Key Points
Chemokine ligands CXCL1-4, 6, 10, 11, and 13 are upregulated in human quiescent HSCs with CXCR2 and CXCL4 regulating their survival.
Genetic ablation of Cxcr2 or Cxcl4 in murine models induces initial expansion but eventual exhaustion of HSC in transplantation assays.
Abstract
The regulation of hematopoietic stem cell (HSC) survival and self-renewal within the bone marrow (BM) niche is not well understood. We therefore investigated global transcriptomic profiling of normal human HSC/hematopoietic progenitor cells [HPCs], revealing that several chemokine ligands (CXCL1-4, CXCL6, CXCL10, CXCL11, and CXCL13) were upregulated in human quiescent CD34+Hoescht−Pyronin Y− and primitive CD34+38−, as compared with proliferating CD34+Hoechst+Pyronin Y+ and CD34+38+ stem/progenitor cells. This suggested that chemokines might play an important role in the homeostasis of HSCs. In human CD34+ hematopoietic cells, knockdown of CXCL4 or pharmacologic inhibition of the chemokine receptor CXCR2, significantly decreased cell viability and colony forming cell (CFC) potential. Studies on Cxcr2−/− mice demonstrated enhanced BM and spleen cellularity, with significantly increased numbers of HSCs, hematopoietic progenitor cell-1 (HPC-1), HPC-2, and Lin−Sca-1+c-Kit+ subpopulations. Cxcr2−/− stem/progenitor cells showed reduced self-renewal capacity as measured in serial transplantation assays. Parallel studies on Cxcl4 demonstrated reduced numbers of CFC in primary and secondary assays following knockdown in murine c-Kit+ cells, and Cxcl4−/− mice showed a decrease in HSC and reduced self-renewal capacity after secondary transplantation. These data demonstrate that the CXCR2 network and CXCL4 play a role in the maintenance of normal HSC/HPC cell fates, including survival and self-renewal.
Introduction
Hematopoietic stem cells (HSCs) are responsible for the maintenance of the hematopoietic system throughout life and their fate is tightly balanced between self-renewal and differentiation in order to sustain the production of multilineage differentiated cells.1,2 In mice and humans, most adult HSCs are quiescent, yet remain poised for activation in response to bone marrow (BM) injury or growth factor/interferon stimulation.3,4 This balanced state between quiescence, proliferation, and differentiation is tightly controlled by numerous transcriptional networks, modulated by both cell autonomous factors and the surrounding BM niche.5
Chemokines are a family of chemotactic cytokines that bind to specific 7-transmembrane G-protein–coupled receptors.6 Chemokines are classified into 4 main families based on the position of conserved cysteine residues within their N-terminal region; CXC, CC, CX3C, and XC chemokines. Chemokines are functionally divided into 2 groups, homeostatic and inflammatory chemokines. The first are expressed constitutively and are mainly involved in controlling the migration of cells during development and tissue maintenance. Inflammatory chemokines are produced in response to infection or injury and attract inflammatory cells to sites of injury.7 Chemokines have a variety of roles in the development, disease, and hematopoiesis, in addition to their role in chemotactic activity on leukocytes.7,8 HSC proliferation and survival have been shown to be mediated by members of the chemokine family.9,10 Members of the CXC family of chemokines, such as CXCL12 and its receptor CXCR4, play important roles in hematopoietic cell survival, BM localization/retention, and mobilization at early stages of life and during adulthood.11,12
The role of chemokine ligands and their receptors in HSC fates has not been extensively characterized so far. We have shown that CCL3 is a negative regulator of HSC proliferation with potential for therapeutic application.13 A previous study has shown that CXCL1 promotes growth and self-renewal in embryonic stem cells,14 whereas CXCL4 has been shown to inhibit cell cycle entry in endothelial cells, together with inhibition of DNA synthesis.15
Importantly, recent data have shown that CXCL4 expressed by megakaryocytes (MKs) also regulates HSC quiescence.16 In addition, CXCL12 and CXCR4 play an important role in maintaining HSC quiescence and repopulating activity.17,18 HSC survival in vivo is dependent on the expression of CXCL12 from endothelial cells and mesenchymal progenitors of the surrounding BM stroma.19 Finally, it has been shown that chemokines support murine leukemia stem cells in myeloproliferative disorders.20-22
To identify important regulators of HSC/HPC cell fate decisions, we performed transcriptional profiling of human quiescent and actively dividing stem/progenitor cells, and demonstrated that several chemokine ligands (CXCL1-4, CXCL6, CXCL8, CXCL10, CXCL11, and CXCL13) were upregulated in the quiescent fraction.23 These data suggested that chemokine signaling pathways may be involved in the regulation of HSC fates, such as survival or self-renewal. Here, we have further investigated and characterized the requirement for members of the chemokine family in mouse and human HSC/HPC cell survival, focusing on CXCR2 and CXCL4.
Materials and methods
Reagents
SB225002 was purchased from Merk (Nottingham, United Kingdom) and used at the concentrations indicated. All reagent grade chemicals were purchased from Sigma-Aldrich (Poole, United Kingdom), unless otherwise stated.
Human samples
This research was approved by the West of Scotland Research Ethics Committee 4 and all human participants gave written informed consent. Granulocyte colony-stimulating factor-mobilized peripheral blood (PB) stem cell samples were obtained as excess material from normal donors for allogeneic transplantation. CD34-enrichment was performed using CliniMACS (Miltenyi Biotec Inc., Auburn, CA) as previously reported.24 CD34+, Hoescht+/−, Pyronin Y+/−, and CD34+38+/− populations were isolated as previously described.25 Samples were isolated using CD34 (340667; Becton Dickinson, Oxford, United Kingdom) and CD38 (551400; Becton Dickinson) antibodies, Pyronin Y (CAS 92-32-0; San Cruz Biotech, Heidelberg, Germany), and Hoechst (ThermoFisher 33342; Life Technologies, Grand Island, NY).
Mouse strains
All experiments were carried out according to the United Kingdom Home Office regulations using 8- to 12-week-old animals on a C57BL/6 background (CD45.2) with age and sex matched wild-type (WT) controls (Cxcr2−/− and Cxcl4−/−). C57BL/6 animals expressing CD45.1 were used as recipient mice for BM reconstitution experiments. The Cxcl4−/− mice were donated by Mortimer Poncz (The Children’s Hospital of Philadelphia).
Cell isolation
Human CD34+ cells were cultured in serum free medium supplemented with a high growth factor cocktail as described previously.23 Murine BM cells were isolated by crushing whole bones. Cells were filtered after crushing. Mouse BM cells were cultured in Iscove modified Dulbecco medium supplemented with 10% fecal calf serum, penicillin, streptomycin, l-glutamine, and interleukin-3 (20 ng/mL), interleukin-6 (20 ng/mL), and stem cell factor (40 ng/mL) (BioLegend, London, United Kingdom). Prior to lentiviral transduction, mouse BM samples were enriched for c-Kit+ cells using MicroBeads (Miltenyi Biotec Inc.).
Fluorescence-activated cell sorting (FACS)
Flow cytometry was performed using the FACSCantoII and FACSAria cell sorter (Becton Dickinson, Oxford, United Kingdom) and analyzed using FloJo software (Tree Star, Ashland, OR). Murine BM cells were incubated in Fc block prior to antibody staining. Human BM was stained with CD34 (340667; Becton Dickinson) and CD38 (551400; Becton Dickinson). Mouse tissue was stained with a lineage cocktail against CD4 (553649; Becton Dickinson), CD5 (553019; Becton Dickinson), CD8a (553029; Becton Dickinson), Gr-1 (553125; Becton Dickinson), B220 (553086; Becton Dickinson), Ter-119 (553672; Becton Dickinson), and CD11b (553309; Becton Dickinson). The stem/progenitor population was assessed using antibodies against c-Kit (47-1171-82; eBioscience, Hatfield, United Kingdom), Sca-1 (122514; BioLegend), CD150 (115910; BioLegend), and CD48 (103406; BioLegend). For apoptosis assays, cells were stained with Annexin V (556570; Becton Dickinson) and 4,6 diamidino-2-phenylindole (DAPI). Transplantations were monitored using antibodies against CD45.1 and CD45.2 (558701 and 553772, respectively; Becton Dickinson).
RNA isolation and real-time polymerase chain reaction (RT-PCR)
RNA was extracted using the RNeasy RNA Extraction Kit (Qiagen Sciences, Germantown, MD) and reverse transcribed into complementary DNA (Life Technologies). If <5000, cells were sorted using the CellsDirect One-Step qRT-PCR Kit (Life Technologies, Paisley, United Kingdom). RT-PCR was carried out using TaqMan probes and data were acquired using an Applied Biosystems 7900HT Fast RT-PCR machine (Life Technologies) or Fluidigm platform (Fluidigm Corporation), and data analysis was performed using the Relative Quantitation Manager Software (Life Technologies). Human TaqMan probe ID numbers are as follows: human CXCL1: Hs00236937_m1; human CXCL2: Hs00601975_m1; human CXCL4: Hs00427220_g1; human CXCL6: Hs00605742_g1; human CDC6: Hs00153374_m1; human CD38: Hs01120071_m1; and human CXCL8: Hs01115388_m1. Mouse Cxcr2: Mm00438258_m1; mouse Cxcl4: Mm00451315_g1; mouse Cxcl1: Mm04207460_m1; mouse Cxcl2: Mm00436450_m1; mouse Cxcl3: Mm01701838_m1; and mouse Cxcl12: Mm00445553_m1.
Immunofluorescence staining
Cells were fixed and permeabilized with 4% formaldehyde and 0.25% Triton X-100, blocked in 10% goat or donkey serum (Sigma-Aldrich) and incubated overnight in primary antibodies against CXCL1 (1:100, sc-1374; Santa Cruz, Heidelberg, Germany), CXCL2 (1:100, sc-1375; Santa Cruz), CXCL4 (1:100, sc-73638; Santa Cruz), CXCL6 (1:100, sc-5813; Santa Cruz), CXCL8 (1:100, MAB208; R&D Systems, Abingdon, United Kingdom), CXCR2 (1:100, sc-682; Santa Cruz), or immunoglobulin G (AB-108-C; R&D Systems). Cells were incubated with appropriate secondary donkey anti-goat (1:500, ab150129; Abcam, Cambridge, United Kingdom), goat anti-mouse (1:500, ab150113; Abcam), or goat anti-rabbit (1:500, ab150077; Abcam), Alexa Fluor 488, and mounted with DAPI (Vector Laboratories Ltd, Burlingame, CA). Images were acquired using a Zeiss fluorescence microscope and 3-dimensional images were generated using ImageJ.
Colony forming cell (CFC) assay
CFC assays were carried out using MethoCult (H4434 or M3434; Stem Cell Technologies, Grenoble, France). Resulting colonies were counted after ∼10 to 14 days in culture. Colonies were harvested, counted, and reseeded into MethoCult for serial replating assays.
In vivo transplantation
Donor mice were CD45.2+. HSC (Lin−Sca-1+c-Kit+ [LSK], CD150+CD48−) were sorted from donor Cxcr2−/−, Cxcl4−/−, or WT animals, and transplanted by IV injections into CD45.1+ recipient mice (irradiated at 7 Gy) using 100 HSC together with 200 000 CD45.1+ support BM cells per mouse. Mice were given Baytril antibiotic in their drinking water for 2 weeks following irradiation. Animals were bled every 4 weeks posttransplant to assess chimerism and sacrificed at 16 weeks posttransplant. BM, spleen, and thymus were harvested and examined for donor cell contribution. For secondary transplantations, 2000 CD45.2+ LSK cells were sorted from primary recipients, and transplanted with 200 000 CD45.1+ support BM cells into CD45.1+ irradiated recipient mice and followed as above. For homing experiments, sorted LSK donor cells (1 × 105) were transplanted into lethally irradiated recipient mice; 24 hours after transplant, the recipient mice were analyzed.
Lentiviral transduction and electroporation
Mouse Cxcl4 short hairpin RNA (shRNA) (Cat No. RMM4534-NM_019932) were purchased from Thermo Fisher (Life Technologies). shRNA of interest were subcloned from the original pLKO.1 vectors into a pLKO.1 plasmid containing a green fluorescent protein (GFP) tag. Calcium chloride was used to transfect human embryonic kidney-293 cells with specific hairpin or scrambled control using HIV-1 and glycoprotein of vesicular stomatitis virus as accessory plasmids. Primary cells were cultured in viral medium supplemented with TransDux (Cambridge Bioscience, Cambridge, United Kingdom) or Polybrene at 400g for 90 minutes. Viral medium was collected at 24, 48, and 72 hours posttransfection, and the spin inoculation was repeated for 3 rounds of viral infection. Cells were resuspended in appropriate medium for 48 hours before selection of positively transduced cells using positive expression of GFP and FACS.
CXCL4-small interfering RNA (siRNA) (221753; Applied Biosystems) and scrambled siRNA1 (AM4611; Applied Biosystems) (100 nM) were electroporated into cells using the Amaxa Nucleofector Kit V (Lonza, Cambridge, United Kingdom) together with a GFP-containing plasmid following the manufacturer’s instructions. Transduction efficiency ranged between 30% and 50%. After 24 hours, the GFP+ cells were sorted and analyzed.
Differential expression analysis of transcriptional data and network construction
Raw data were obtained via ArrayExpress (E-MTAB-2508); robust multi-array analysis was normalized and analyzed using Rank products.26,27 MetaCore’s “Expand by one interaction” algorithm was used to expand the gene network around the 8 chemokines differentially expressed in quiescent vs dividing CD34+ cells23 ; only those genes exhibiting differential expression (false discovery rate = 0.05) between quiescent and dividing normal CD34+ cells were retained.
Statistical analysis
Statistical analyses were performed using the Student t test. *P < .05 was defined as statistically significant. **P < .01 and ***P < .001 were considered highly statistically significant.
Results
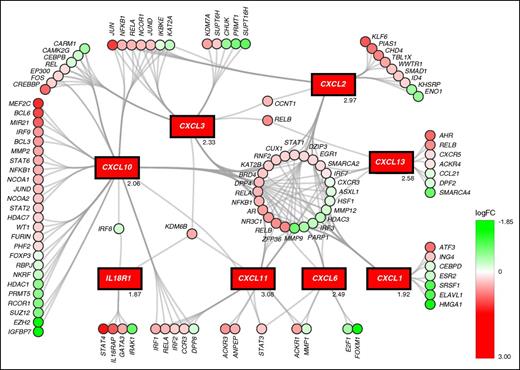
Chemokine ligands are upregulated in human CD34+38− compared with CD34+38+ cells
We previously analyzed the transcriptional differences between human quiescent (CD34+, Hoechst−, and Pyronin Y−) cells and their more proliferative (CD34+, Hoechst+, and Pyronin Y+) counterparts. We observed that several chemokine ligands, including CXCL1-3, CXCL6, CXCL10, CXCL11, and CXCL13 were upregulated in the quiescent population, suggesting a possible role for these factors in the regulation of HSC maintenance.23 No probes for CXCL4 were included in the microarray. Here we have further analyzed these data with a focus on the chemokine family. Figure 1 shows the upregulated chemokines and genes predicted to be connected with the chemokine network in human HSCs. In quiescent cells, the chemokine network highlighted regulation of factors relevant for proliferative status, such as regulators of cell cycle E2F1 (downregulated) and EGR1 (upregulated).28 To validate chemokine regulation and examine a possible mechanism by which these ligands are linked to HSC cell cycle status, BM stem/progenitor cells from normal, healthy volunteers were sorted into primitive (CD34+38−) and more proliferative progenitor cell (CD34+38+) populations. Cell cycle and differentiation status of these 2 populations was confirmed by the differential expression of CDC6 and CD38, respectively (see supplemental Figure 1, available on the Blood Web site). As predicted, CDC6 and CD38 were downregulated in CD34+38− compared with CD34+38+ cells, indicating that CD34+38− cells were mainly quiescent and undifferentiated. Chemokine genes were selected for RT-PCR validation based on the level of upregulation in the microarray with the addition of CXCL4, which has recently been shown to be relevant in the regulation of HSC quiescence.16 The expression of CXCL1, CXCL2, CXCL4, CXCL6, and CXCL8 was significantly increased in CD34+38− compared with CD34+38+ cells (Figure 2A; *P < .05; **P < .01), suggesting a possible role for these chemokines in maintenance of the quiescent HSC pool.
Chemokine ligands are upregulated in quiescent vs dividing CD34+ cells. The transcriptional differential regulation connected to chemokine expression (shown in rectangles) in human quiescent (CD34+, Hoechst−, and Pyronin Y−) vs dividing (CD34+, Hoechst+, and Pyronin Y+) cells. Upregulation is shown in red and downregulation in green; color intensity indicates the extent of differential regulation as indicated in the color key. logFC values for the chemokines are given on the bottom-right of the relevant box. logFC, log fold change.
Chemokine ligands are upregulated in quiescent vs dividing CD34+ cells. The transcriptional differential regulation connected to chemokine expression (shown in rectangles) in human quiescent (CD34+, Hoechst−, and Pyronin Y−) vs dividing (CD34+, Hoechst+, and Pyronin Y+) cells. Upregulation is shown in red and downregulation in green; color intensity indicates the extent of differential regulation as indicated in the color key. logFC values for the chemokines are given on the bottom-right of the relevant box. logFC, log fold change.
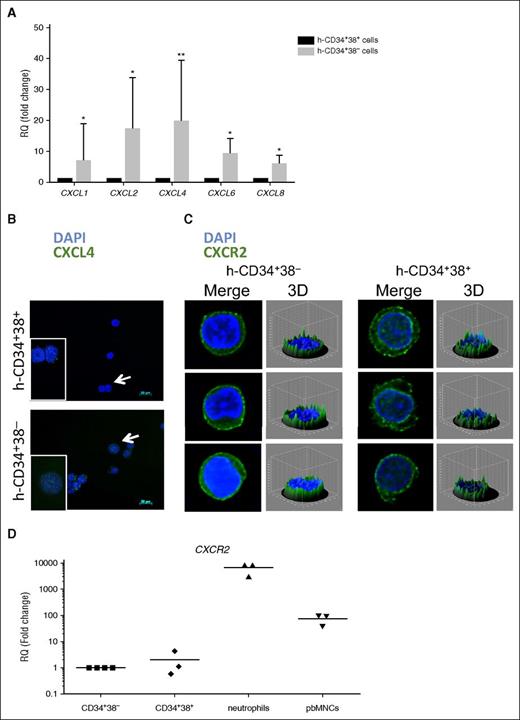
CXCL4 and CXCR2 are expressed on CD34+38− and CD34+38+ cells. (A) RT-PCR in human CD34+38− and CD34+38+ cells shows gene expression differences for the chemokine ligands CXCL1, CXCL2, CXCL4, CXCL6, and CXCL8. Fold change was calculated relative to the reference gene (GAPDH) according to the ΔΔCT method. n = 5. *P < .05; **P < .01. All error bars indicate standard error of the mean (SEM). (B) Human CD34+38− (top) and CD34+38+ (bottom) cells show expression for CXCL4 (white arrows indicate cells in enlarged picture), and (C) CXCR2. Nuclei were stained using DAPI and images were acquired using a Zeiss microscope; 3-D figures generated with ImageJ software are shown for CXCR2 staining (n = 3). (D) Real-time quantitative PCR in human CD34+38−, CD34+38+, neutrophils, and PB mononuclear cells shows the gene expression profile for CXCR2 (n = 3). The fold change was calculated relative to the reference gene (GAPDH) according to the ΔΔCT method. 3-D, 3-dimensional; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; h, human; RQ, relative quantitation.
CXCL4 and CXCR2 are expressed on CD34+38− and CD34+38+ cells. (A) RT-PCR in human CD34+38− and CD34+38+ cells shows gene expression differences for the chemokine ligands CXCL1, CXCL2, CXCL4, CXCL6, and CXCL8. Fold change was calculated relative to the reference gene (GAPDH) according to the ΔΔCT method. n = 5. *P < .05; **P < .01. All error bars indicate standard error of the mean (SEM). (B) Human CD34+38− (top) and CD34+38+ (bottom) cells show expression for CXCL4 (white arrows indicate cells in enlarged picture), and (C) CXCR2. Nuclei were stained using DAPI and images were acquired using a Zeiss microscope; 3-D figures generated with ImageJ software are shown for CXCR2 staining (n = 3). (D) Real-time quantitative PCR in human CD34+38−, CD34+38+, neutrophils, and PB mononuclear cells shows the gene expression profile for CXCR2 (n = 3). The fold change was calculated relative to the reference gene (GAPDH) according to the ΔΔCT method. 3-D, 3-dimensional; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; h, human; RQ, relative quantitation.
Immunofluorescence staining showed that human CXCL4 protein was detectable in both CD34+38− and CD34+38+ cells, but was higher in expression in CD34+38− cells (Figure 2B). Similarly, immunofluorescence staining was carried out in human CD34+38+ and CD34+38− cells to investigate the expression of CXCL1, CXCL2, CXCL6, and CXCL8 (supplemental Figure 1B). Although CXCL6 and CXCL8 were barely detectable at the protein level in either population, CXCL1 and CXCL2 showed clear expression in CD34+38+ cells, but in contrast to what was observed at the gene level, no upregulation was detected in the CD34+38− cells. Because CXCR2 is the best characterized of the receptors for chemokine ligands, we examined its expression at the messenger RNA (mRNA) (Figure 2D) and protein level (Figure 2C and supplemental Figure 1B) in the CD34+38− and CD34+38+ fractions. CXCR2 mRNA and protein were expressed at similar levels in both cell populations. The CXCR2 mRNA level was compared with that of neutrophils and PB mononuclear cells that were used as positive controls. Based on their roles in cell growth or self-renewal in other cellular contexts, CXCL4 and CXCR2 were selected for functional studies.15,16
CXCL4 and CXCR2 support viability and colony forming potential of human CD34+ cells
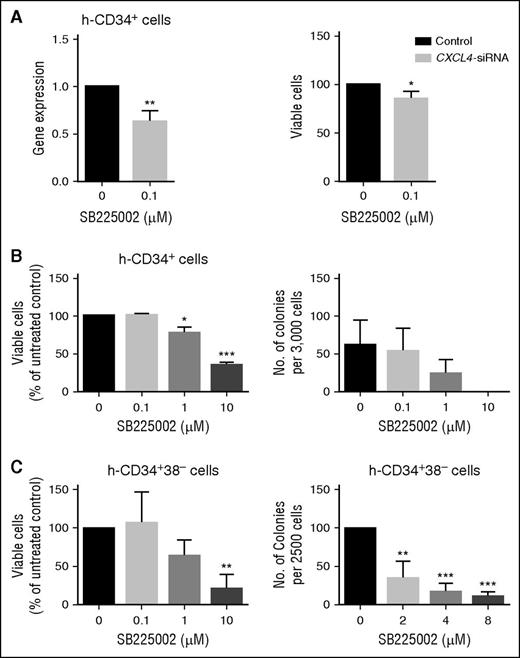
We investigated the role of the most differentially expressed chemokine CXCL4, in CD34+ cells. In keeping with the <50% reduction in gene expression achieved, siRNA-mediated knock-down of CXCL4 resulted in a modest but significant decrease in cell viability compared with nontargeting siRNA (Figure 3A). To investigate the role of CXCL4’s known chemokine receptor CXCR2, CD34+ cells were treated with the CXCR2 inhibitor SB225002. SB225002 is a potent and selective antagonist of CXCR2, which results in inhibition of ligand-mediated signal transduction.29 CD34+ cells were treated for 72 hours. A reduction in cell viability and colony formation was observed in a drug concentration-dependent manner compared with the vehicle-treated control (Figure 3B) (drug concentration based on published literature30 ). Similarly, CD34+38− cells treated for 72 hours with SB225002 showed a marked decrease in cell viability and colony formation ability in comparison with control (Figure 3C). In addition, treatment of CD34+38− cells with SB225002 at 1 μM led to an obvious increase in the percentage of cells in gap 0 (G0) of the cell cycle (supplemental Figure 1C). Taken together, these data suggest that inhibition of chemokine signaling, in particular that of CXCL4 and CXCR2, impairs human stem/progenitor cell function, and imply that these factors may be required for survival and maintenance of these cells.
Inhibition of CXCL4 ligands and CXCR2 signaling reduces cell viability and colony formation in vitro. (A) CD34+ enriched cells were transduced with CXCL4 siRNA, and relative scrambled control and level of knock-down (left) and viability (right) measured (n = 3). (B) CD34+ cells were treated with increasing concentrations of SB225002 and viable cells analyzed using Annexin V/DAPI staining (left) (n = 3). CFC count was performed in the same cells treated with SB225002 at the concentration indicated (right) (n = 3). (C) CD34+38− enriched cells were treated with increasing concentrations of SB225002 and viable cells analyzed using Annexin V/DAPI staining (left) (n = 3). CFC count was performed in cells treated with SB225002 at the concentration indicated (right) (n = 3). Statistical analysis was performed using a paired two-tailed Student t test (*P < .05; **P < .01; ***P < .001). All error bars indicate SEM of the mean. h, human.
Inhibition of CXCL4 ligands and CXCR2 signaling reduces cell viability and colony formation in vitro. (A) CD34+ enriched cells were transduced with CXCL4 siRNA, and relative scrambled control and level of knock-down (left) and viability (right) measured (n = 3). (B) CD34+ cells were treated with increasing concentrations of SB225002 and viable cells analyzed using Annexin V/DAPI staining (left) (n = 3). CFC count was performed in the same cells treated with SB225002 at the concentration indicated (right) (n = 3). (C) CD34+38− enriched cells were treated with increasing concentrations of SB225002 and viable cells analyzed using Annexin V/DAPI staining (left) (n = 3). CFC count was performed in cells treated with SB225002 at the concentration indicated (right) (n = 3). Statistical analysis was performed using a paired two-tailed Student t test (*P < .05; **P < .01; ***P < .001). All error bars indicate SEM of the mean. h, human.
Chemokine expression in murine stem and progenitor cells
To investigate a role for the chemokine family in the maintenance of HSCs in their physiological BM microenvironment, parallel murine models were employed. We first interrogated a publicly available gene expression data set and determined that Cxcl4 was expressed in murine LSK cells.31 Following cell sorting, real-time quantitative PCR was used to assess the expression of Cxcl1, Cxcl2, Cxcl3, Cxcl4, Cxcl12, and Cxcr2 across HSC (LSKCD150+CD48−), multipotent progenitors (MPPs; LSKCD150−CD48−), and primitive HPCs (HPC-1; LSKCD150−CD48+ and HPC-2; and LSKCD150+CD48+) derived from adult murine BM (see sorting strategy in supplemental Figure 2A; data in supplemental Figure 2B; and Calaminus et al32 ). As shown, Cxcl4 and Cxcr2 were expressed in the HSC fraction.
Cxcr2−/− HSCs have impaired self-renewal capacity
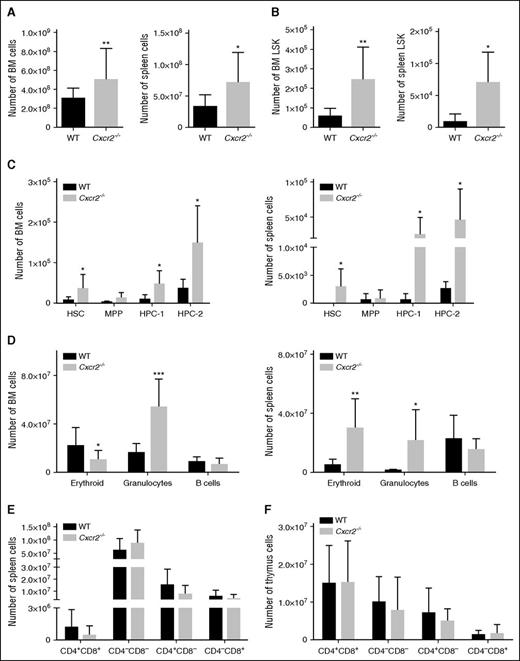
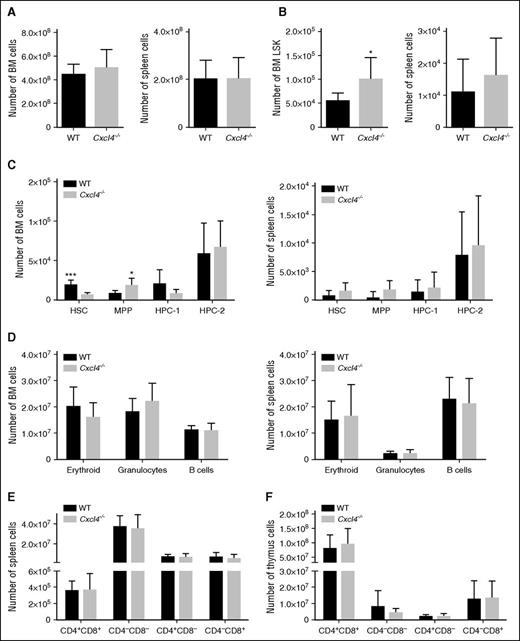
We next investigated the requirement for Cxcr2 in mouse HSC function. Cxcr2−/− mice have previously been characterized and are known to show a considerable expansion of myeloid cells in the BM, spleen, and PB, an effect that was shown to be caused by the deletion of Cxcr2 from the BM microenvironment.33,34 However, to our knowledge, a comprehensive analysis of the hematopoietic stem and progenitor compartments, and their functionality, has not previously been investigated in these mice. Total BM and spleen cellularity were increased in the Cxcr2−/− mice (*P < .05; **P < .01) (Figure 4A). Immunophenotypic analyses revealed that the absolute numbers of LSK cells were increased in the BM and spleen of Cxcr2−/− mice (*P < .05; **P < .01) (Figure 4B). More detailed analyses of the LSK compartment using CD150 and CD48 markers indicated a significant increase in the HSC, HPC-1, and HPC-2 populations in the Cxcr2−/− mice, both in the BM and in the spleen (*P < .05) (Figure 4C). A decrease in the erythroid (*P < .05) and an increase in the myeloid compartments (***P < .001) of Cxcr2−/− mice were also observed in the BM, consistent with the previous report (Figure 4D, left).34 In the spleen of Cxcr2−/− mice, an increase in both erythroid and the granulocytic compartments was seen (*P < .05; **P < .01) (Figure 4D, right). No differences were detected in the T-cell compartments (CD4 and CD8) between the Cxcr2−/− and the WT mice, either in the spleen or in the thymus (Figure 4E-F). When cell cycle was analyzed in long-term HSCs from WT and Cxcr2−/− mice, a decrease in the percentage of Cxcr2−/− cells in G0 phase, together with an increase in those in G1 phase was detected (supplemental Figure 3A). No changes in apoptosis were detected (supplemental Figure 3C).
Cxcr2−/− mice show an expansion of the stem cell compartment. (A) BM (left) and spleen (right) from WT mice (n = 6) or Cxcr2−/− mice (n = 6) were harvested and assessed for total cellularity. (B) Numbers of LSK cells for BM (left) and spleen (right) from WT mice or Cxcr2−/− mice. (C) Numbers of HSC, MPP, HPC-1, and HPC-2 in the BM (left) and spleen (right) from WT mice or Cxcr2−/− mice are shown. (D) Numbers for BM (left) and spleen (right) erythroid, granulocytic, and B-cell populations from WT and Cxcr2−/− mice are shown. (E) Numbers for T-cell populations in the WT and Cxcr2−/− spleen identified with CD4 and CD8 markers are shown. (F) Numbers for T-lineage populations in the WT and Cxcr2−/− thymi identified with CD4 and CD8 markers are shown. Statistical analysis was performed using an unpaired two-tailed Student t test. All error bars indicate SEM of the mean (*P < .05; **P < .01; ***P < .001) (n = 6).
Cxcr2−/− mice show an expansion of the stem cell compartment. (A) BM (left) and spleen (right) from WT mice (n = 6) or Cxcr2−/− mice (n = 6) were harvested and assessed for total cellularity. (B) Numbers of LSK cells for BM (left) and spleen (right) from WT mice or Cxcr2−/− mice. (C) Numbers of HSC, MPP, HPC-1, and HPC-2 in the BM (left) and spleen (right) from WT mice or Cxcr2−/− mice are shown. (D) Numbers for BM (left) and spleen (right) erythroid, granulocytic, and B-cell populations from WT and Cxcr2−/− mice are shown. (E) Numbers for T-cell populations in the WT and Cxcr2−/− spleen identified with CD4 and CD8 markers are shown. (F) Numbers for T-lineage populations in the WT and Cxcr2−/− thymi identified with CD4 and CD8 markers are shown. Statistical analysis was performed using an unpaired two-tailed Student t test. All error bars indicate SEM of the mean (*P < .05; **P < .01; ***P < .001) (n = 6).
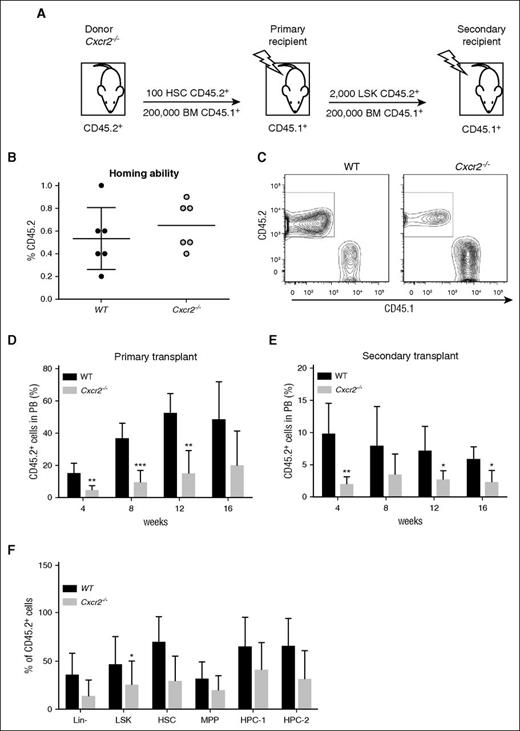
In previous examples of genes that are critical for HSC maintenance, an expansion in stem and progenitor cell numbers has often been associated with an alteration in the balance between self-renewal and differentiation, leading to stem cell exhaustion.35 To determine whether the difference observed in overall HSC frequency in Cxcr2−/− mice translated into a change in self-renewal capacity, Cxcr2−/− HSCs were investigated in serial transplantation assays (Figure 5A). No difference in homing ability was detected between the WT and Cxcr2−/− HSCs at 24 hours following transplantation (Figure 5B). HSCs from WT or Cxcr2−/− mice (CD45.2 background) were transplanted into irradiated recipients (CD45.1 background), together with CD45.1+ support BM cells. Chimerism between donor CD45.2 and recipient CD45.1 was analyzed (Figure 5C). A decrease in the percentage of CD45.2+ chimerism was observed in the PB up to 16 weeks posttransplant for Cxcr2−/− HSCs in comparison with the control (**P < .01; ***P < .001) (Figure 5D). To further investigate the self-renewal capacity of Cxcr2−/− HSCs, secondary transplantation assays were carried out at 16 weeks post-primary transplant. BM from the recipients of the primary transplants was harvested and donor-derived LSK were transplanted into irradiated secondary recipients together with support BM. A decrease in the percentage of donor-derived cells was found in the PB of secondary recipients transplanted with Cxcr2−/− LSK in comparison with the controls out to 16 weeks (**P < .01; *P < .05) (Figure 5E). To elucidate whether the phenotype observed in Cxcr2−/− mice was due to an HSC autonomous effect or conferred by the surrounding niche (non-autonomous effect), we analyzed the phenotype of donor-derived BM cells after primary transplantation in WT recipient mice (Figure 5F). In contrast to what was observed in mutant LSK prior to transplant (significant increase as compared with WT; Figure 4B), a significant decrease was observed in the percentage of donor mutant LSK compared with the WT following transplantation, suggesting that signals from the microenvironment may play a role in the phenotype observed. Collectively, the results show that Cxcr2 plays a key role in HSC maintenance.
Cxcr2−/− HSCs show a reduction in engraftment in primary and secondary BM transplantation assays. (A) Experimental layout for CD45.2+ HSCs from WT or Cxcr2−/− mice (n = 3 per strain) transplanted into irradiated CD45.1+ recipients (n = 6). (B) Graph showing engraftment ability of CD45.2+ WT or Cxcr2−/− HSC after 24 hours from transplant in CD45.1+ recipients. (C) Graph showing chimerism between CD45.2+ and CD45.1+ cells. (D) Engraftment was analyzed in the blood every 4 weeks posttransplant up to 16 weeks. Data are presented as the mean percentage of CD45.2+ cells within the PB. (E) After the primary recipients were sacrificed, CD45.2+ LSK cells were transplanted into irradiated recipients. Engraftment was analyzed in the blood every 4 weeks posttransplant up to 16 weeks. (F) Percentage of CD45.2+ cells was analyzed in different BM populations after 16 weeks from primary transplant, and compared between WT and Cxcr2−/− mice. Statistical analysis was performed using an unpaired two-tailed Student t test. All error bars indicate SEM of the mean (*P < .05; **P < .01; ***P < .001) (n = 6).
Cxcr2−/− HSCs show a reduction in engraftment in primary and secondary BM transplantation assays. (A) Experimental layout for CD45.2+ HSCs from WT or Cxcr2−/− mice (n = 3 per strain) transplanted into irradiated CD45.1+ recipients (n = 6). (B) Graph showing engraftment ability of CD45.2+ WT or Cxcr2−/− HSC after 24 hours from transplant in CD45.1+ recipients. (C) Graph showing chimerism between CD45.2+ and CD45.1+ cells. (D) Engraftment was analyzed in the blood every 4 weeks posttransplant up to 16 weeks. Data are presented as the mean percentage of CD45.2+ cells within the PB. (E) After the primary recipients were sacrificed, CD45.2+ LSK cells were transplanted into irradiated recipients. Engraftment was analyzed in the blood every 4 weeks posttransplant up to 16 weeks. (F) Percentage of CD45.2+ cells was analyzed in different BM populations after 16 weeks from primary transplant, and compared between WT and Cxcr2−/− mice. Statistical analysis was performed using an unpaired two-tailed Student t test. All error bars indicate SEM of the mean (*P < .05; **P < .01; ***P < .001) (n = 6).
Cxcl4 contributes to the regulation of self-renewal of HSCs and progenitor cells
To support the in vitro data on human CXCL4 (Figures 1-3) and to complement the Cxcr2−/− mouse model, we analyzed hematopoiesis in Cxcl4−/− mice.36 Previous studies have shown that Cxcl4−/− mice exhibited an increased number of HSC and increased HSC proliferation.16 We observed that Cxcl4−/− mice showed normal spleen and BM cellularity (Figure 6A), whereas the LSK numbers in BM were increased (*P < .05) (Figure 6B). Furthermore, we found that Cxcl4−/− mice had a significant decrease in the HSC and an increase in MPP in the BM, but not in the spleen (*P < .05; ***P < .001) (Figure 6C). Mature cell subsets, including erythroid, granulocytic, and B and T cells, were not significantly altered in the BM, spleen, or thymus of Cxcl4−/− mice (Figure 6D-F). When cell cycle (LSK cells) was analyzed in the WT and Cxcl4−/− mice, no significant difference was detected between phases (supplemental Figure 3B). No changes in apoptosis were detected (supplemental Figure 3D).
Cxcl4−/− mice display a reduction in HSC compartment. (A) BM (left) and spleen (right) from WT mice (n = 6) or Cxcl4−/− mice (n = 6) were harvested and total cellularity assessed. (B) Numbers of LSK cells for BM (left) and spleen (right) from WT mice or Cxcl4−/− mice. (C) Numbers of HSC, MPP, HPC-1, and HPC-2 in the BM (left) and spleen (right) from WT mice or Cxcl4−/− mice are shown. (D) Numbers for BM (left) and spleen (right) erythroid, granulocyte, and B-cell compartments from WT and Cxcl4−/− mice are shown. (E) Numbers for T-cell populations in the WT and Cxcl4−/− spleen identified with CD4 and CD8 markers are shown. (F) Numbers for T-lineage populations in the WT and Cxcl4−/− thymi were identified with CD4 and CD8 markers. Statistical analysis was performed using an unpaired two-tailed Student t test. All error bars indicate SEM of the mean (*P < .05; ***P < .001).
Cxcl4−/− mice display a reduction in HSC compartment. (A) BM (left) and spleen (right) from WT mice (n = 6) or Cxcl4−/− mice (n = 6) were harvested and total cellularity assessed. (B) Numbers of LSK cells for BM (left) and spleen (right) from WT mice or Cxcl4−/− mice. (C) Numbers of HSC, MPP, HPC-1, and HPC-2 in the BM (left) and spleen (right) from WT mice or Cxcl4−/− mice are shown. (D) Numbers for BM (left) and spleen (right) erythroid, granulocyte, and B-cell compartments from WT and Cxcl4−/− mice are shown. (E) Numbers for T-cell populations in the WT and Cxcl4−/− spleen identified with CD4 and CD8 markers are shown. (F) Numbers for T-lineage populations in the WT and Cxcl4−/− thymi were identified with CD4 and CD8 markers. Statistical analysis was performed using an unpaired two-tailed Student t test. All error bars indicate SEM of the mean (*P < .05; ***P < .001).
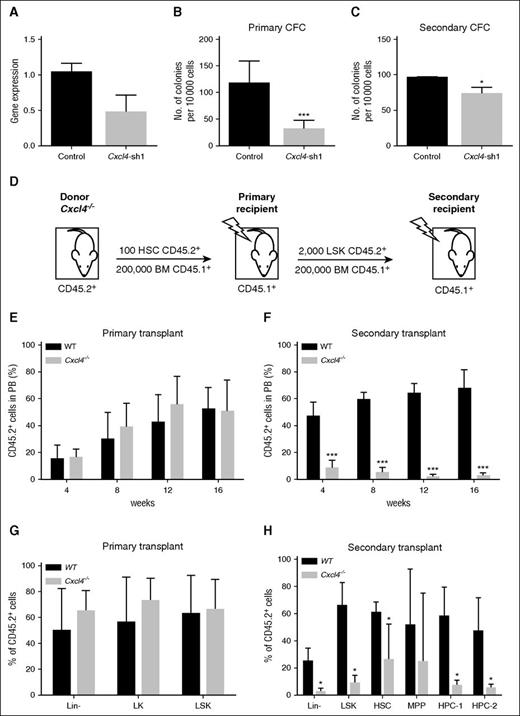
To investigate changes in self-renewal capacity, a specific Cxcl4 shRNA construct was used to reduce Cxcl4 expression in mouse c-Kit+ BM cells using a lentiviral transduction system. A reduction in Cxcl4 gene expression was found in Cxcl4 knockdown cells (control, n = 2; Cxcl4-sh1, n = 3) (Figure 7A). To determine whether Cxcl4 played a role in progenitor cell self-renewal, Cxcl4 knockdown cells were plated into methylcellulose and then replated after 7 days. A significant reduction in CFC was detected in both the primary (***P < .001) and secondary cultures (*P < .05) (Figure 7B-C). Cxcl4−/− HSCs were then investigated for their ability to reconstitute myeloablated hosts in primary and secondary transplantation assays, as previously carried out for Cxcr2−/− HSCs (Figures 5 and 7D). No changes in the percentage of CD45.2+ donor chimerism were observed in the PB up to 16 weeks post-primary transplant for the Cxcl4−/− HSCs in comparison with the control (Figure 7E). However, secondary transplantation assays showed a significant decrease in the percentage of donor-derived cells in the PB of recipients transplanted with Cxcl4−/− LSK in comparison with the controls out to 16 weeks (***P < .001) (Figure 7F), indicating that Cxcl4 also contributes to HSC maintenance.
Inhibition of Cxcl4 reduces colony formation in vitro and Cxcl4−/− HSCs show a reduction in engraftment in secondary BM transplantation assays. (A) WT BM cells enriched for c-Kit+ were transduced with a Cxcl4-shRNA vector or control, and mRNA level analyzed by RT-PCR (n = 2). (B-C) Positively transduced cells were selected using GFP, and plated into primary (B) and secondary (C) CFC assays. Data are presented as the mean colony numbers from Cxcl4-shRNA transduced cells or the control. (D) Experimental layout for CD45.2+ HSCs from WT or Cxcl4−/− mice transplanted into irradiated CD45.1+ recipients (n = 5 per strain). (E) Engraftment for primary transplant was analyzed in the blood every 4 weeks posttransplant up to 16 weeks. (F) After the primary recipients were sacrificed, CD45.2+ LSK cells were transplanted into irradiated recipients (n = 5). Engraftment was analyzed in the blood every 4 weeks posttransplant up to 16 weeks. (G) A percentage of CD45.2+ cells was analyzed in different BM populations after 16 weeks from primary transplant, and compared between WT and Cxcl4−/− mice. (H) Percentage of CD45.2+ cells was analyzed in different BM populations after 16 weeks from secondary transplant, and compared between WT and Cxcl4−/− mice. Data are presented as the mean percentage of CD45.2+ cells within the PB. *P < .05; ***P < .001.
Inhibition of Cxcl4 reduces colony formation in vitro and Cxcl4−/− HSCs show a reduction in engraftment in secondary BM transplantation assays. (A) WT BM cells enriched for c-Kit+ were transduced with a Cxcl4-shRNA vector or control, and mRNA level analyzed by RT-PCR (n = 2). (B-C) Positively transduced cells were selected using GFP, and plated into primary (B) and secondary (C) CFC assays. Data are presented as the mean colony numbers from Cxcl4-shRNA transduced cells or the control. (D) Experimental layout for CD45.2+ HSCs from WT or Cxcl4−/− mice transplanted into irradiated CD45.1+ recipients (n = 5 per strain). (E) Engraftment for primary transplant was analyzed in the blood every 4 weeks posttransplant up to 16 weeks. (F) After the primary recipients were sacrificed, CD45.2+ LSK cells were transplanted into irradiated recipients (n = 5). Engraftment was analyzed in the blood every 4 weeks posttransplant up to 16 weeks. (G) A percentage of CD45.2+ cells was analyzed in different BM populations after 16 weeks from primary transplant, and compared between WT and Cxcl4−/− mice. (H) Percentage of CD45.2+ cells was analyzed in different BM populations after 16 weeks from secondary transplant, and compared between WT and Cxcl4−/− mice. Data are presented as the mean percentage of CD45.2+ cells within the PB. *P < .05; ***P < .001.
Similar to the analysis performed for Cxcr2−/−, we analyzed the phenotype of Cxcl4−/− donor-derived BM cells after primary transplantation into WT recipient mice, but no difference was seen between the WT and the mutant percentage of donor cells (Figure 7G). However, significant decreases were observed in the percentages of donor mutant cells after secondary transplantation in all populations except MPP, when compared with the WT counterparts (Figure 7H). In this case, the effect observed in the Cxcl4−/− HSCs before transplantation (significant decrease as compared with WT; Figure 6C) was maintained after transplantation into a WT microenvironment.
Discussion
Our results indicate that chemokines play an important role in the regulation of HSC survival and self-renewal. HSC homeostasis is a critical process required for the correct functioning of HSCs and their progeny. Here we propose that chemokine signaling pathways are involved in the regulation of HSCs, and cooperate to maintain the quiescent and self-renewing state typical of these cells. Our findings suggest that some chemokines, in particular CXCL4 and the receptor CXCR2, play a key role in the maintenance of the HSC pool. We have observed that Cxcr2 and Cxcl4 proved to be critical for HSC reconstitution, suggesting a role in self-renewal.
CXCL1-4, together with CXCL6, CXCL8, CXCL10, CXCL11, and CXCL13, were all significantly upregulated in the quiescent/primitive fraction of human stem/progenitor cells and, upon modest knockdown of CXCL4, the survival of primitive cells was impaired. It has been shown previously that human hematopoietic CD34+ cells and endothelial cells respond to exogenous CXCL4, with effects on cell viability, adhesion, and stem cell expansion.15,37-40
CXCL4 signaling is complex and in certain biological contexts its functions appear to be mediated by the alternatively spliced receptor CXCR3 (CXCR3B).41 However, to date, expression of CXCR3B has not been described on human or murine HSCs. It has also been suggested that CXCL4 functions may be mediated through another, unnamed receptor/mechanism, with CXCL4 binding to integrin receptors, which have indeed been shown to be important for HSC behavior.42
Interactions between CXCL12 and CXCR4 are of key importance for the maintenance of HSCs in humans.41-43 AMD3100, a selective CXCR4 antagonist (plerixafor), antagonizes the binding of CXCL12 to CXCR4, leading to a rapid and reversible mobilization of HSCs into the peripheral circulation.44 Currently, plerixafor is used for mobilization of stem cells in patients with non-Hodgkin lymphoma or multiple myeloma.45-47
It has been shown that microenvironmental Cxcl4 derived from MKs regulates HSC cell cycle activity. Using transgenic inducible diphtheria toxin receptor mice (where MKs are depleted by inducible diphtheria toxin receptor expression) crossed with Cxcl4-cre mice, Bruns et al16 showed that short-term (7 days) depletion of Cxcl4 led to an increase in HSC proliferation, a 4.6-fold increase in HSC numbers, and enhanced reconstitution following a single transplantation. However, more extended Cxcl4 depletion (for 6 weeks) resulted in an attenuated effect, and the authors suggested that the reduced quiescence driven by loss of Cxcl4 may lead in time to HSC exhaustion as previously described.4,48 We showed that complete loss of Cxcl4 caused a decrease in HSC numbers, followed by a decrease in self-renewal capacity detected after secondary transplantation. It is therefore likely that Cxcl4 inhibition does lead at first to a temporary increase in HSC numbers (seen by Bruns et al), and that this effect is then followed by HSC exhaustion and depletion (due to increased proliferation and differentiation) and by a decrease in self-renewal capacity as we observed in our serial transplantation assay.
By interrogating a previously published transcriptional array, we observed that in mice Cxcl4 was the only chemokine ligand upregulated in the HSC compartment (unpublished observations and Månsson et al31 ), providing a rationale to investigate this factor in vivo. Our results indicated that mice lacking Cxcl4 showed changes in LSK, MPP, and HSC numbers, and reduced secondary transplantation capacity, possibly due to changes in the microenvironment as well as in autocrine HSC signaling itself. Although several chemokines act through CXCR2, CXCL4 is not known to do so and thus this receptor does not provide a unifying explanation for the similarity in function of CXCL4 and CXCR2. One possibility is that CXCR2 ligands and CXCL4 function as a heterodimer within the BM, which mediates CXCR2-dependent signaling. Such a mechanism is supported by the clear evidence of complex heterodimeric interactions between chemokines, which lead to biological outcomes different from those of either of the individual component chemokines.49-51
Similar to what happens in other systems,52-55 our results showed that the CXCR2 signaling pathway played an important role in HSC maintenance. CXCR2 inhibition with the specific CXCR2 inhibitor SB225002 replicated the results for knockdown of CXCL4. Importantly, despite previous studies having shown that human primitive HSCs do not express CXCR2, we were able to clearly detect CXCR2 protein expression in human CD34+38− and CD34+38+ cells by immunofluorescence.56,57 To further characterize the known effect on the myeloid compartment in Cxcr2−/− mice,34 we investigated how its deletion affected the primitive HSC compartment. Cxcr2−/− mice exhibit a marked expansion of viable LSK, HSC, and HPC in the BM and spleen, whereas analysis of serial transplantation data showed significantly reduced long-term repopulating ability of these cells. Taken together, these results indicate that Cxcr2 deficiency leads to a marked perturbation in normal hematopoietic homeostasis.
Our previous research has shown that differences in chemokine expression between quiescent and primitive cells is maintained in normal as well as in leukemic (chronic myeloid leukemia [CML]) HSCs.23 Recently, it has been shown that CCL3 expression is required for the development of CML in mice, and that inflammatory chemokines such as CCL3, promote leukemia development.21 Therefore, high levels of chemokine expression in primitive HSC/leukemic stem cells may suggest that chemokine signaling pathways play a role in the development and maintenance of hematologic cancers of stem cell origin. With Zhang et al, we have previously indicated that CML leukemic stem cells show a decreased homing ability due to a lower CXCL12 expression in CML BM compared with HSC, and treatment with the tyrosine kinase inhibitor imatinib reverted the abnormal cytokine levels and HSC growth.20 Similarly, multipotent stromal cells derived from mouse models of myeloproliferative neoplasia have been shown to remodel the BM niche into a self-reinforcing leukemic niche, through expression of the chemokine CCL3.22 In light of our novel findings presented here, although chemokines could represent a novel therapeutic target in myeloproliferative disorders, prudence would be advised because some chemokine family members regulate survival and self-renewal in normal HSCs, and their modulation may be detrimental in the longer term.
Collectively, these data indicate that chemokines play an important role in human and mouse HSC survival, and maintenance. These studies represent a starting point for elucidation of the role of the chemokine family in hematopoietic homeostasis, both in normal stem cells and in leukemia; however, further investigations into the mechanisms through which the chemokines act at the level of HSCs are required to fully understand HSC behavior in the BM niche.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank normal BM donors, A. Hair for sample processing, J. Cassels for cell sorting, G. V. Helgason for constructive suggestions, and Mortimer Poncz for the Cxcl4−/− mice.
This study was supported by grants from Leukaemia and Lymphoma Research (11017, 13035, and 08071), Kay Kendall Leukaemia Fund (690 and 501), The Howat Foundation, Friends of Paul O’Gorman, Cancer Research UK (C11074/A11008), Biotechnology and Biological Sciences Research Council (BB/F016050/1), and the Medical Research Council (G0600782 and MR/K014854/1). Support from Cancer Research UK Glasgow Centre (C596/A18076) and the Biological Service Unit facilities at the Cancer Research UK Beatson Institute (C596/A17196) are also acknowledged. This study was also supported by the Glasgow Experimental Cancer Medicine Centre (Cancer Research UK and the Chief Scientist’s Office, Scotland). K.R.K. is a Cancer Research UK Senior Cancer Research Fellow.
Authorship
Contribution: A.S., L.P., F.P., and L.E.M.H. designed and performed research, analyzed data, and wrote the manuscript; M.S., K.D., and M.D. performed experiments and reviewed the manuscript; S.A.A., S.C., and A.V.G. assisted with some in vitro experiments; R.K., O.S., A.M.M., L.M., K.R.K., and G.J.G. provided material, interpreted data, and reviewed the manuscript; and T.L.H. designed the study, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for is A.S. is Institute of Biodiversity, Animal Health and Comparative Medicine, College of Medical, Veterinary and Life Sciences, University of Glasgow, Glasgow, United Kingdom.
Correspondence: Tessa L. Holyoake, Paul O’Gorman Leukaemia Research Centre, Gartnavel General Hospital, 21 Shelley Rd, Glasgow G12 0ZD, United Kingdom; e-mail: tessa.holyoake@glasgow.ac.uk.
References
Author notes
A.S. and L.P. are joint first authors.
F.P. and T.L.H. are joint senior authors.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal