; *P < .05 (1-way ANOVA). (C) Average GC size (left), numbers (middle) and GC/spleen area ratios obtained by staining with anti-PNA at least 3 independent spleen sections for each mouse. Pix, pixels. n = number of analyzed mice, average ± SD; *P < .05 (1-way ANOVA). (D) Representative flow cytometry analysis of splenic GC DZ and LZ cells as defined by CXCR4 and CD86 expression. (E) DZ/LZ cell ratios determined as in panel D in FBXO11 WT, HET, and KO mice. n = number of analyzed mice, average ± SD; *P < .05 (1-way ANOVA). (F) DZ/LZ cell ratios in Iμ-HA-BCL6 mice and WT littermates. n = number of analyzed mice, average ± SD; *P < .05 (1-way ANOVA).
To further dissect the impact of FBXO11 deletion on GC B cells, splenic GC B cells (B220+, CD95+, PNA+) were subdivided in DZ and LZ B cells according to the expression of CD86 and CXCR42,22 (Figure 2D). Although WT mice displayed the expected 2:1 ratio between DZ and LZ populations, FBXO11 HET and KO mice showed higher DZ/LZ ratios (Figure 2E). Notably, a similar phenotype was observed in Iµ-HA-BCL6 mice (Figure 2F) that express a human influenza hemagglutinin (HA)–tagged, full-length murine BCL6 coding sequence under the control of the immunoglobulin heavy chain Iµ promoter.11 An increase in the number of GC B cells, as well as a higher DZ/LZ ratio, was still observed in the KO mice 20 days after immunization (supplemental Figure 2). Overall, these data show that deletion of FBXO11 in GC B cells impacts the GC formation leading to an increased number of GC B cells that are skewed toward the DZ compartment. This phenotype is similar to that of mice constitutively expressing BCL6 (Figure 2E-F).
To assess whether CSR was affected in the mice lacking FBXO11, we analyzed the ability of B cells to switch to IgG1. Mice with reduced or no expression of FBXO11 displayed a higher percentage of IgG1-switched GC B cells (supplemental Figure 3A), whereas the percentage of IgG1-switched within the total B-cell compartment was identical to WT mice (supplemental Figure 3B). These results suggest that the switching to IgG1 is not altered, but the ability of the switched cells to exit the GC is impaired. In agreement with this notion, the levels of IgG1 specific for the NP hapten were slightly reduced in the serum of the HET and KO mice 14 days after immunization with NP (supplemental Figure 3C), whereas the serum titers were comparable 28 days after immunization (supplemental Figure 3D). Affinity maturation was normal in these mice, suggesting that SHM is not affected by the deletion of FBXO11 (supplemental Figure 3E).
Taken together, these findings indicate that defective FBXO11 expression causes abnormal GC expansion that is compatible with normal CSR and affinity maturation.
Deletion of FBXO11 increases BCL6 protein levels in vivo
BCL6 has been shown to be targeted by the SCF complex for ubiquitylation and subsequent proteosomal degradation.12 In addition, the ability of FBXO11 to degrade BCL6 was impaired in DLBCL cell lines carrying deletions and/or inactivating mutations targeting the FBXO11 gene.12 In order to investigate the effect of FBXO11 ablation on BCL6 expression under physiologic conditions, normal GC B cells were isolated from WT, HET, and KO mice on day 10 upon immunization with SRBCs. Immunoblotting analyses confirmed the lack of FBXO11 protein expression in the KO mice, whereas the HET mice showed, as expected, half of the amount as compared with WT mice (Figure 3A). Consistent with the data previously obtained in transformed cells, the deletion of FBXO11 in vivo led to increased levels of BCL6 protein. The KO mice showed approximately twofold increase as compared with the WT mice, and the HET mice displayed an increase in BCL6 that was intermediate between the WT and the KO (Figure 3A). These data were further confirmed by cytofluorimetric analyses measuring the mean fluorescent intensity of BCL6 in GC B cells (Figure 3B). In addition, consistent with the increase in BCL6 protein expression and with the fact that BCL6 acts as its own transcriptional repressor, BCL6 transcription was reduced (supplemental Figure 4). Taken together, these results confirm the role of FBXO11 in the regulation of BCL6 in normal cells in vivo.
FBXO11 deletion increases BCL6 protein levels in vivo. (A) Immunoblot analysis of FBXO11, BCL6, and actin in GC B cells isolated from spleens of FBXO11+/+-Cγ1cretg/+ (WT), FBXO11fl/+-Cγ1cretg/+ (HET), and FBXO11fl/fl-Cγ1cretg/+ (KO) mice. Protein expression levels were quantitated by densitometric analysis, normalized to actin, and fold changes are displayed relatively to WT. (B) Average BCL6 mean fluorescence intensity measured by flow cytometry analysis and displayed as fold change relative to WT mice. n = number of analyzed mice, average ± SD; *P < .05 (1-way ANOVA).
FBXO11 deletion increases BCL6 protein levels in vivo. (A) Immunoblot analysis of FBXO11, BCL6, and actin in GC B cells isolated from spleens of FBXO11+/+-Cγ1cretg/+ (WT), FBXO11fl/+-Cγ1cretg/+ (HET), and FBXO11fl/fl-Cγ1cretg/+ (KO) mice. Protein expression levels were quantitated by densitometric analysis, normalized to actin, and fold changes are displayed relatively to WT. (B) Average BCL6 mean fluorescence intensity measured by flow cytometry analysis and displayed as fold change relative to WT mice. n = number of analyzed mice, average ± SD; *P < .05 (1-way ANOVA).
FBXO11 silencing impairs downregulation of BCL6 upon IgM treatment
BCL6 protein degradation is mediated by multiple signaling pathways and is essential during the GC reaction to ensure proper differentiation. In order to identify the role of FBXO11 in BCL6 degradation upon BCR engagement, a well-known pathway involved in the negative regulation of BCL6,7 we analyzed the effects of BCR signaling on BCL6 protein expression in the absence of FBXO11. Toward this goal, given that normal GC B cells have an extremely short life span ex-vivo, we relied on transformed GC-derived cells and generated cell lines that express 3 different shRNAs targeting FBXO11 in a doxycycline-inducible manner. Consistent with previously published observations,12 silencing of FBXO11 led to an increase in BCL6 protein expression (Figure 4). BCR was engaged 24 hours after inducing the expression of shRNA against FBXO11 or, as a control, against Renilla. Activation of the BCR signaling pathway resulted in 5- to 10-fold downregulation of BCL6 protein in Daudi cells expressing the control shRNA, whereas cells with reduced FBXO11 protein expression displayed only ≤2.5-fold reduction of BCL6 (Figure 4). The experiment was performed multiple times (n = 2-3) with each of the 3 shRNAs targeting FBXO11 confirming a significant delay in BCR-mediated degradation of BCL6 upon FBXO11 silencing (binomial distribution under the null hypothesis of no differential degradation: sh#2 and sh#5, P = .0019; sh#7, P = .016). The fact that upon BCR engagement BCL6 degradation was delayed in the absence of FBXO11 suggests that FBXO11 is involved in the pathway leading to BCR-mediated degradation of BCL6.7
FBXO11 silencing impairs downregulation of BCL6 upon IgM treatment. Immunoblot analysis of FBXO11, BCL6, and actin protein expression in a Burkitt lymphoma cell line (Daudi) treated with IgM in the presence or absence of FBXO11 expression. Three different shRNAs (#2, top; #5, middle; #7, bottom) were used to silence FBXO11 expression. Data were quantitated by densitometric analysis, normalized to actin levels, and graphed relative to untreated cells (right). A representative experiment is displayed out of 2 to 3 independent assays that were performed for each shFBXO11.
FBXO11 silencing impairs downregulation of BCL6 upon IgM treatment. Immunoblot analysis of FBXO11, BCL6, and actin protein expression in a Burkitt lymphoma cell line (Daudi) treated with IgM in the presence or absence of FBXO11 expression. Three different shRNAs (#2, top; #5, middle; #7, bottom) were used to silence FBXO11 expression. Data were quantitated by densitometric analysis, normalized to actin levels, and graphed relative to untreated cells (right). A representative experiment is displayed out of 2 to 3 independent assays that were performed for each shFBXO11.
FBXO11 silencing in mice leads to lymphoproliferative disease
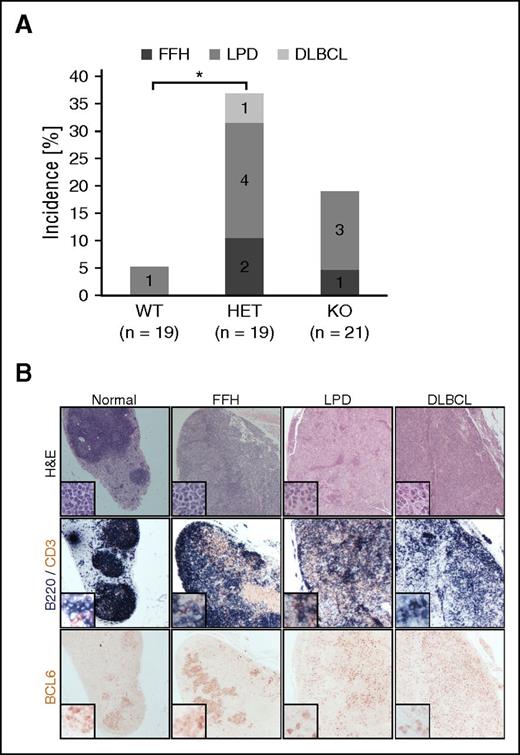
To assess the role of FBXO11 inactivation in lymphomagenesis in vivo, a tumor cohort including ∼20 mice/genotype was established. The mice were immunized with SRBCs 6 times in their life span and euthanized at the age of 17 to 18 months. Inspection of spleen weight did not show any difference in the cohorts. Overall, 1/19 (5%) of the WT, 7/19 (36.8%) of the HET, and 4/21 (19%) of the KO mice developed B-cell lymphoproliferations (Figure 5A), including: (1) FFH, defined by expanded lymphoid follicles showing large, reactive GCs often with increased centroblastic proliferation with blurring of the DZ/LZ demarcation and attenuated mantle zones; (2) LPD resulting in alteration of the lymph node or splenic architecture by vaguely nodular, interconnected lymphoid aggregates or follicles of variable size, often accompanied by diffuse infiltration of large lymphocytes that display centroblastic morphology and the immunophenotype of GC B cells (B220+, PAX5+, BCL6+, IRF4−); and (3) DLBCL that show obliteration of nodal architecture by an infiltrate of large centroblast-like lymphocytes (Figure 5B).
FBXO11 deletion in mice leads to lymphoproliferative disease. (A) Incidence of florid follicular hyperplasia (FFH), lymphoproliferative disease (LPD), and DLBCL in 17- to 18-month-old FBXO11+/+-Cγ1cretg/+ (WT), FBXO11fl/+-Cγ1cretg/+ (HET), and FBXO11fl/fl-Cγ1cretg/+ (KO) mice. N = number of analyzed mice; *P < .05 (1-way ANOVA). (B) Lymph node sections representative of no pathology (normal), FFH, LPD, and DLBCL were stained with hematoxilin/eosin (H&E), B220 (blue) and CD3 (brown), or BCL6 as indicated. Images were acquired using 10× and (for the insets) 20× objectives leading to an overall magnification of ×100 and ×200, respectively.
FBXO11 deletion in mice leads to lymphoproliferative disease. (A) Incidence of florid follicular hyperplasia (FFH), lymphoproliferative disease (LPD), and DLBCL in 17- to 18-month-old FBXO11+/+-Cγ1cretg/+ (WT), FBXO11fl/+-Cγ1cretg/+ (HET), and FBXO11fl/fl-Cγ1cretg/+ (KO) mice. N = number of analyzed mice; *P < .05 (1-way ANOVA). (B) Lymph node sections representative of no pathology (normal), FFH, LPD, and DLBCL were stained with hematoxilin/eosin (H&E), B220 (blue) and CD3 (brown), or BCL6 as indicated. Images were acquired using 10× and (for the insets) 20× objectives leading to an overall magnification of ×100 and ×200, respectively.
Discussion
Based on previous results identifying a role of FBXO11 in the regulation of BCL6 in cell cultures,12 this study was aimed at defining the role of FBXO11 in the development of GC and GC-derived transformation in vivo. Our results confirm and extend the results of Duan et al by elucidating a signaling pathway involving FBXO11 in BCL6 regulation.12 Most notably, our findings indicate a role for FBXO11 in GC homeostasis and lymphomagenesis.
Mice with conditional inactivation of FBXO11 display enlarged GC, a phenotype that has significant similarities with that observed in mice with constitutive expression of the GC master regulator BCL611 or with conditional deletion of the plasma cell master regulator PRDM1/Blimp1.23 These phenotypes most likely reflect a variable disturbance in GC exit and the inability of GC B cells to efficiently enter the plasmablastic differentiation pathway. A unifying interpretation of these phenotypes suggests that FBXO11 inactivation may deregulate BCL6 at the level of protein stability in a manner, at least in part, equivalent to BCL6 transcriptional deregulation, which, in turn, may abnormally modulate PRDM1 expression partially mimicking PRDM1 deletion. However, it is conceivable that FBXO11 may have additional targets involved in GC homeostasis, which can be identified as proteins interacting with the SCF ubiquitin ligase complex in vivo.
The results herein suggest that FBXO11 is involved in the MAPK-induced phosphorylation and ubiquitin/proteasome-mediated degradation of BCL6 that occur upon BCR engagement.7 This finding is apparently in contrast with the results by Duan et al, which have excluded the involvement of FBXO11 in this pathway based on the observation that the interaction between BCL6 and FBXO11 occurs even in the absence of BCR signaling and does not require phosphorylation of BCL6 by MAPK.12 Our data show that FBXO11 ablation affects BCL6 degradation upon BCR engagement consistent with the fact that phosphorylated BCL6, which is primed for degradation, also interacts with FBXO11. Thus, our findings suggest that FBXO11 works both to maintain a steady-state expression level of (nonphosphorylated) BCL6 in GC B cells that have not been activated and to enforce BCL6 degradation in response to activation stimuli, including BCR signaling (leading to BCL6 phosphorylation), that occur at the end of the GC reaction.
Our results indicate that FBXO11 inactivation is associated with lymphoproliferative pathologies in vivo. This finding conclusively demonstrates that FBXO11 is a tumor suppressor, as previously suggested based on in vitro evidence.12 The lymphoproliferative phenotypes are variable, ranging from cell expansion altering the lymphoid organ architecture to full-blown DLBCL, and are only partially penetrant. In particular, these phenotypes are less penetrant than previously observed in mice mimicking transcriptional deregulation of BCL6.11 These observations suggest that FBXO11 inactivation is not sufficient for lymphomagenesis, which may require additional alterations involving BCL6 as well as other pathways critical for GC development. One interesting observation is that the frequency of lymphoproliferative phenotypes is significantly higher in HET vs homozygous FBXO11-deficient GC B cells. This result, which is consistent with the monollalelic inactivating mutations observed in DLBCL,12,16 suggests that some residual FBXO11 activity may provide an advantage during transformation, possibly because of the less favorable selection of cells expressing higher levels of BCL6 and/or other FBXO11 targets. Finally, the observation that FBXO11 deficiency affects BCL6 in vivo identifies a further mechanism of BCL6 deregulation associated with DLBCL pathogenesis, in addition to direct transcriptional deregulation by chromosomal translocation or MEF2B mutation10 and defective acetylation-mediated inactivation caused by genetic loss of the CREBBP/EP300 acetyltransferases.9
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank P. Brescia for helping with cytofluorimetric data analysis; R. Rabadan for consultation on statistics; C. Scuoppo for helping with the design of the FBXO11-shRNAs; M. Wu, H. Tang, T. Mo, and J. Zhong for technical support; and the Flow Cytometry Shared Resource of the Herbert Irving Comprehensive Cancer Center at Columbia University for assistance with the cell-sorting procedures.
F.H.S. was supported by a fellowship of the Dr. Werner Jackstaedt Foundation; and C.S. by a fellowship of the Mildred-Scheel-Stiftung, Deutsche Krebshilfe, Germany.
Authorship
Contribution: C.S., K.B., and R.D.-F. designed the study, analyzed results, and wrote the manuscript; C.S., K.B., L.A., Q.S., F.H.S., B.T., and M.B. performed experiments; D.D.-S. and G.B. performed the histopathological analyses; W.G. and N.K. generated the FBXO11fl/fl mice; and all authors read and approved the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for C.S. is Department of Internal Medicine III, Ulm University, Ulm, Germany.
The current affiliation for D.D.-S. is Department of Oncological Sciences and Department of Pathology, Icahn School of Medicine at Mount Sinai, New York, NY.
Correspondence: Riccardo Dalla-Favera, Institute for Cancer Genetics, Columbia University, 1130 St. Nicholas Ave, New York, NY 10032; e-mail: rd10@cumc.columbia.edu.
References
Author notes
K.B. and R.D.-F. contributed equally to this study.


![Figure 2. FBXO11 deletion leads to an increase in GC B cells and affects the DZ/LZ ratio. (A) Flow cytometry analysis of splenic mononuclear cells from age-matched FBXO11+/+-Cγ1cretg/+ (WT), FBXO11fl/+-Cγ1cretg/+ (HET), and FBXO11fl/fl-Cγ1cretg/+ (KO) for lectin PNA and the B-cell marker B220. Mice were immunized with SRBCs 10 days prior to the analysis. (B) Percentages of B220+/PNA+ splenic GC B cells. n = number of analyzed mice, average ± standard deviation [SD]; *P < .05 (1-way ANOVA). (C) Average GC size (left), numbers (middle) and GC/spleen area ratios obtained by staining with anti-PNA at least 3 independent spleen sections for each mouse. Pix, pixels. n = number of analyzed mice, average ± SD; *P < .05 (1-way ANOVA). (D) Representative flow cytometry analysis of splenic GC DZ and LZ cells as defined by CXCR4 and CD86 expression. (E) DZ/LZ cell ratios determined as in panel D in FBXO11 WT, HET, and KO mice. n = number of analyzed mice, average ± SD; *P < .05 (1-way ANOVA). (F) DZ/LZ cell ratios in Iμ-HA-BCL6 mice and WT littermates. n = number of analyzed mice, average ± SD; *P < .05 (1-way ANOVA).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/128/5/10.1182_blood-2015-11-684357/4/m_660f2.jpeg?Expires=1767896345&Signature=ZHkm~ef5tXLtyKbTCLDGZBn2ksiGZ8DZC0KIZVbwWY8SVIkjcrEme9tGLc5p3X29Ca99nXsTI3lgaO6sFu3Um1gn38ylnhwuTrNUfTQ3W6dd1o~Qu89WamFFZFwVro4JUjeoLaAHzfhBJ3KZ9w8TxXpme5tKaKZaky3vabU4uiX~1paJvo2cuOpd~ItJ30oYORr2Dv2KkmUJPzUJLn4q9i7nXgh75QyaLDhvkQRlh8NbiXnnCVe~xuWB3dff4ZaMS5Hq7UFWm2mIQBwHA8nF7e90AE9oBKMKYVzBcUa2~EG7bLPkoNYax6VmvRdjn0~FxNq5is9SpwRrp1Lj6zCPRw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal