Key Points
Aberrant splicing is an underrecognized mechanism causing VWD and is affected by shear stress.
Alternative splicing of endothelial VWF occurs in the normal population.
Abstract
Approximately 10% of von Willebrand factor (VWF) gene mutations are thought to alter messenger RNA (mRNA) splicing through disruption of consensus splice sites. This mechanism is likely underrecognized and affected by mutations outside consensus splice sites. During VWF synthesis, splicing abnormalities lead to qualitative defects or quantitative deficiencies in VWF. This study investigated the pathologic mechanism acting in 3 von Willebrand disease (VWD) families with putative splicing mutations using patient-derived blood outgrowth endothelial cells (BOECs) and a heterologous human embryonic kidney (HEK 293(T)) cell model. The exonic mutation c.3538G>A causes 3 in-frame splicing variants (23del, 26del, and 23/26del) which cannot bind platelets, blood coagulation factor VIII, or collagen, causing VWD through dominant-negative intracellular retention of coexpressed wild-type (WT) VWF, and increased trafficking to lysosomes. Individuals heterozygous for the c.5842+1G>C mutation produce exon 33 skipping, exons 33-34 skipping, and WT VWF transcripts. Pathogenic intracellular retention of VWF lacking exons 33-34 causes their VWD. The branch site mutation c.6599-20A>T causes type 1 VWD through mRNA degradation of exon 38 skipping transcripts. Splicing ratios of aberrant transcripts and coexpressed WT were altered in the BOECs with exposure to shear stress. This study provides evidence of mutations outside consensus splice sites disrupting splicing and introduces the concept that VWF splicing is affected by shear stress on endothelial cells.
Introduction
von Willebrand disease (VWD) is the most common bleeding disorder in humans, present in 1:100 individuals, with a symptomatic prevalence of 1:1000.1,2 It is characterized by prolonged and excessive mucocutaneous bleeding which can be quantified using a bleeding assessment tool (BAT) to determine an individual’s bleeding score (BS).3 There are 3 main types of VWD: type 1 with mild to moderate quantitative deficiencies in von Willebrand factor (VWF), type 2 reflecting qualitative VWF defects, and type 3 where VWF is virtually absent.4 Autosomal inheritance, commonly of VWF mutations, often leads to pathologic family bleeding histories; however, issues of incomplete penetrance and variable expressivity complicate diagnosis, especially in type 1 VWD.4
VWD is caused by a variety of mutations, although in over 30% of type 1 cases the pathogenic mutation has not been identified.5 Approximately 10% of individuals diagnosed with type 1 VWD are suspected to have mutations in their VWF gene which disrupt the normal splicing of VWF messenger RNA (mRNA), leading to aberrant or abolished protein production.5 This estimate primarily considers canonical splice site mutations and, unfortunately, often the underlying molecular basis of these mutations is not thoroughly investigated and the pathologic mechanisms remain uncharacterized. Therefore, it is likely that mutations further removed from canonical splice sites affect VWF splicing and this pathologic splicing is underrecognized.
Splicing is a vital and strictly regulated cellular process involving intron removal and exon ligation in primary pre-mRNA transcripts before translation of the mRNA.6 Although many genes have naturally occurring splice variants, incorrect splicing can result in the removal of important exons or integration of nonsensical intronic sequences into the mRNA transcript. This can lead to degradation of the malformed mRNA, production of defective or truncated proteins, and can have pathologic outcomes. Initially, only mutations of the most highly conserved nucleotides in intron-exon boundaries were thought to disrupt normal splicing; however, recent studies have implicated exonic and intronic mutations in this pathologic process.7
This study investigates 3 VWD families with putative splicing mutations located within exonic, intronic, and consensus splice site regions of VWF to provide support for splicing acting as an underrecognized mechanism causing VWD. Patient-derived blood outgrowth endothelial cells (BOECs) and heterologous human embryonic kidney (HEK 293(T)) cell models were used to elucidate the pathologic mechanisms acting in these families.
Methods
Patients: genotyping and phenotyping
Patients were selected from the Canadian Type 1 VWD Study5 and Canadian Type 3 VWD Study8 when in silico splicing analysis indicated that their mutations may disrupt or abolish recognition of a VWF splice site, and BOECs were accessible. All participants gave informed consent, and this study was approved by the Research Ethics Board of Queen’s University, Kingston, Canada. The severity of these patients’ bleeding symptoms was evaluated using the Condensed MCMDM-1VWD BAT where a BS of ≥4 denotes abnormal bleeding.9 Peripheral blood samples were taken to obtain plasma, genomic DNA, and BOECs from these patients. Putative splicing mutations were confirmed though Sanger DNA sequencing (supplemental Table 1, available on the Blood Web site). The patients’ VWD diagnoses were confirmed through a VWF antigen (VWF:Ag) enzyme-linked immunosorbent assay (ELISA)10 and a ristocetin cofactor assay (VWF:RCo).11 VWF multimer analysis was completed on a 1.7% sodium dodecyl sulfate–polyacrylamide gel electrophoresis gel,12 and factor VIII (FVIII) coagulant activity (FVIII:C) was measured in a 1-stage assay.13 Type 1 and 3 collagen and FVIII (Advate) binding were also assessed by ELISA.14,15
BOEC isolation and culture
BOECs were isolated from a 48-mL peripheral blood sample from patients and 3 normal healthy controls, in CPT vacutainer tubes (BD Biosciences) which were centrifuged for 30 minutes at 1600 relative centrifugal force.16-18 The plasma and mononuclear cells were resuspended in phosphate-buffered saline supplemented with fetal bovine serum. After several cycles of centrifugation and resuspension, cells were seeded onto collagen-coated 6-well tissue-culture plates at a density of 3 × 107 to 5 × 107 cells per well using endothelial cell growth medium-2 media supplemented with 10% fetal bovine serum, antibiotics, and the endothelial cell growth medium-2 bullet kit. BOEC colonies appeared between day 9 and 23 in culture and were characterized through flow cytometry for CD31+, CD144+, CD14−, and CD45− surface markers.17
Immunofluorescent confocal microscopy
BOECs were imaged for intracellular VWF (rabbit anti-human A0084, 1:500; DAKO) and actin (phalloidin-tetramethylrhodamine; Sigma-Aldrich). Biosynthesis and trafficking of the aberrant splice forms were evaluated through immunofluorescent staining of subcellular compartments 48 hours after Lipofectamine (Invitrogen) transient transfection into HEK293 cells as per the manufacturer’s recommendations. Primary antibodies include: mouse anti-protein disulphide-isomerase (PDI) (endoplasmic reticulum [ER], 1:100; Abcam), mouse anti-GOLPH4 (cis-Golgi, 1:100; Santa Cruz Biotechnology), goat anti-Rab 27a (pseudo-Weibel Palade body [WPB], 1:50; Santa Cruz Biotechnology), mouse anti-Lamp1 (lysosome, 1:100; R&D Systems), and VWF (rabbit anti-VWF, 1:500; DAKO). Secondary antibodies include: Alexa Fluor 488 goat anti-rabbit immunoglobulin G (IgG; Invitrogen), Alexa Fluor 568 donkey anti-goat IgG (Invitrogen), and rhodamine donkey anti-goat IgG (Santa Cruz Biotechnology).19,20 Imaging was conducted using a Quorum Wave Effects Spinning Disc confocal microscope at ×60 magnification, captured with a Hamamatsu Orca high-resolution camera and ImageJ software analysis. VWF colocalization with organelles was quantified using MetaMorph microscopy automation and ImageJ analysis software (Molecular Devices) to calculate the Manders colocalization coefficient (MCC).21
Shear stress
BOECs were plated to confluence on 0.2-µ Luer slides (Ibidi) and subjected to laminar shear stress at 50 dynes/cm2 for 48 hours using an Ibidi pump system. RNA was extracted with a RNAqueous- Micro kit (Ambion) to assess the effect of shear stress on splicing.22
Transcript identification and quantification
BOEC RNA was isolated from the patients using the QIAamp RNA Blood Mini kit (Qiagen) and reverse transcribed to visualize variation in RNA splicing products (supplemental Table 2). Splicing was evaluated through reverse transcription polymerase chain reaction of the VWF gene in 9 overlapping segments (exons 1-9, 8-17, 16-21, 20-28, 28-31, 30-36, 35-41, 40-45, 44-52) (supplemental Table 3). These segments were visualized on 1% agarose gels, and bands of variant sizes were sequenced to identify splicing alterations. The ratio of wild-type (WT) and aberrantly spliced transcripts was quantified with the Superscript III Platinum SYBR Green 1-step quantitative reverse transcription polymerase chain reaction kit (Invitrogen) as per the manufacturer’s protocol.
Identification of circulating aberrant splice forms
Western blotting was used to detect the presence of heteromultimers in the patient plasma. Plasma was reduced and run on a 10% gel (4561039S; Bio-Rad ) for 2 hours at 100 V, transferred to a polyvinylidene fluoride membrane for 90 minutes at 50 V, and blocked with a 5% skim milk solution overnight at 4°C. VWF was visualized using a polyclonal rabbit anti-human VWF/horseradish peroxidase antibody (P0226; DAKO) and chemiluminescent detection. Recombinant WT and aberrantly spliced VWF from HEK293T cells were used to determine protein sizing.
Plasmids and transient transfections
Expression vectors of each identified aberrant splice form were created using QuikChange II site-directed mutagenesis (Agilent Technologies) of a pCIneoVWF-ESN expression vector (supplemental Table 4). Expression vectors were transiently transfected using calcium chloride into HEK293T cells to evaluate VWF synthesis and expression and HEK293 cells for intracellular localization studies.23,24 To ensure that alterations in VWF expression levels were caused by the mutant plasmids and not the decreased amount of WT complementary DNA in the transfection cocktail, a blank vector not coding for VWF (mock) was also evaluated.
Constitutive and stimulated VWF secretion
The capacity of the BOECs to produce and secrete VWF constitutively and after stimulation by the secretagogue phorbol 12-myristate 13-acetate (PMA) at 160 nM for 1 hour was evaluated by VWF:Ag ELISA of media and lysates after 24 hours in Opti-MEM reduced serum media (Life Technologies).18 HEK293T cells were used to evaluate constitutive and stimulated secretion of identified splicing mutants 48 hours after transfection with and without cotransfected WT VWF expression vectors.24
In vitro characterization of aberrant VWF splice forms
Recombinant mutant VWF was concentrated from transfection supernatants using Centricon Plus-70 centrifugal filter units (EMD Millipore) and evaluated for its ability to bind platelets (VWF:RCo), collagen (ELISA), and FVIII (Advate ELISA).14,15 Multimerization of aberrant splice forms was visualized as described in “Patients: genotyping and phenotyping.”12 Proteasomal degradation of the aberrant proteins was assessed through proteasomal inhibition by MG-132.25
Data analysis
Statistical analyses were performed using the unpaired Student t test. MCC was used to assess colocalization.21
Results
Patient genotypes and phenotypes
Six individuals from 3 families suspected of having a splicing mutation underlying their VWD were enrolled (Figure 1A-C). Phenotypic testing was conducted to confirm the diagnosis of VWD in these patients by VWF:Ag and VWF:RCo levels lower than 0.5 IU/mL, and abnormal BS ≥4 using the MCMDM-1VWD BAT (Table 1).9
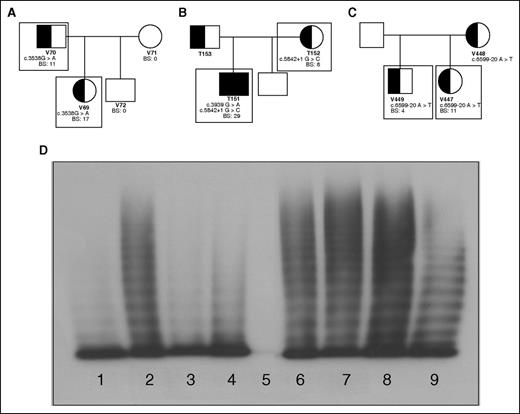
Family pedigrees; patient genotypic and phenotypic characteristics. Patients included in this study are outlined with boxes. (A) Family 1 with an exonic mutation, c.3538G>A, causing type 2A VWD in a father and daughter. (B) Family 2 with a consensus splice site mutation, c.5842+1G>C, causing mild type 1 VWD mother and type 3 VWD when coinherited with c.3939G>A in her son. (C) Family 3 with an intronic mutation c.5699-20A>T causing type 1 VWD in 2 siblings. (D) Plasma multimers from affected individuals. (1) Abnormal type 2A VWD control; (2) NPP control; (3) patient V69 with reduced HMWM; (4) patient V70 with reduced HMWM; (5) patient T151 showing no multimers; (6) patient T152 with full-range multimers; (7) patient V447 with full-range multimers; (8) patient V449 with full-range multimers.
Family pedigrees; patient genotypic and phenotypic characteristics. Patients included in this study are outlined with boxes. (A) Family 1 with an exonic mutation, c.3538G>A, causing type 2A VWD in a father and daughter. (B) Family 2 with a consensus splice site mutation, c.5842+1G>C, causing mild type 1 VWD mother and type 3 VWD when coinherited with c.3939G>A in her son. (C) Family 3 with an intronic mutation c.5699-20A>T causing type 1 VWD in 2 siblings. (D) Plasma multimers from affected individuals. (1) Abnormal type 2A VWD control; (2) NPP control; (3) patient V69 with reduced HMWM; (4) patient V70 with reduced HMWM; (5) patient T151 showing no multimers; (6) patient T152 with full-range multimers; (7) patient V447 with full-range multimers; (8) patient V449 with full-range multimers.
Patient VWD phenotypes
| Mutation . | Patient ID (sex) . | Bleeding Score . | VWF:Ag, IU/mL . | VWF:RCo, IU/mL . | FVIII:C, IU/mL . | VWF:collagen binding, IU/mL . | VWF:FVIII binding, IU/mL . | Multimers . |
|---|---|---|---|---|---|---|---|---|
| c.3538G>A | V69 (F) | 17 | 0.13 | 0.12 | 0.15 | 0.09 | 0.07 | Lack HMWM |
| V70 (M) | 11 | 0.27 | 0.17 | 0.45 | 0.13 | 0.20 | Lack HMWM | |
| c.5842+1G>Cc.3939G>A | T151 (M) | 29 | 0.02 | 0.04 | 0.01 | 0.04 | 0.03 | None |
| c.5842+1G>C | T152 (F) | 8 | 0.49 | 0.42 | 1.16 | 0.61 | 0.911 | Normal |
| c.6599-20 A>T | V447 (F) | 11 | 0.41 | 0.38 | 0.51 | 0.55 | 0.71 | Normal |
| V449 (M) | 4 | 0.44 | 0.44 | 0.75 | 0.50 | 0.70 | Normal |
| Mutation . | Patient ID (sex) . | Bleeding Score . | VWF:Ag, IU/mL . | VWF:RCo, IU/mL . | FVIII:C, IU/mL . | VWF:collagen binding, IU/mL . | VWF:FVIII binding, IU/mL . | Multimers . |
|---|---|---|---|---|---|---|---|---|
| c.3538G>A | V69 (F) | 17 | 0.13 | 0.12 | 0.15 | 0.09 | 0.07 | Lack HMWM |
| V70 (M) | 11 | 0.27 | 0.17 | 0.45 | 0.13 | 0.20 | Lack HMWM | |
| c.5842+1G>Cc.3939G>A | T151 (M) | 29 | 0.02 | 0.04 | 0.01 | 0.04 | 0.03 | None |
| c.5842+1G>C | T152 (F) | 8 | 0.49 | 0.42 | 1.16 | 0.61 | 0.911 | Normal |
| c.6599-20 A>T | V447 (F) | 11 | 0.41 | 0.38 | 0.51 | 0.55 | 0.71 | Normal |
| V449 (M) | 4 | 0.44 | 0.44 | 0.75 | 0.50 | 0.70 | Normal |
Average VWF:Ag, VWF:RCo, and FVIII:C from all available patient plasma levels. BS ≥ 4 indicate symptomatic abnormal bleeding. The normal ranges for VWF:Ag, VWF:RCo, FVIII:C, collagen binding, and FVIII binding are between 0.50 and 1.50 IU/mL.
F, female; ID, identification; M, male.
The first family (with patients V69 and V70) exhibited significant mucocutaneous bleeding with positive BSs and correspondingly low VWF and FVIII plasma levels (Table 1). Both patients were heterozygous for the exonic mutation c.3538G>A in exon 26 of VWF. Their plasma VWF had a decreased capacity to bind collagen and FVIII (Table 1). They also lacked high-molecular-weight multimers (HMWMs) characteristic of type 2A VWD (Figure 1D; Table 1).
In family 2, heterozygosity for the consensus splice site mutation c.5842+1G>C in intron 34 was identified in the type 1 VWD patient (T152) and her type 3 son (T151) who also inherited the nonsense mutation c.3939G>A, contributing to his increased bleeding tendency and decreased factor levels (Figure 1B). Both patients had bleeding phenotypes, factor levels, and multimerization reflective of their VWD status (Table 1; Figure 1D). Collagen binding was minimal in T151; however, T152’s plasma VWF reflected normal collagen and FVIII-binding abilities (Table 1).
Family 3 patients, V447 and V449, were heterozygous for the intron 37 mutation c.6599-20A>T (Figure 1C) with mild reductions in VWF:Ag and VWF:RCo (Table 1). This mutation is thought to be a branch site mutation and therefore integral to proper splicing surrounding intron 37. Both individuals have phenotypically mild type 1 VWD and their plasma VWF retains its full collagen and FVIII-binding capacity (Table 1) and normal multimers (Figure 1D).
Ex vivo BOEC analyses
BOECs were isolated from affected individuals as an ex vivo model to evaluate the mechanisms behind their VWD; however, BOEC culture was unsuccessful from patient V69. BOECs were positive for endothelial markers CD31 and CD144, and negative for monocyte markers CD14 and CD45. Constitutive VWF expression levels from the BOECs were reflective of VWF levels in the patients’ plasma and decreased compared with normal BOECs (Figure 2). The type 1 BOECs were responsive to PMA stimulus, which replicates the patients’ responses to DDAVP in vivo; however, the type 3 BOECs from T151 showed a negligible response, as expected (Figure 2). BOECs from V70 showed intracellular retention of VWF (P < .001), T151 and V447 had reduced intracellular VWF relative to the normal BOECs (P < .001; P < .05), and T152 and V449 had lysate levels in the normal range (supplemental Figure 1).
Constitutive and induced VWF expression from BOECs. BOECs constitutive VWF expression from unstimulated cells as well as PMA (160 nM, 1 hour) stimulated release of BOEC WPBs (N = 3; *P < .05; **P < .01; ***P < .001). Underlined significance stars indicate significant increase in VWF release with PMA stimulation. Significance stars without underlining indicate a significant difference in expression compared with unstimulated BOECs from 3 normal controls. The average expression from the normal BOECs was set to 1 and the patient BOECs were presented relative to the normals.
Constitutive and induced VWF expression from BOECs. BOECs constitutive VWF expression from unstimulated cells as well as PMA (160 nM, 1 hour) stimulated release of BOEC WPBs (N = 3; *P < .05; **P < .01; ***P < .001). Underlined significance stars indicate significant increase in VWF release with PMA stimulation. Significance stars without underlining indicate a significant difference in expression compared with unstimulated BOECs from 3 normal controls. The average expression from the normal BOECs was set to 1 and the patient BOECs were presented relative to the normals.
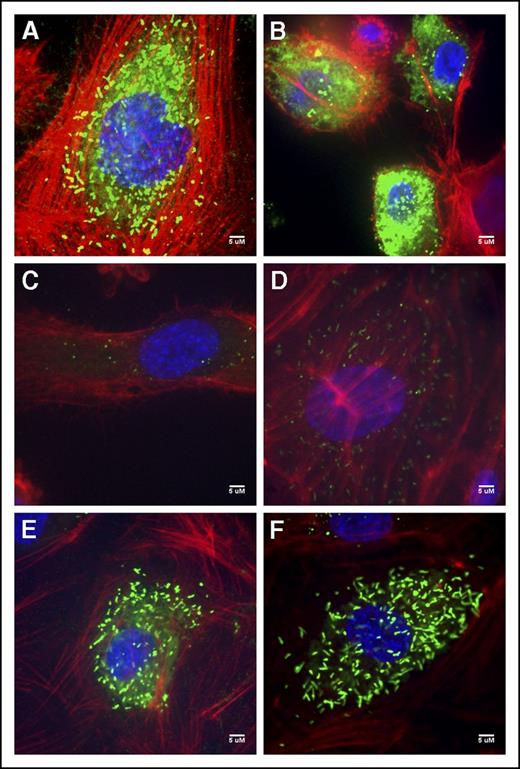
Immunofluorescent staining of the V70 BOECs exhibited diffuse staining as well as punctate and rounded WPB; many of the cells also exemplified the intense VWF accumulation quantified within the cell lysates (Figure 3B; supplemental Figure 1). T151’s BOECs contained few rounded WPBs and some diffuse staining (Figure 3C). The type 1 T152 BOECs showed both rounded WPBs and the rod-shaped WPBs characteristic of these organelles (Figure 3D). BOECs of patients V447 and V449 were unremarkable from normal BOECs (Figure 3A), exhibiting elongated WPBs and some diffuse staining particularly within V447’s BOECs (Figure 3E-F).
Normal and patient BOEC VWF immunofluorescent confocal images. (A) Normal BOECs with rod-shaped WPBs. (B) Patient V70 (c.3538G>A) with punctate VWF as well as diffuse VWF staining throughout the cells. (C) Type 3 patient T151 (c.3939G>A, c.5842+1G>C) with few punctate WPBs. (D) Patient T152 (c.5842+1G>C) BOECs show not only more classic, but also rounded WPBs. (E) Patient V447’s (c.6599-20A>T) VWF appears diffuse within the cell as well as localized to punctate WPBs. (F) VWF staining in patient V449’s (c.6599-20A>T) appear relatively normal with many rod-shaped WPBs.). Imaging was conducted at room temperature using a Quorum Wave Effects Spinning Disc confocal microscope at ×60 magnification and captured with a Hamamatsu Orca high-resolution camera and ImageJ software analysis. Scale bar, 5 μM.
Normal and patient BOEC VWF immunofluorescent confocal images. (A) Normal BOECs with rod-shaped WPBs. (B) Patient V70 (c.3538G>A) with punctate VWF as well as diffuse VWF staining throughout the cells. (C) Type 3 patient T151 (c.3939G>A, c.5842+1G>C) with few punctate WPBs. (D) Patient T152 (c.5842+1G>C) BOECs show not only more classic, but also rounded WPBs. (E) Patient V447’s (c.6599-20A>T) VWF appears diffuse within the cell as well as localized to punctate WPBs. (F) VWF staining in patient V449’s (c.6599-20A>T) appear relatively normal with many rod-shaped WPBs.). Imaging was conducted at room temperature using a Quorum Wave Effects Spinning Disc confocal microscope at ×60 magnification and captured with a Hamamatsu Orca high-resolution camera and ImageJ software analysis. Scale bar, 5 μM.
RNA investigation
Total RNA was isolated from the BOECs, and VWF mRNA was reverse transcribed to identify any aberrant band patterns reflective of variant splicing in the patient RNA, particularly around the region of their individual mutations. Patient V70 BOEC RNA contained 4 VWF splice forms, WT VWF, skipping of exon 23 (23del), skipping of exon 26 (26del), and combined skipping of exons 23 and 26 (23/26del). In static culture, these aberrant transcripts comprised the majority of the patient’s VWF mRNA (Table 2). These additional transcripts are in-frame changes which should result in production of the aberrant VWF protein.
Quantification of variant VWF splice forms
| Patient (mutation) . | Splice form . | % of BOECs VWF RNA under static conditions . | % of BOEC VWF RNA under 50 dynes/cm2 shear stress . |
|---|---|---|---|
| V70 (c.3538G>A) | WT | 2 ± 0.6 | 98 ± 1** |
| 23del | 22 ± 7 | 0 ± 0** | |
| 26del | 64 ± 3 | 2 ± 1** | |
| 23/26del | 11 ± 8 | 0 ± 0.5* | |
| T151 (c.5842+1 G>C; c.3939G>A) | WT | 41 ± 8 | n/a |
| 33del | 15 ± 3 | n/a | |
| 33-34del | 44 ± 4 | n/a | |
| T152 (c.5842+1 G>C) | WT | 51 ± 7 | 22 ± 9** |
| 33del | 22 ± 11 | 68 ± 13** | |
| 33-34del | 27 ± 5 | 10 ± 1** | |
| V447 (c.6599-20 A>T) | WT | 97 ± 31 | 40 ± 3* |
| 38del | 2 ± 0.85 | 60 ± 5* | |
| V449 (c.6599F-20 A>T) | WT | 97 ± 31 | 2 ± 2* |
| 38del | 3 ± 4 | 98 ± 22* | |
| Normal BOECs (N = 3) | WT | 86 ± 0.9 | 65 ± 3* |
| 23del | 0 ± 0 | 0 ± 0 | |
| 26del | 0 ± 0 | 0 ± 0 | |
| 23/26del | 0 ± 0 | 0 ± 0 | |
| 33del | 13 ± 0.3 | 34 ± 3* | |
| 33-34del | 0 ± 0 | 0 ± 0 | |
| 38del | 0 ± 0 | 0 ± 0 |
| Patient (mutation) . | Splice form . | % of BOECs VWF RNA under static conditions . | % of BOEC VWF RNA under 50 dynes/cm2 shear stress . |
|---|---|---|---|
| V70 (c.3538G>A) | WT | 2 ± 0.6 | 98 ± 1** |
| 23del | 22 ± 7 | 0 ± 0** | |
| 26del | 64 ± 3 | 2 ± 1** | |
| 23/26del | 11 ± 8 | 0 ± 0.5* | |
| T151 (c.5842+1 G>C; c.3939G>A) | WT | 41 ± 8 | n/a |
| 33del | 15 ± 3 | n/a | |
| 33-34del | 44 ± 4 | n/a | |
| T152 (c.5842+1 G>C) | WT | 51 ± 7 | 22 ± 9** |
| 33del | 22 ± 11 | 68 ± 13** | |
| 33-34del | 27 ± 5 | 10 ± 1** | |
| V447 (c.6599-20 A>T) | WT | 97 ± 31 | 40 ± 3* |
| 38del | 2 ± 0.85 | 60 ± 5* | |
| V449 (c.6599F-20 A>T) | WT | 97 ± 31 | 2 ± 2* |
| 38del | 3 ± 4 | 98 ± 22* | |
| Normal BOECs (N = 3) | WT | 86 ± 0.9 | 65 ± 3* |
| 23del | 0 ± 0 | 0 ± 0 | |
| 26del | 0 ± 0 | 0 ± 0 | |
| 23/26del | 0 ± 0 | 0 ± 0 | |
| 33del | 13 ± 0.3 | 34 ± 3* | |
| 33-34del | 0 ± 0 | 0 ± 0 | |
| 38del | 0 ± 0 | 0 ± 0 |
N = 3; *P < .05; **P < .01; ***P < .001 comparing transcripts ratios between BOECs grown under static conditions vs under 48-hour subjection to shear stress at 50 dynes/cm2. n/a represents BOECs with proliferative and adhesive properties that did not permit evaluation under high shear rates.
VWF mRNA from patients T151 and T152 was composed of 2 additional splice forms alongside WT VWF. Skipping of exon 33 (33del) was observed which causes a frameshift introducing a premature termination codon (PTC) in exon 34. Additionally, in-frame skipping of both exons 33 and 34 (33-34del) was found (Table 2). Analysis of heterozygous single-nucleotide polymorphisms indicated that the 33del transcript was produced from T152’s normal VWF allele, and was found to comprise 13% ± 0.3% of the VWF RNA in the normal control BOECs in static culture (Table 2).
Family 3 (V447 and V449) BOECs contained primarily WT VWF with minimal levels of a transcript skipping exon 38 (38del) which would introduce a PTC in exon 40. This PTC would target these transcripts for nonsense mediated decay rather than protein translation possibly explaining its low abundance in the patient RNA (Table 2).
When subjected to shear stress at 50 dynes/cm2 for 48 hours, the proportions of the VWF splice forms of BOECs changed. Patient V70’s transcripts shifted substantially in favor of WT, comprising 98% ± 1% of their VWF after stimulation (Table 2). Patient T152’s skipping exon 33 frameshift transcript increased significantly under shear (P < .01) (Table 2). This increase of exon 33 skipping was also observed in normal controls (P < .05). Limited adherence of T151’s BOECs under laminar flow prevented analysis of their RNA after exposure to high shear stress. Once subjected to high shear stress, the proportions of WT:38del transcripts shifted in favor of exon 38 skipping in patients V447 and V449 (P < .05) (Table 2).
Identification of circulating aberrant splice forms
Western blot analysis for circulating aberrant proteins was undertaken. The aberrant splice forms predicted by family 1’s VWF RNA profile were all in-frame changes of small exons, which produce proteins that are 2% to 4% smaller than WT VWF and, thus, distinct aberrant bands were not discernible from WT (Figure 4A). The recombinant 23del was significantly retained in the HEK293T cells and was not visualized from concentrated media therefore immunoblotting of 23del from cell lysates was performed. An additional band at ∼180 kDa was observed in patients T151 and T152, corresponding with the size of the aberrant splice form 33del; however, the in-frame 33-34del splice form did not sufficiently segregate from WT VWF due to a 3% weight difference (Figure 4B). Family 3 displayed no additional bands at the size expected for aberrant 38del VWF (Figure 4C).
Western blot analysis of plasma and recombinant VWF. (A) Family 1 plasma shows thick bands unable to separate potential low concentrations of mutants from similarly sized WT VWF. Lane 1, NPP. Lane 2, Patient V69 plasma. Lane 3, Patient V70 plasma. Lane 4, Recombinant WT VWF. Lane 5, Recombinant 23del VWF. Lane 6, Recombinant 26del VWF. Lane 7, Recombinant 23/26del VWF. Lane 8, Recombinant 23del lysate. (B) Family 2 plasma exhibits an additional band which size corresponds to 33del; 33-34del did not significantly separate from WT VWF. Lane 1, Recombinant WT VWF. Lane 2, Patient T151 plasma. Lane 3, Patient T152 plasma. Lane 4, NPP. Lane 5, Recombinant WT VWF. Lane 6, Recombinant 33del VWF. Lane 7, Recombinant 33-34del VWF. (C) Family 3 plasma shows no additional bands other than WT VWF. Lane 1, NPP. Lane 2, Patient V447 plasma. Lane 3, Patient V449 plasma. Lane 4, Recombinant WT VWF. Lane 5, Recombinant 38del VWF.
Western blot analysis of plasma and recombinant VWF. (A) Family 1 plasma shows thick bands unable to separate potential low concentrations of mutants from similarly sized WT VWF. Lane 1, NPP. Lane 2, Patient V69 plasma. Lane 3, Patient V70 plasma. Lane 4, Recombinant WT VWF. Lane 5, Recombinant 23del VWF. Lane 6, Recombinant 26del VWF. Lane 7, Recombinant 23/26del VWF. Lane 8, Recombinant 23del lysate. (B) Family 2 plasma exhibits an additional band which size corresponds to 33del; 33-34del did not significantly separate from WT VWF. Lane 1, Recombinant WT VWF. Lane 2, Patient T151 plasma. Lane 3, Patient T152 plasma. Lane 4, NPP. Lane 5, Recombinant WT VWF. Lane 6, Recombinant 33del VWF. Lane 7, Recombinant 33-34del VWF. (C) Family 3 plasma shows no additional bands other than WT VWF. Lane 1, NPP. Lane 2, Patient V447 plasma. Lane 3, Patient V449 plasma. Lane 4, Recombinant WT VWF. Lane 5, Recombinant 38del VWF.
Aberrant splice form characterization
Expression vectors reflecting each of the in-frame aberrant splice forms that were predicted to undergo translation (23del, 26del, 23/26del, 33-34del) were transiently transfected into HEK293Tcells to determine the functionality of these mutant proteins and their interaction with coproduced WT VWF. Functional testing was also performed on the 33del splice variant as the transcript was found in the normal BOECs as well as circulating protein in the patient plasma and may be a product of alternative splicing. VWF expression was significantly reduced in all mutant homozygous and heterozygous transfections with the exception of 33del (Figure 5A). Recombinant 23del and 23/26del demonstrated a dominant-negative effect on the expression of cotransfected WT VWF with expression levels below 35% of WT expression when cotransfected (50:50) with WT VWF (Figure 5A) and intracellular retention of the VWF (supplemental Figure 2). When transfected in the absence of WT VWF, none of these aberrant splice forms showed a positive VWF release in response to stimulus with PMA; however, this stimulated release occurred in cells cotransfected with 26del, 23/26del, 33del, or 33-34del alongside WT VWF (Figure 5A). Normal VWF expression and PMA-stimulated secretion was exhibited when WT VWF was cotransfected with the mock expression vector (Figure 5A).
WT and VWF variant expression and functionality from transfected HEK293T cells. (A) Constitutive expression was derived from unstimulated cells and release of VWF from pseudo-WPB was measured after stimulation with PMA. 100% indicates homozygous transfections; 50% indicates transfections in the mock heterozygous state where HEK293T cells were transfected with 50% WT vector and 50% mutant vector. Black significance stars without a bar below represent a significant difference in constitutive expression of the mutants compared with WT. Significance stars with a bar below indicate a significant increase in secretion between constitutive and stimulated HEK293T cells (N = 3; *P < .05; **P < .01; ***P < .001) The absence of VWF from the 100% mock, and restoration of normal VWF levels in the 50% mock, show that expression reductions across the other variables are in fact due to the respective mutant VWF. (B) Functionality of the VWF splice variants from expression in transiently transfected HEK293Tcells measured by VWF:RCo, FVIII:B ELISA, and collagen-binding ELISA. 100% indicates homozygous transfections; 50% indicates transfections in the mock heterozygous state where HEK293T cells were transfected with 50% WT vector and 50% mutant vector (N = 3). Significance stars compare the binding properties of the splicing mutants to WT VWF (*P < .05; **P < .01).
WT and VWF variant expression and functionality from transfected HEK293T cells. (A) Constitutive expression was derived from unstimulated cells and release of VWF from pseudo-WPB was measured after stimulation with PMA. 100% indicates homozygous transfections; 50% indicates transfections in the mock heterozygous state where HEK293T cells were transfected with 50% WT vector and 50% mutant vector. Black significance stars without a bar below represent a significant difference in constitutive expression of the mutants compared with WT. Significance stars with a bar below indicate a significant increase in secretion between constitutive and stimulated HEK293T cells (N = 3; *P < .05; **P < .01; ***P < .001) The absence of VWF from the 100% mock, and restoration of normal VWF levels in the 50% mock, show that expression reductions across the other variables are in fact due to the respective mutant VWF. (B) Functionality of the VWF splice variants from expression in transiently transfected HEK293Tcells measured by VWF:RCo, FVIII:B ELISA, and collagen-binding ELISA. 100% indicates homozygous transfections; 50% indicates transfections in the mock heterozygous state where HEK293T cells were transfected with 50% WT vector and 50% mutant vector (N = 3). Significance stars compare the binding properties of the splicing mutants to WT VWF (*P < .05; **P < .01).
As expected, VWF function was generally negatively affected in these aberrant splice forms. Binding to platelets (VWF:RCo), FVIII, and collagen is negligible in 23del, 26del, and 23/26del aberrant splice forms when produced in the absence of WT VWF (Figure 5B). When these splicing mutants were cotransfected with WT VWF, the functionality of the VWF secreted from the HEK293T cells increased: heterozygous 23del regained marginal FVIII binding and a full capacity to bind collagen (Figure 5B), 26del regained platelet and collagen-binding abilities with a marginal increase in FVIII binding; additionally, 23/26del regained moderate functionality by all 3 measures (Figure 5B). Interestingly, despite an abolishment of platelet and collagen binding, the 33del splice form had an increased FVIII-binding affinity, which was tempered by coexpressed WT VWF (Figure 5B). The in-frame 33-34del mutant retained moderate platelet and collagen binding and half the FVIII-binding capacity of WT VWF; these functions were only marginally ameliorated with WT coexpression (Figure 5B). No proteasomal degradation was observed from any of the splicing variants, with lysate VWF levels remaining consistent between unstimulated cells and after proteasome inhibition by MG-132 (supplemental Figure 3).
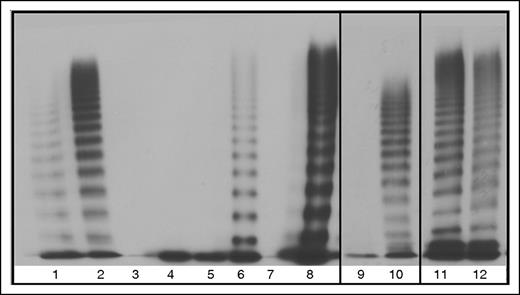
Multimerization was greatly affected by aberrant splicing of the VWF which can be seen by the lack of multimers in all of the homozygous mutant transfections, with the exception of 33-34del which formed band-shifted multimers due to the exons spliced out of each VWF monomer (Figure 6). Slight reparation of the band shift is seen in the heterozygous 33-34del multimers, indicating heteromultimerization of the mutant and WT VWF (Figure 6). Restoration of VWF multimerization was observed for HEK293T cells cotransfected with WT and 23/26del or 33del which may indicate minimal interaction between the unmultimerized aberrantly spliced VWF and cotransfected WT (Figure 6). Heterozygous 26del showed a possible decrease in HMWM which may indicate this aberrant protein may interact with WT VWF limiting multimerization capacity (Figure 6). No multimerization was observed in heterozygous 23del transfections, indicating that 23del VWF exerts a dominant-negative effect on coexpressed WT VWF multimerization and secretion.
Multimers from recombinant WT and mutant VWF splice variants secreted from HEK293T cells. 100% indicates homozygous transfections; 50% indicates transfections in the mock heterozygous state where HEK293T cells were transfected with 50% WT vector and 50% mutant vector. Lane 1, Abnormal 2A control. Lane 2, NPP control. Lane 3, 100% 23del. Lane 4, 50% 23del. Lane 5, 100% 26del. Lane 6, 50% 26del. Lane 7, 100% 23/26del. Lane 8, 50% 23/26del. Lane 9, 100% 33del. Lane 10, 50% 33 del. Lane 11, 100% 33-34del. Lane 12, 50% 33-34del.
Multimers from recombinant WT and mutant VWF splice variants secreted from HEK293T cells. 100% indicates homozygous transfections; 50% indicates transfections in the mock heterozygous state where HEK293T cells were transfected with 50% WT vector and 50% mutant vector. Lane 1, Abnormal 2A control. Lane 2, NPP control. Lane 3, 100% 23del. Lane 4, 50% 23del. Lane 5, 100% 26del. Lane 6, 50% 26del. Lane 7, 100% 23/26del. Lane 8, 50% 23/26del. Lane 9, 100% 33del. Lane 10, 50% 33 del. Lane 11, 100% 33-34del. Lane 12, 50% 33-34del.
Trafficking of aberrant splice forms
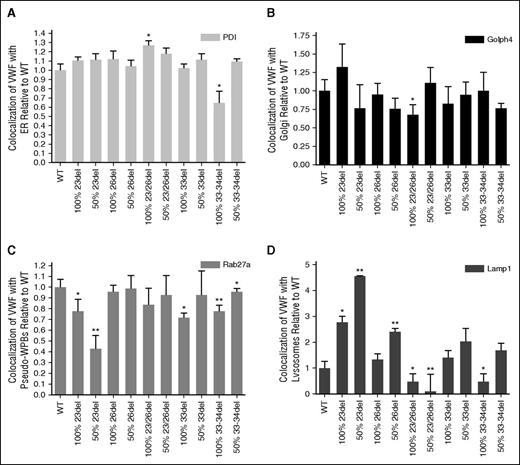
Costaining of transfected HEK293 cells for aberrantly spliced VWF and markers of organelles involved in VWF biosynthesis enabled the evaluation of mutant trafficking to explain the decreased expression of these mutants (supplemental Figures 4-14). Aberrantly spliced 23/26del showed significant increases in colocalization with the ER lumen marker PDI (P < .05; Figure 7A). Although many of the mutant transfections showed a trend of decreased colocalization with the Golgi body marker Golph4, this was only significant in the homozygous 23/26del transfection (P < .05; Figure 7B). Recombinant 23del and 33-34del showed decreased colocalization with the pseudo-WPB marker Rab27a in both homozygous and heterozygous transfections (Figure 7C). Similar decreases in pseudo-WPB localization occurred in the homozygous 33del transfections (P < .05; Figure 7C). Lysosomal colocalization was the most variable. Homozygous (P < .05) and heterozygous 23del and heterozygous 26del had significantly increased lysosomal colocalization compared with WT VWF (P < .01). Colocalization with the Lamp1 marker was decreased in heterozygous 23/26del and homozygous 33-34del splicing mutants (P < .05; Figure 7D).
Quantification of WT and mutant VWF colocalization with organelle markers. Staining for ER with mouse anti-PDI (ER, 1:100; Abcam) (A), mouse anti-GOLPH4 (cis-Golgi, 1:100; Santa Cruz Biotechnology) (B), goat anti-Rab 27a (pseudo-WPB, 1:50; Santa Cruz Biotechnology) (C), mouse anti-Lamp1 (lysosome, 1:100; R&D Systems) (D), and VWF (rabbit anti-VWF, 1:500; DAKO). Secondary antibodies: Alexa Fluor 488 goat anti-rabbit IgG (Invitrogen), Alexa Fluor 568 donkey anti-goat IgG (Invitrogen), and rhodamine donkey anti-goat IgG (Santa Cruz Biotechnology). 100% indicates homozygous transfections; 50% indicates transfections in the mock heterozygous state where HEK293T cells were transfected with 50% WT vector and 50% mutant vector. Imaging was conducted at room temperature using a Quorum Wave Effects Spinning Disc confocal microscope at ×60 magnification, captured with a Hamamatsu Orca high-resolution camera and ImageJ software analysis. Colocalization of the VWF and the organelles was quantified using MetaMorph microscopy automation and image analysis software (Molecular Devices) to calculate the MCC (N = 40-50 cells per marker; *P < .05; **P < .01).
Quantification of WT and mutant VWF colocalization with organelle markers. Staining for ER with mouse anti-PDI (ER, 1:100; Abcam) (A), mouse anti-GOLPH4 (cis-Golgi, 1:100; Santa Cruz Biotechnology) (B), goat anti-Rab 27a (pseudo-WPB, 1:50; Santa Cruz Biotechnology) (C), mouse anti-Lamp1 (lysosome, 1:100; R&D Systems) (D), and VWF (rabbit anti-VWF, 1:500; DAKO). Secondary antibodies: Alexa Fluor 488 goat anti-rabbit IgG (Invitrogen), Alexa Fluor 568 donkey anti-goat IgG (Invitrogen), and rhodamine donkey anti-goat IgG (Santa Cruz Biotechnology). 100% indicates homozygous transfections; 50% indicates transfections in the mock heterozygous state where HEK293T cells were transfected with 50% WT vector and 50% mutant vector. Imaging was conducted at room temperature using a Quorum Wave Effects Spinning Disc confocal microscope at ×60 magnification, captured with a Hamamatsu Orca high-resolution camera and ImageJ software analysis. Colocalization of the VWF and the organelles was quantified using MetaMorph microscopy automation and image analysis software (Molecular Devices) to calculate the MCC (N = 40-50 cells per marker; *P < .05; **P < .01).
Discussion
This study characterized VWF splicing mutations causing VWD in 3 families and, although the mechanism of disease varied between splice variants, links could be made between the behavior of the aberrant splice product, mutation location, the BOECs, and the patient phenotype.
Heterozygosity for c.3538G>A causes type 2A VWD in family 1, creating 3 aberrant splice forms: exon 23 skipping, exon 26 skipping, and exon 23 and 26 skipping simultaneously. This removal of exons in the D3 domain influences VWF multimerization and FVIII binding in the HEK293T model and patient plasma. The observed losses of VWF function in vitro may indicate downstream misfolding of the encoded protein which disrupts accessibility to these functional sites on VWF. The collagen-binding ability of VWF is often reflective of VWF multimerization; yet, the recombinant product expressed in heterozygous 23del transfections displayed collagen binding without corresponding multimerization. This is unusual and may be explained by a conformational change in potential aberrantly spliced heterodimers enabling low-molecular-weight VWF adherence to collagen in this static assay.26 Unfortunately, because the aberrant splice forms predicted by family 1’s VWF RNA profile were in-frame deletions of small exons, resulting proteins were marginally (2%-4%) smaller than WT VWF and clear aberrant bands were not distinguishable from WT. The exact pathogenic mechanism acting in these patients’ endothelium may fluctuate between intracellular retention (23del and 23/26del) and intracellular degradation (26del) depending on the proportions of the aberrant transcripts being produced.
Decreased secretion of 33-34del VWF in the D4N region of VWF was observed from c.5842+1G>C in family 2. The idea that this region of the D4 assembly of VWF is important for VWF secretion is supported by Zhou et al.27 The preserved platelet, collagen, and FVIII binding of this splice form and its ability to multimerize with WT VWF may explain the mild type 1 phenotype seen in T152. Heteromultimers of this aberrant splice form were expected in the patient plasma; however, due to the minimal size difference (∼3%) between 33-34del and WT VWF, these forms were indistinguishable by western blot. Single exon 33 skipping was transcribed from the normal allele in family 2 and was subsequently observed in normal controls as well. This transcript introduces a PTC and therefore was expected to be targeted for nonsense-mediated decay; however, an additional band of ∼180 kDa was observed in patients T151 and T152, corresponding with the size of the aberrant splice form 33del. Overproduction of 33del in these patients may be overwhelming the RNA degradation system in these individuals, resulting in production of this aberrant splice form which is not observed in the normal pooled plasma (NPP) control. The lack of proteomic evidence of 33del in the NPP control on the western blot may indicate that in the normal population this transcript is degraded by nonsense-mediated decay or is produced in low abundance relative to WT VWF. This process of production and degradation of VWF skipping exon 33 may serve to regulate VWF expression levels in vivo.
Nonsense-mediated decay has been described as a common outcome for PTC-containing VWF transcripts.6,28,29 This mechanism is likely responsible for degradation of aberrant 38del transcripts caused by heterozygosity for the intronic mutation c.6599-20A>T, leading to pathologic haploinsufficiency causing mild type 1 VWD in family 3. This prediction is strengthened by the observation of only WT VWF in the western blot analysis of family 3’s plasma.
This study has shown exonic, consensus splice site, and intronic mutations causing VWD through different mechanisms such as RNA degradation, intracellular retention, and intracellular degradation. Although some splice products producing prematurely truncated VWF are retained in the ER30 others can be constitutively secreted if translated (exon 33 skipping). Similar variability in splice variant outcomes have been observed through other studies evaluating VWF splicing mutations.6,28,29,31,32
Significant shifts in aberrant splice form ratios were observed between BOECs from static culture and those exposed to high laminar flow. For example, high laminar flow increased the abundance of the 33del transcript which may indicate that splicing errors are increased in stressed endothelium. The patterns observed indicate that exposure to shear stress may mediate VWF splicing through shear-responsive motifs in the VWF promotor.33 These shear-induced alterations in splicing appear to decrease production of aberrant VWF as in-frame splice forms such as 23del, 23/26del, 26del, and 33-34del, and while PTC-containing transcripts (33del and 38del) are increased, their destruction by nonsense-mediated decay would limit their interference with WT VWF production. This insight into the effect of shear stress on endothelial VWF splicing is limited as only high physiologic laminar flow, as found in large arteries, was investigated.34 Shifting splice form ratios may also be prompted by other biomechanical and chemical stimuli. The exact pathogenic mechanism acting in these patients’ endothelium may fluctuate depending on the extent to which each aberrant transcript is being produced. This may also indicate fluctuating severity and presentation of the patient phenotype; in particular for family 1, there may be shifts between type 2A and type 1 VWD presentation. These alterations in splice form ratios from unrecognized splicing mutations may, in part, explain the variable expressivity and incomplete penetrance characteristic of type 1 VWD.
This study exemplifies how different mutations can affect splicing of VWF. The translation of the exon 33 skipping variant in family 2 suggests there may be a function for this protein other than modulation of VWF expression when transcribed at higher levels and bypassing full RNA degradation. As this transcript is shown to increase with high levels of laminar flow, this aberrant splice form may be produced in the normal population under these conditions and its increased FVIII binding may indicate a function as a FVIII chaperone if translated. This increased FVIII-binding affinity warrants further investigation into its biosynthesis and function in vivo. The implication of shear stress influencing VWF splicing patterns in endothelial cells should be more thoroughly explored. Particularly, in conjunction with the presence of the exon 33 transcript, this suggests an investigation into alternative splicing of VWF may be warranted as this process occurs in ∼95% of mammalian multiexon genes.35,36
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank David Lillicrap for the use of his Ibidi pump system, the Queen’s Cancer Research Institute for the use of the Quorum Wave Effects Spinning Disc confocal microscope and MetaMorph Microscopy Automation Software, and A. Tuttle for multimer analysis. Patients were recruited from Foothills Medical Centre, Alberta Health Services, Calgary, Health Sciences Centre, St. Johns, and Centre Hospitalier Universitaire St. Justine.
This work was supported by the Queen’s Pathology Award, Queen’s Graduate Awards, Ontario Graduate Scholarships, Robert Kisilevsky Research Education Awards, and Queen Elizabeth II Scholarship in Science & Technology. P.D.J. has received the Bayer Early Career Investigator Award and support from the Zimmerman Program for Molecular and Cellular Biology of von Willebrand Disease by the National Institutes of Health, National Heart, Lung, and Blood Institute program project grant HL081588.
Authorship
Contribution: L.H. designed and performed experiments, analyzed data, and wrote the paper; M.L.B. performed experiments and edited the paper; M.-C.P., M.-F.S., and G.-E.R. provided patients and samples for these studies and reviewed the paper.; and P.D.J. designed experiments, edited the paper, and funded this research.
Conflict-of-interest disclosure: M.-C.P. received research funding from CSL Behring and is on advisory boards for CSL Behring, Baxalta, Biogen, Bayer, and Novo Nordisk with honorarium. P.D.J. receives research funding from: Bayer, CSL Behring, and Octapharma, honoraria from: Baxalta, Biogen, Octapharma, and CSL Behring, and is on advisory boards for: CSL Behring, Baxalta & Biogen. The remaining authors declare no competing financial interests.
Correspondence: Paula D. James, Department of Medicine, Queen’s University, Etherington Hall, Room 2015, Kingston, ON K7L 2V6, Canada; e-mail: jamesp@queensu.ca.