Key Points
miR-125b reduces mitochondrial respiration and promotes elongation of mitochondrial network through BIK and MTP18 silencing, respectively.
The miR-125b/BIK/MTP18 axis promotes adaptation of monocytes to inflammation.
Metabolic changes drive monocyte differentiation and fate. Although abnormal mitochondria metabolism and innate immune responses participate in the pathogenesis of many inflammatory disorders, molecular events regulating mitochondrial activity to control life and death in monocytes remain poorly understood. We show here that, in human monocytes, microRNA-125b (miR-125b) attenuates the mitochondrial respiration through the silencing of the BH3-only proapoptotic protein BIK and promotes the elongation of the mitochondrial network through the targeting of the mitochondrial fission process 1 protein MTP18, leading to apoptosis. Proinflammatory activation of monocyte-derived macrophages is associated with a concomitant increase in miR-125b expression and decrease in BIK and MTP18 expression, which lead to reduced oxidative phosphorylation and enhanced mitochondrial fusion. In a chronic inflammatory systemic disorder, CD14+ blood monocytes display reduced miR-125b expression as compared with healthy controls, inversely correlated with BIK and MTP18 messenger RNA expression. Our findings not only identify BIK and MTP18 as novel targets for miR-125b that control mitochondrial metabolism and dynamics, respectively, but also reveal a novel function for miR-125b in regulating metabolic adaptation of monocytes to inflammation. Together, these data unravel new molecular mechanisms for a proapoptotic role of miR-125b in monocytes and identify potential targets for interfering with excessive inflammatory activation of monocytes in inflammatory disorders.
Introduction
Excessive and prolonged activation of monocytes/macrophages inflicts sustained tissue damage and targets organ inflammation. How the complex networks of survival and apoptotic regulators are integrated to tightly control the life span of infiltrating monocytes to avoid harmful reactions and allow healing and maintenance of tissue homeostasis remains elusive. Mitochondria play a pivotal role in innate immune cell homeostasis by supplying energy and integrating converging signals that lead to apoptosis or inflammation.1,2 In particular, the mitochondrial network is crucial for cell homeostasis, because the balance between fusion and fission is essential for mitochondria biogenesis and removal, the optimized redistribution of mitochondria in response to local demand of ATP production, and the progression of apoptosis.3 Furthermore, mitochondria dysfunction is frequently associated with pathological conditions, including cancer and inflammatory disorders.4,5 The mechanisms that control mitochondrial activity and dynamics are, however, not fully understood.
MicroRNAs (miRNAs or miRs) are endogenous short non-coding RNA molecules that control gene expression mainly at the posttranscriptional level by pairing to the target transcripts.6 They participate in all the biological functions that have been investigated so far, and dysfunction of miRNAs is implicated in major human disorders, including cancer and autoimmunity.7,-9 The function of most of the mammalian miRNAs has yet to be determined. There is growing interest in exploring the regulation of mitochondria by miRNAs. Few miRNAs have been demonstrated to control mitochondrial metabolism and dynamics, as well as mitochondria-mediated apoptosis. In addition, the outer membrane of mitochondria is one of the destinations of miRNAs.10
Among the miRNAs that have been assigned apoptotic and immune functions and that regulate nuclear encoded mitochondria-associated proteins, miR-125b is of particular interest.11 It is a highly conserved miRNA that has multiple targets, including proteins regulating apoptosis, innate immunity, inflammation, and differentiation, and its dysregulation has been reported to occur in multiple cancer types.12,,,,-17 Depending on the cellular context, miR-125b shows different patterns of expression and effects, acting as a tumor suppressor gene or oncogene through the regulation of multiple genes involved in the mitochondrial pathways of apoptosis.18,,-21 In hematopoietic stem cells, miR-125b is highly expressed, enhancing self-renewal and survival while providing resistance to apoptosis and blocking differentiation, and is upregulated in lymphoblastic and myeloblastic leukemia.22 Importantly, miR-125b plays a critical role in myeloid biology,23 with its expression being low in common myeloid progenitors,22 induced upon inflammatory stimuli in macrophages,24,25 and deregulated in myeloid malignancies.15,26 Lastly, miR-125b regulates adaptation of cell metabolism to cancer transformation27 and is one of the master miRNAs involved in the Toll-like receptor 4 (TLR4) signaling pathway during the development of endotoxin tolerance.25,28
To provide novel insight into the integrated genetic regulatory network specifying cell fate, we have searched for novel target genes of miR-125b and further explored its role in the context of human monocytes, including the mechanisms on mitochondria metabolism.
Materials and methods
Cell culture and reagents
THP-1 and HEK293 cell lines were respectively grown in RPMI 1640 and in Dulbecco’s modified Eagle medium, both supplemented with 10% fetal calf serum, 1% penicillin/streptomycin, and l-glutamine. Transfection was performed using Lipofectamine 2000 (Invitrogen). For small interfering RNA (siRNA), pre-miRNA, and antago-miRNA, cells were transfected at 50 nM and harvested 48 hours later. For M1-polarized macrophages, THP-1 cells were treated 3 days with phorbol 12-myristate 13-acetate (PMA; 0.1 μg/mL), followed by 24 hours of stimulation with interferon γ (IFN-γ; 20 ng/mL) plus lipopolysaccharide (LPS; 0.1 μg/mL).
Blood CD14+CD16− monocytes were selected from peripheral blood mononuclear cells by magnetic separation (Dynabeads Untouched Human Monocytes Kit; Invitrogen), and grown in complemented RPMI 1640 media (purity >97%). For rheumatoid arthritis (RA) patients and healthy controls, informed consent was provided in accordance with procedures approved by the local human ethics committee (Comité de Protection des Personnes Sud Méditerrannée IV; NCT02909998). Fresh peripheral blood was obtained from 6 healthy donors with no history of autoimmune diseases (75% women) and from 8 age- and sex-matched patients with RA (Table 1).
Characteristics of the RA patients
| Characteristics . | RA . |
|---|---|
| No. of samples | 8 |
| Women, % | 75 |
| Mean age ± SD, y | 65.9 ± 7.7 |
| Positive ACPA, % | 62.50 |
| Positive RF, % | 62.50 |
| DAS28 | 5.3 ± 2.0 |
| Drug use, % | |
| Methotrexate | 62.50 |
| Corticoid | 50 |
| Rituximab | 1.25 |
| Characteristics . | RA . |
|---|---|
| No. of samples | 8 |
| Women, % | 75 |
| Mean age ± SD, y | 65.9 ± 7.7 |
| Positive ACPA, % | 62.50 |
| Positive RF, % | 62.50 |
| DAS28 | 5.3 ± 2.0 |
| Drug use, % | |
| Methotrexate | 62.50 |
| Corticoid | 50 |
| Rituximab | 1.25 |
ACPA, Anti-citrullinated protein antibodies; DAS28, disease activity score referring to the 28 joints examined; RF, rheumatoid factor.
Gene expression analysis
Total RNAs were extracted using the miRNeasy Mini Kit with a Qiacube (QIAGEN). Expression levels of miRNAs and messenger RNAs (mRNAs) were quantified using TaqMan MicroRNA Assays and Gene Expression Assays (Life Technologies), respectively. The expression of RNU6B and GAPDH was used as endogenous control for miRNA and mRNA data normalization, respectively. Specific primers were used for quantification (supplemental Table 1, available on the Blood Web site). The TaqMan Array Human Inflammation Panel (Life Technologies) was used according to the manufacturer‘s protocols, and relative gene expression was calculated using the comparative threshold cycle method.
Luciferase assay
HEK293 cells were transfected with 50 ng of either psiCHECK-2 vector encoding BIK 3′ untranslated region (3′UTR) or luciferase reporter plasmid GoClone Reporter (SwitchGear Genomics) encoding MTP18 3′UTR (see supplemental Experimental Procedures), together with 1 to 100 nM of pre- or antago-miR-125b. Luciferase activity was measured after 48 hours using the Dual-Luciferase Reporter Assay System (Promega). Normalization was performed with the number of living cells.
Protein extraction and western blotting
Cells were lysed with CelLytic MT (Sigma-Aldrich). Proteins were separated on NuPAGE 12% Bis-TRIS gels (10 μg per lane) and transferred to nitrocellulose membranes, which were blocked with primary antibody. Anti-BIK antibody was purchased from Santa-Cruz Biotechnology, and anti–mitochondrial fission process 1 protein (anti-MTP18) antibody was kindly provided by Daniel Tondera (Silence Therapeutics, Berlin, Germany). Supplemental Table 2 describes the antibodies and dilutions used for the immunoblot analysis. Proteins were visualized and quantitated using the Odyssey Infrared Imaging System (LI-COR Biosciences). Autophagy and autophagic flux were assessed according to previously reported methods.29
Real-time monitoring of the mitochondrial energy metabolism
Cells were suspended 48 hours after transfection in XF assay buffer supplemented with 1 mM pyruvate and 12 mM d-glucose at pH 7.4 and then plated (250 000 cells per well) in 100 μL on cellTak-coated assay plates (BD Biosciences). The oxygen consumption rate was measured with an XF24 extracellular flux analyzer (Seahorse Bioscience).30 Test compounds (Sigma-Aldrich) were injected during the assay at the following final concentrations: 0.9 μM oligomycin, 0.5 μM carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone (FCCP), 0.9 μM rotenone, and 0.9 μM antimycin A (see supplemental Experimental Procedures). Normalization was performed with the number of living cells.
Fluorescence microscopy
THP-1 and CD14+ cells were seeded at 50% confluence, treated 12 hours with PMA (0.1 μg/mL), and fixed with 4% PFA, permeabilized in 0.2% Triton X-100, and incubated in blocking solution (DAKO) before staining with TOM20 and secondary antibody Alexa 488. Cells were visualized using a Leica TCS SP5 confocal laser-scanning microscope (model DM 6000S) and the LAS AF acquisition software (Wetzlar, Germany). Images were captured on the Cellomics ArrayScan VTi platform (Thermo Scientific) using the cell health profiling algorithm to quantify the percentage of cells containing TOM20 mitochondrial staining.
Cell viability and apoptosis assay
Cytotoxic effects and caspase 3/7 activation were measured 48 hours posttransfection using CellTiter-Glo Reagent (Promega) and the Caspase-Glo 3/7 Assay (Promega), respectively.
Microarray hybridization and data analysis
Quality control and quantification of total RNA was ensured with Bioanalyzer (Agilent Technologies) and NanoDrop ND-1000 (Thermo Scientific), respectively. Generation of complementary RNA, sample hybridization (using Affymetrix HG U133 plus 2.0 arrays), and scanning with a GeneChip Scanner 3000 (Affymetrix) were performed as described previously.31 Data from 3 pairs of conditions were analyzed using the BioRetis database (supplemental Experimental Procedures). The chip data discussed in this publication have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus database and are accessible through accession number GSE62693.
Statistical analysis
GraphPad software was used to perform unpaired nonparametric Mann-Whitney or Student t tests according to the experimental design for 2-group comparisons. One-way analysis of variance after the Tukey multiple comparisons posttest analysis was performed for statistical tests of more than 2 groups. Correlations were evaluated with Spearman correlation analyses. P values <.05 were considered statistically significant.
Results
miR-125b controls the expression of genes involved in inflammatory and apoptotic functions
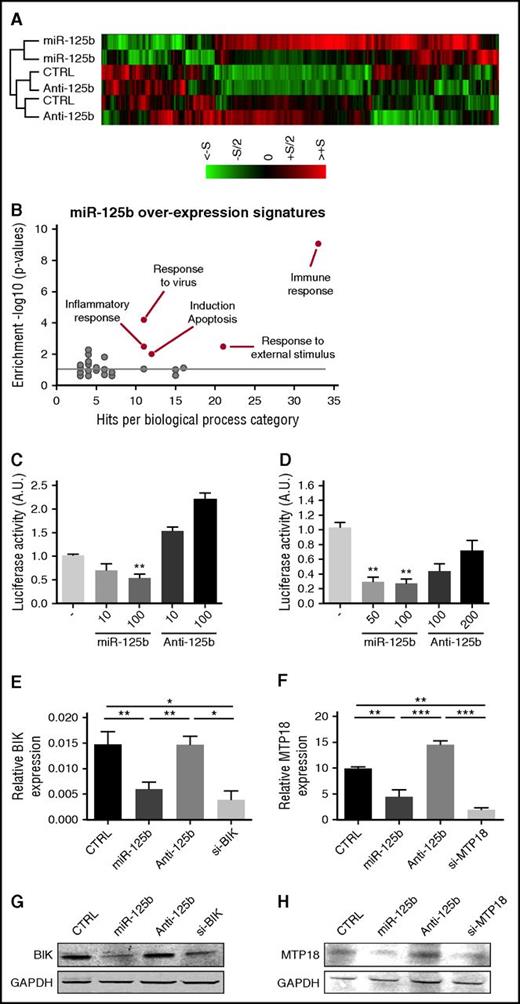
To identify novel genes and pathways regulated by miR-125b in human monocytes, we profiled the gene expression changes of the human monocytic THP-1 cell line after miR-125b gain- and loss-of-function experiments (Figure 1). Analysis revealed that 419 genes were significantly upregulated or downregulated in cells transfected with miR-125b mimics as compared with control conditions. We selected mRNAs with fold change >1.5, P < .05, and false discovery rate <5% (Figure 1A). Analysis using the Database for Annotation, Visualization, and Integrated Discovery32 revealed high enrichment of specific biological processes, including apoptosis, inflammation, and immune responses (Figure 1B). Selecting the specific downregulated mRNAs in cells overexpressing miR-125b, we identified 4 putative miR-125b gene targets predicted by 2 major target prediction algorithms (http://www.microRNA.org and http://www.TargetScan.org). All putative target genes were involved in the apoptosis pathway: TMEM77 (alias DRAM2), HTATIP2 (alias TIP30), MTP18 (alias MTFP1), and BIK.
miR-125b represses the expression of BIK and MTP18. (A) Transcriptomic analysis of miR-125b-overexpressing THP-1 cells (miR-125b) as compared with negative control miRNA-precursor-transfected cells (CTRL) and miR-125b-antagomir-transfected cells (Anti-125b). Hierarchical clustering of the 419 genes with P < .05 and fold change of at least 1.5 was generated from 2 independent experiments (S = 2). (B) Analysis of differentially expressed genes in miR-125b-overexpressing THP-1 cells vs negative controls using the functional annotation of the Database for Annotation, Visualization, and Integrated Discovery, National Institute of Allergy and Infectious Diseases. (C-D) Luciferase activity levels upon cotransfection of HEK293 cells with a luciferase construct (50 ng) containing 3′UTR of BIK (C) or MTP18 (D) together with control miRNA, miR-125b mimics, or antagomir (n = 3). The results are shown 48 hours after transfection and expressed as mean ± SD. Data are representative of 2 independent experiments. **P < .01 compared with control condition. (E-H) mRNA (E-F) and protein (G-H) levels of BIK (E,G) and MTP18 (F,H) in THP-1 cells transfected with control miRNA (CTRL), miR-125b mimics (miR-125b), miR-125b antagomir (Anti-125b), or siRNA targeting human BIK (si-BIK) or MTP18 (si-MTP18) (50 nM). Results are expressed as mean ± SD of 4 independent experiments. *P < .05; **P < .01; ***P < .001. A.U., arbitrary units; SD, standard deviation.
miR-125b represses the expression of BIK and MTP18. (A) Transcriptomic analysis of miR-125b-overexpressing THP-1 cells (miR-125b) as compared with negative control miRNA-precursor-transfected cells (CTRL) and miR-125b-antagomir-transfected cells (Anti-125b). Hierarchical clustering of the 419 genes with P < .05 and fold change of at least 1.5 was generated from 2 independent experiments (S = 2). (B) Analysis of differentially expressed genes in miR-125b-overexpressing THP-1 cells vs negative controls using the functional annotation of the Database for Annotation, Visualization, and Integrated Discovery, National Institute of Allergy and Infectious Diseases. (C-D) Luciferase activity levels upon cotransfection of HEK293 cells with a luciferase construct (50 ng) containing 3′UTR of BIK (C) or MTP18 (D) together with control miRNA, miR-125b mimics, or antagomir (n = 3). The results are shown 48 hours after transfection and expressed as mean ± SD. Data are representative of 2 independent experiments. **P < .01 compared with control condition. (E-H) mRNA (E-F) and protein (G-H) levels of BIK (E,G) and MTP18 (F,H) in THP-1 cells transfected with control miRNA (CTRL), miR-125b mimics (miR-125b), miR-125b antagomir (Anti-125b), or siRNA targeting human BIK (si-BIK) or MTP18 (si-MTP18) (50 nM). Results are expressed as mean ± SD of 4 independent experiments. *P < .05; **P < .01; ***P < .001. A.U., arbitrary units; SD, standard deviation.
BIK and MTP18 are 2 novel targets for miR-125b
The 3′UTR of the 4 predicted targets were cloned downstream from the luciferase gene, HEK-293 cells were cotransfected with the synthetic miR-125b precursor or antagonist, and relative luciferase activities were determined (Figure 1C-H; supplemental Figure 1). Overexpression of miR-125b significantly decreased the luciferase activity of the BIK and MTP18 reporter systems only. For both cases, increasing doses of miR-125b antisense dose-dependently reversed the inhibitory effect of basal miR-125b expression on the reporter system. To prove that miR-125b regulates endogenous BIK and MTP18 mRNA expression in monocytes, we transfected THP-1 cells with synthetic miR-125b precursor or antagonist. Data revealed that BIK and MTP18 mRNA and protein levels were significantly reduced by enforced expression of miR-125b to levels comparable with BIK and MTP18 siRNA-mediated knockdown (Figure 1E-H).
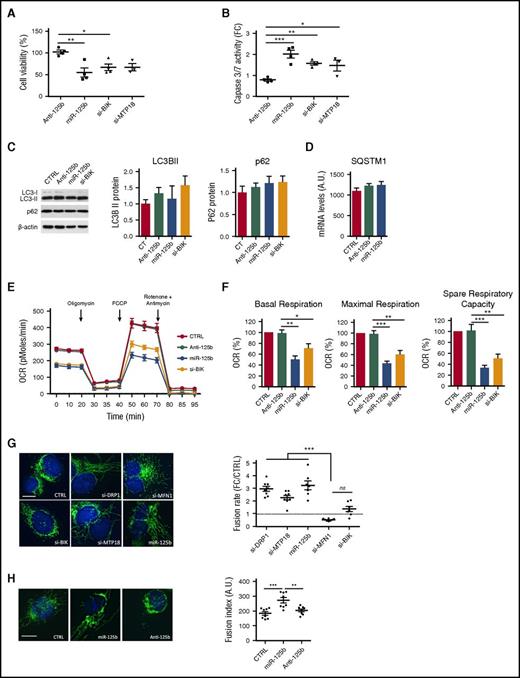
miR-125b promotes apoptosis of monocytes
BIK is a BH3-only proapoptotic protein localized in the endoplasmic reticulum (ER) that sensitizes mitochondrial apoptosis.33 MTP18 is localized in the mitochondria and induces mitochondrial fission and apoptosis.34 Because miR-125b targets 2 proapoptotic genes, we studied the effect of miR-125b on THP-1 apoptosis. Unexpectedly, enforced expression of miR-125b, as well as BIK or MTP18 siRNAs, induced apoptosis as determined by decreased cell viability and increased caspase 3/7 activation (Figure 2A-B). To further understand the functional role of miR-125b in apoptosis, we quantified several other anti- and proapoptotic genes that have been previously identified as directly targeted by miR-125b. The quantification of mRNA and protein levels revealed that the expression of BAK1, BMF, and BCL-2 was not controlled by miR-125b in THP-1 (supplemental Figure 2).
miR-125b modulates mitochondrial metabolism and dynamics through BIK and MTP18 silencing, respectively. THP-1 cells were transfected with si-DRP1, si-MPT18, si-MFN1, si-BIK, control miRNA (CTRL), miR-125b mimics (miR-125b), or miR-125b antagomir (Anti-125b) and analyzed 48 hours later. (A-B) Cell apoptosis was assessed by measuring cell viability and caspase 3/7 activity. The data are presented as percentage (A) or a fold change (FC) (B) of the cell transfected with control miRNA. Data represent mean ± SD of 4 independent experiments. (C-D) Autophagy was monitored using LC3/p62 immunoblotting to track the conversion of LC3-I into LC3-II and the expression of p62 for autophagic activity. Representative western blot of LC3-I/LC3-II, p62, and β-actin is shown (left), and quantitative analysis of LC3-II and p62 are plotted as mean ± SD of 3 exposures of 2 independent experiments (center and right) (C). Quantification of SQSTM1 mRNA (D) was performed using quantitative reverse transcription polymerase chain reaction. Data represent 2 technical independent experiments. (E) The OCR was measured in real time under basal conditions: oligomycin, ATP-synthetase-inhibited rate; FCCP, uncoupled rate; and rotenone + antimycin A, inhibited rate. The OCR was normalized by number of cells in each condition (n = 6 per group). (F) Representative graphs of quantification of various parameters of mitochondrial respiratory profiles, presented as mean ± SD of 4 independent experiments. (G-H) Monitoring of mitochondrial fusion and fission in cells stained with anti-TOM20 antibodies (n = 8). Representative images of mitochondria stained with TOM20 (left; bars represent 10 μm) and quantification of fluorescence using Cellomics ArrayScan VTi platform (right; n = 20 per experimental replicate) are shown. Results are expressed as mean ± SD. *P < .05; **P < .01; ***P < .001.
miR-125b modulates mitochondrial metabolism and dynamics through BIK and MTP18 silencing, respectively. THP-1 cells were transfected with si-DRP1, si-MPT18, si-MFN1, si-BIK, control miRNA (CTRL), miR-125b mimics (miR-125b), or miR-125b antagomir (Anti-125b) and analyzed 48 hours later. (A-B) Cell apoptosis was assessed by measuring cell viability and caspase 3/7 activity. The data are presented as percentage (A) or a fold change (FC) (B) of the cell transfected with control miRNA. Data represent mean ± SD of 4 independent experiments. (C-D) Autophagy was monitored using LC3/p62 immunoblotting to track the conversion of LC3-I into LC3-II and the expression of p62 for autophagic activity. Representative western blot of LC3-I/LC3-II, p62, and β-actin is shown (left), and quantitative analysis of LC3-II and p62 are plotted as mean ± SD of 3 exposures of 2 independent experiments (center and right) (C). Quantification of SQSTM1 mRNA (D) was performed using quantitative reverse transcription polymerase chain reaction. Data represent 2 technical independent experiments. (E) The OCR was measured in real time under basal conditions: oligomycin, ATP-synthetase-inhibited rate; FCCP, uncoupled rate; and rotenone + antimycin A, inhibited rate. The OCR was normalized by number of cells in each condition (n = 6 per group). (F) Representative graphs of quantification of various parameters of mitochondrial respiratory profiles, presented as mean ± SD of 4 independent experiments. (G-H) Monitoring of mitochondrial fusion and fission in cells stained with anti-TOM20 antibodies (n = 8). Representative images of mitochondria stained with TOM20 (left; bars represent 10 μm) and quantification of fluorescence using Cellomics ArrayScan VTi platform (right; n = 20 per experimental replicate) are shown. Results are expressed as mean ± SD. *P < .05; **P < .01; ***P < .001.
Because BIK can induce autophagy by displacing Bcl-2 from Beclin 1 and NAF-1,35,,-38 we investigated the effect of miR-125b on autophagy. We first analyzed the accumulation of LC3-II, which reflects the number of autophagosomes.39 Neither the overexpression of miR-125b nor the silencing of BIK modified the levels of LC3-II as compared with controls (Figure 2C). Second, to assess modulation of the autophagic flux, we analyzed the expression of the autophagic cargo SQSTM1/p62. The expression levels of SQSTM1/p62 mRNA and protein were unaltered in all conditions tested (Figure 2C-D). Overall, these results strongly suggest that miR-125b does not modulate autophagy in THP-1 cells.
miR-125b dampens mitochondrial respiration rate
To further explore the underlying mechanisms by which miR-125b-mediated silencing of BIK promotes apoptosis, we monitored the mitochondrial metabolism through quantification of the oxygen consumption rate (OCR) using the Seahorse technology, in a basal state and after the addition of oligomycin (to block adenosine triphosphate [ATP] synthesis), FCCP (to uncouple ATP synthesis from the electron transport chain), and rotenone and antimycin A (to block complex I and III of the electron transport chain, respectively). Ectopic expression of miR-125b and BIK silencing both significantly reduced basal OCRs and decreased maximal mitochondria respiration and spare respiratory capacity when compared with control conditions (Figure 2E-F). MTP18 loss-of-function did not alter basal respiration, ATP production, or respiratory capacities (supplemental Figure 3). Rescue of BIK expression restored mitochondrial respiration parameters (supplemental Figure 4D-F). Overall, our data suggest that miR-125b induces apoptosis in THP-1 by regulating mitochondrial respiration, at least in part through BIK silencing, but not through MTP18 silencing.
Increase of mitochondrial fusion by miR-125b
Because BIK activates mitochondrial fragmentation with little release of cytochrome c to the cytosol,40,41 and MTP18 loss-of-function promotes fusion of mitochondria,42 we assessed the effects of miR-125b ectopic expression on mitochondrial dynamics as compared with BIK and MTP18 silencing. DRP1 and MFN1 knockdown were used as positive controls for mitochondria fusion and fission, respectively. Using staining with an antibody against the outer membrane (TOM20), we evidenced changes in the pattern of mitochondrial networks after enforced expression of miR-125b (Figure 2G-H). The hyperfused mitochondrial structure observed was comparable to mitochondrial networks observed with siDRP1 or siMTP18. Neither BIK silencing (Figure 2G) nor miR-125b inhibition (Figure 2H) affected mitochondrial dynamics. Importantly, neither mitochondrial content nor membrane potential was affected by miR-125b, as evidenced by unmodified MitoTracker Red accumulation in mitochondria and staining of the mitochondrial protein TOM20, respectively (supplemental Figure 5). Lastly, rescue experiments cotransfecting cells with miR-125b mimics and either BIK or MTP18 expression plasmids were performed. Although cotransfection of miR-125b mimics together with mock- or BIK-expressing plasmids significantly increased the mitochondrial fusion rate, rescue of MTP18 expression restored fusion to levels comparable to the control condition (supplemental Figure 4G). In addition, rescue of MTP18 expression significantly increased the mitochondrial fission rate as compared with the 3 other conditions (supplemental Figure 4H). These results suggest that miR-125b regulates mitochondrial fusion, at least in part through MTP18 silencing, but not through BIK silencing.
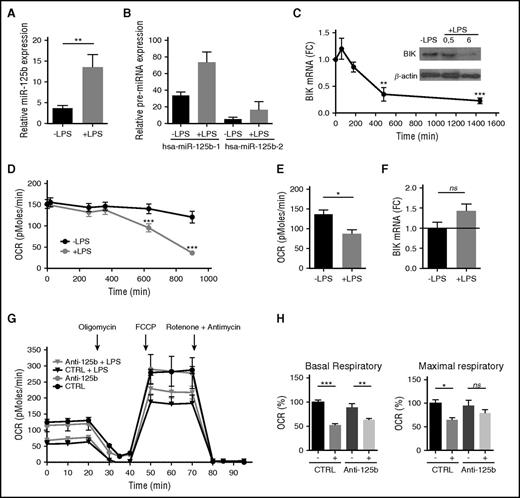
TLR4 engagement induces the miR-125b-mediated silencing of BIK
LPS is a potent activator of monocytes that controls survival and apoptosis through TLR4 engagement. TLR4 is not only a component of innate immunity but also a modulator in metabolic systems. To further address the functional role for miR-125b in regulating the mitochondrial metabolism in monocytes, we investigated the link between TLR4 stimulation, BIK downregulation, and metabolic switch. First, consistent with previous studies,25 miR-125b was significantly upregulated by TLR4 engagement in THP-1 cells (Figure 3A). Both paralogs coding for the same mature miR-125b sequence and located on 2 different polycistronic miRNA clusters (hsa-miR-125b-1 and hsa-miR-125b-2 on chromosomes 11 and 21, respectively)43 were overexpressed upon LPS challenge. Although the induction of miR-125b-2 expression was higher than miR-125b-1 after LPS stimulation, there was 4 times more miR-125b-1 expressed in the cells than miR-125b-2 (Figure 3B). These data suggest that the majority of the mature forms of miR-125b detected were probably processed from the hsa-miR-125b-1. Second, consistent with an induction of miR-125b by LPS and a control of BIK expression by miR-125b, we found that LPS-induced activation of THP-1 cells promoted a decrease of BIK mRNA and protein (Figure 3C), as well as changes in mitochondrial respiration (Figure 3D). Similar to what we observed after miR-125b overexpression or BIK knockdown (Figure 2E-F), LPS significantly reduced oxygen consumption from mitochondria (Figure 3D-E). Importantly, when the LPS-induced miR-125b increase was prevented using introduction of synthetic miRNA antagonists, BIK expression (Figure 3F) and mitochondrial maximal respiration (Figure 3G-H) were restored to near normal levels. Thus, blocking the LPS-induced miR-125b increase preserved the reserve respiratory capacity of the cells. Together, these data suggest that miR-125b regulates metabolic adaptation of monocytes to inflammation.
TLR4 engagement mimics BIK silencing. (A-B) Expression levels of mature miR-125b (A) or pre-miR125b-1 and pre-miR-125b-2 (B) in THP-1 monocytes without stimulation or after 4 hours of LPS stimulation (1 μg/mL). RNU6B and GAPDH expression was used as endogenous control for miRNA and pre-mRNA data normalization. Mean ± SD of 2 to 8 duplicates are shown. (C) Kinetics of BIK mRNA expression in THP-1 cells after LPS stimulation (1 μg/mL). RNA quantification presents fold change (FC) to time 0. Error bars represent the SD of 3 to 7 independent experiments. BIK protein levels were analyzed by immunoblot at the indicated time points. (D) Kinetics of the OCR measured in THP-1 after LPS stimulation (1 μg/mL). All data are mean ± SD (n = 6), representative of 3 independent experiments. (E) The OCR measurement on THP-1 after 12 hours of LPS stimulation. Error bars represent the mean ± SD of 2 to 3 replicates of 3 independent experiments. (F) BIK mRNA expression levels 48 hours after transfection with miR-125b antagomir and 4 hours of LPS stimulation (1 μg/mL). Data are presented as FC with THP-1 transfected with control pre-miRNA. Error bars represent the SD of 4 independent experiments. (G) The OCR measured on THP-1 after 48 hours of transfection with control miRNA (CTRL) or miR-125b antagomir (Anti-125b) (50 nM), including 12 hours of LPS treatment (1 μg/mL), normalized by number of cells. OCRs were measured in real time under basal conditions: oligomycin, ATP-synthetase-inhibited rate; FCCP, uncoupled rate; and rotenone + antimycin A, inhibited rate. Data are representative of 4 independent experiments. (H) Parameters of respiratory profiles are presented as mean ± SD for 4 independent experiments. *P < .05; **P < .01; ***P < .001. ns, Not significant.
TLR4 engagement mimics BIK silencing. (A-B) Expression levels of mature miR-125b (A) or pre-miR125b-1 and pre-miR-125b-2 (B) in THP-1 monocytes without stimulation or after 4 hours of LPS stimulation (1 μg/mL). RNU6B and GAPDH expression was used as endogenous control for miRNA and pre-mRNA data normalization. Mean ± SD of 2 to 8 duplicates are shown. (C) Kinetics of BIK mRNA expression in THP-1 cells after LPS stimulation (1 μg/mL). RNA quantification presents fold change (FC) to time 0. Error bars represent the SD of 3 to 7 independent experiments. BIK protein levels were analyzed by immunoblot at the indicated time points. (D) Kinetics of the OCR measured in THP-1 after LPS stimulation (1 μg/mL). All data are mean ± SD (n = 6), representative of 3 independent experiments. (E) The OCR measurement on THP-1 after 12 hours of LPS stimulation. Error bars represent the mean ± SD of 2 to 3 replicates of 3 independent experiments. (F) BIK mRNA expression levels 48 hours after transfection with miR-125b antagomir and 4 hours of LPS stimulation (1 μg/mL). Data are presented as FC with THP-1 transfected with control pre-miRNA. Error bars represent the SD of 4 independent experiments. (G) The OCR measured on THP-1 after 48 hours of transfection with control miRNA (CTRL) or miR-125b antagomir (Anti-125b) (50 nM), including 12 hours of LPS treatment (1 μg/mL), normalized by number of cells. OCRs were measured in real time under basal conditions: oligomycin, ATP-synthetase-inhibited rate; FCCP, uncoupled rate; and rotenone + antimycin A, inhibited rate. Data are representative of 4 independent experiments. (H) Parameters of respiratory profiles are presented as mean ± SD for 4 independent experiments. *P < .05; **P < .01; ***P < .001. ns, Not significant.
miR-125b promotes classical proinflammatory activation of THP-1 cells
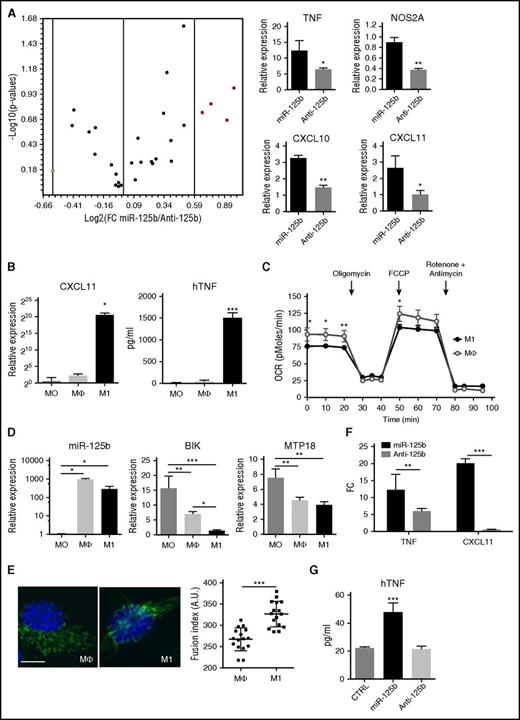
To further explore the mechanisms of miR-125b-induced metabolic adaptation to inflammation, we quantified a set of genes implicated in the inflammatory response (Figure 4A). After enforced expression of miR-125b, a variety of genes was elevated as compared with miR-125b inhibition, particularly chemokines and proinflammatory mediators: CXCL10, CXCL11, TNF, and NOS2A genes. All these genes provide a likely mechanism for the THP-1 cells that occurs after increased miR-125b expression, because they are indicative of the M1-type polarization known to induce distinct genetic and metabolic profiles, switching metabolism from oxidative phosphorylation (OXPHOS) to glycolysis and triggering specific proinflammatory cytokines.44
miR-125b promotes M1 macrophage polarization. (A) Total RNA was isolated from THP-1 transfected with control miRNA, miR-125b mimics (miR-125b), or miR-125b antagomir (Anti-125b). After reverse transcription, analysis of the expression level of 96 inflammatory genes was performed using a TaqMan-based Low Density Array Human Inflammation Panel. The volcano plot (top) displays statistical significance vs fold-change (FC) pre-miR-125b/antagomiR-125b on the y- and x-axes, respectively. Red dots represent M1 specific genes: TNF, NOS2A, CXCL10, and CXCL11. Graphs (bottom) represent RNA expression levels of these genes as mean ± SD of 3 technical experiments. (B) THP-1 monocytes (MO) were differentiated into macrophages (Mϕ) after 3 days of culture with PMA (0.1 μg/mL), and then polarized into M1 macrophages using IFN-γ and LPS stimulation for 24 hours. M1 polarization phenotype was monitored by quantification of CXCL11 mRNA expression using quantitative reverse transcription polymerase chain reaction (left), and of human TNF (hTNF) secretion using enzyme-linked immunosorbent assay (right). (C) Real-time analysis of the OCR for THP-1 monocytes differentiated into macrophages, polarized or not (open circles) toward M1 phenotype (filled circles). (D) Expression levels of miR-125b, BIK, and MTP18 mRNA in monocytes (MO), macrophages (Mϕ), or M1-polarized macrophages (M1) using real-time polymerase chain reaction. Error bars represent the mean ± SD of 3 independent experiments. (E) Unpolarized (Mϕ) or M1-polarized macrophages (M1) were fixed and stained with anti-TOM20 antibodies. Representative images (left; bar represents 10 μm) and quantification of captured images (right) are shown. (F-G) Expression levels of CXCL11 and TNF mRNAs (F) and human TNF secretion (G) in THP-1 cells transfected with control miRNA (CTRL), miR-125b mimics, or miR-125b antagomir. RNA quantification is presented as fold change to the controls, and error bars represent the SD of 4 independent experiments. *P < .05; **P < .01; ***P < .001. TFN, tumor necrosis factor.
miR-125b promotes M1 macrophage polarization. (A) Total RNA was isolated from THP-1 transfected with control miRNA, miR-125b mimics (miR-125b), or miR-125b antagomir (Anti-125b). After reverse transcription, analysis of the expression level of 96 inflammatory genes was performed using a TaqMan-based Low Density Array Human Inflammation Panel. The volcano plot (top) displays statistical significance vs fold-change (FC) pre-miR-125b/antagomiR-125b on the y- and x-axes, respectively. Red dots represent M1 specific genes: TNF, NOS2A, CXCL10, and CXCL11. Graphs (bottom) represent RNA expression levels of these genes as mean ± SD of 3 technical experiments. (B) THP-1 monocytes (MO) were differentiated into macrophages (Mϕ) after 3 days of culture with PMA (0.1 μg/mL), and then polarized into M1 macrophages using IFN-γ and LPS stimulation for 24 hours. M1 polarization phenotype was monitored by quantification of CXCL11 mRNA expression using quantitative reverse transcription polymerase chain reaction (left), and of human TNF (hTNF) secretion using enzyme-linked immunosorbent assay (right). (C) Real-time analysis of the OCR for THP-1 monocytes differentiated into macrophages, polarized or not (open circles) toward M1 phenotype (filled circles). (D) Expression levels of miR-125b, BIK, and MTP18 mRNA in monocytes (MO), macrophages (Mϕ), or M1-polarized macrophages (M1) using real-time polymerase chain reaction. Error bars represent the mean ± SD of 3 independent experiments. (E) Unpolarized (Mϕ) or M1-polarized macrophages (M1) were fixed and stained with anti-TOM20 antibodies. Representative images (left; bar represents 10 μm) and quantification of captured images (right) are shown. (F-G) Expression levels of CXCL11 and TNF mRNAs (F) and human TNF secretion (G) in THP-1 cells transfected with control miRNA (CTRL), miR-125b mimics, or miR-125b antagomir. RNA quantification is presented as fold change to the controls, and error bars represent the SD of 4 independent experiments. *P < .05; **P < .01; ***P < .001. TFN, tumor necrosis factor.
To induce classic (M1) polarization, PMA-differentiated THP-1 macrophages (Mϕ) were incubated in the presence of IFN-γ plus LPS.45 Polarization was first confirmed using a panel of established markers (Figure 4B) and then confirmed by real-time monitoring of the mitochondrial energy metabolism (Figure 4C). M1-polarized THP-1 macrophages showed increased expression of TNF, CXCL10, and CXCL11, as well as a decreased OCR, compared with M0 or Mϕ. As previously reported for mouse monocytes,14 we observed enhanced miR-125b expression in THP-1 macrophages (Mϕ) that is maintained when polarized into M1-like macrophages (Figure 4D). These data suggest that miR-125b may be involved in the induction and maintenance of the activated nature of macrophages.
Because the expression of BIK and MTP18 in M1-like macrophages was inversely correlated to that of miR-125b (Figure 4D), we determined the effects of M1 polarization on mitochondria respiration and dynamics. We showed that M1 polarization reproduces both the effect of miR-125b overexpression or BIK silencing on the mitochondrial respiration of macrophages (Figures 2E and 3G) and the effect of miR-125b overexpression or MTP18 silencing on the mitochondrial dynamics (Figure 2C), namely, a reduced OCR (Figure 4C) and increased fusion of mitochondrial structures (Figure 4E), respectively. Ectopic expression of miR-125b promoted M1-like polarization, as determined by increased expression of M1 markers compared with control miRNA or knockdown of miR-125b (Figure 4F-G). Importantly, rescue of BIK and MTP18 expression inhibited miR-125b-induced TNF and CXCL11 transcription (supplemental Figure 4I-J). These data show that miR-125b contributes to M1-like macrophage polarization, mimicking the IFN-γ/LPS stimulation effect, and suggest that BIK and MTP-18 may mediate the regulatory role of miR-125b on M1-like macrophage activation.
The miR-125b/BIK/MTP18 axis plays a role in the adaptation of human CD14+ monocytes to inflammation
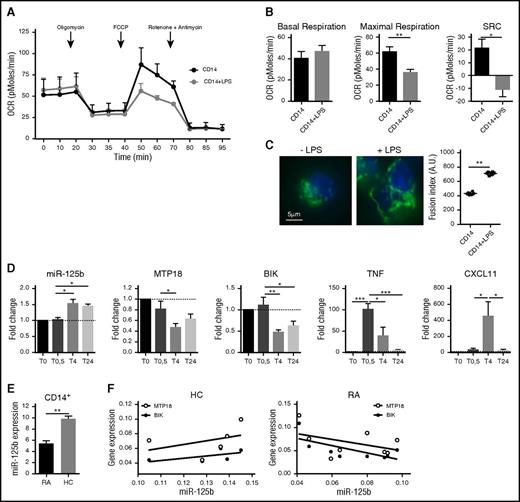
Lastly, we tested the mechanisms of miR-125b-mediated metabolic adaptation to inflammation in human primary monocytes. We showed that purified CD14+ monocytes challenged with LPS displayed a reduced OCR (Figure 5A-B) and increased mitochondrial fusion (Figure 5C). Furthermore, LPS induced the rapid increase in the miR-125b expression level, which is associated with reduced MTP18 and BIK expression levels, as well as parallel increased expression of the proinflammatory TNF cytokine and the CXCL11 chemokine (Figure 5D). These data support the hypothesis that metabolic adaptation of CD14+ monocytes to inflammatory stimulus involves the control of mitochondrial respiration and dynamics through the miR-125b/BIK/MTP18 axis.
miR-125b and mitochondrial metabolism in human CD14+ monocytes under inflammatory conditions. CD14+ monocytes were negatively selected from peripheral blood mononuclear cells by magnetic separation isolated from either healthy donors (A-D) or patients with RA (E-G). (A) Kinetics of the OCR measured in CD14+ after LPS stimulation (0.1 μg/mL), in real time under basal conditions: oligomycin, ATP-synthetase-inhibited rate; FCCP, uncoupled rate; and rotenone + antimycin A, inhibited rate. Data are representative of 3 independent experiments. (B) Quantification of respiratory profile parameters presents 6 technical replicates. (C) Monitoring of mitochondrial fusion in CD14+ stained with anti-TOM20 antibodies (n = 3). Representative images of mitochondria stained with TOM20 (left; bar represents 5 μm) and quantification of fluorescence using Cellomics ArrayScan VTi platform (right; n = 20 per experimental replicate) are shown. (D) Kinetics of expression levels of miR-125b, MTP18, BIK, TNF, and CXCL11 mRNA in CD14+ monocytes after LPS stimulation using real-time polymerase chain reaction. RNA quantification presents fold change to time 0 (T0), and error bars represent the SD of 3 independent experiments. (E) miR-125b expression was quantified in CD14+ peripheral blood monocytes isolated from healthy controls (HC; n = 5) or patients with RA (n = 8). (F) Correlation study of miR-125b and BIK or MTP18 expression levels in CD14+ monocytes from healthy controls (n = 5) or patients with RA (n = 8) was performed using a Spearman correlation analysis. All results are expressed as mean ± SD. *P < .05; **P < .01; ***P < .001. SRC, spare respiratory capacity.
miR-125b and mitochondrial metabolism in human CD14+ monocytes under inflammatory conditions. CD14+ monocytes were negatively selected from peripheral blood mononuclear cells by magnetic separation isolated from either healthy donors (A-D) or patients with RA (E-G). (A) Kinetics of the OCR measured in CD14+ after LPS stimulation (0.1 μg/mL), in real time under basal conditions: oligomycin, ATP-synthetase-inhibited rate; FCCP, uncoupled rate; and rotenone + antimycin A, inhibited rate. Data are representative of 3 independent experiments. (B) Quantification of respiratory profile parameters presents 6 technical replicates. (C) Monitoring of mitochondrial fusion in CD14+ stained with anti-TOM20 antibodies (n = 3). Representative images of mitochondria stained with TOM20 (left; bar represents 5 μm) and quantification of fluorescence using Cellomics ArrayScan VTi platform (right; n = 20 per experimental replicate) are shown. (D) Kinetics of expression levels of miR-125b, MTP18, BIK, TNF, and CXCL11 mRNA in CD14+ monocytes after LPS stimulation using real-time polymerase chain reaction. RNA quantification presents fold change to time 0 (T0), and error bars represent the SD of 3 independent experiments. (E) miR-125b expression was quantified in CD14+ peripheral blood monocytes isolated from healthy controls (HC; n = 5) or patients with RA (n = 8). (F) Correlation study of miR-125b and BIK or MTP18 expression levels in CD14+ monocytes from healthy controls (n = 5) or patients with RA (n = 8) was performed using a Spearman correlation analysis. All results are expressed as mean ± SD. *P < .05; **P < .01; ***P < .001. SRC, spare respiratory capacity.
We then analyzed the expression profile relationships between miR-125b and its target genes on monocytes in the context of a systemic inflammatory disorder. Because CD14+ monocytes are important contributors to inflammation in patients with RA through the production of proinflammatory cytokines, we determined whether miR-125b-mediated regulation of BIK and MTP18 expression was affected on activated monocytes. In CD14+ blood monocytes from RA subjects, miR-125b exhibited decreased expression as compared to healthy controls (Figure 5E) and was inversely correlated with BIK (r = 0.46; P = .032) and MTP18 mRNA (r = 0.19) expression, whereas no correlation was found in CD14+ monocytes isolated from healthy donors (Figure 5F). Cofactors could not account for the observed modifications (data not shown). The observation of a reduced expression of the proapoptotic miR-125b in CD14+ monocytes from RA patients is consistent with previous findings showing that CD14+ monocytes are resistant to oxidative stress and to apoptosis, and thus abundantly present in RA tissues.46,47
Discussion
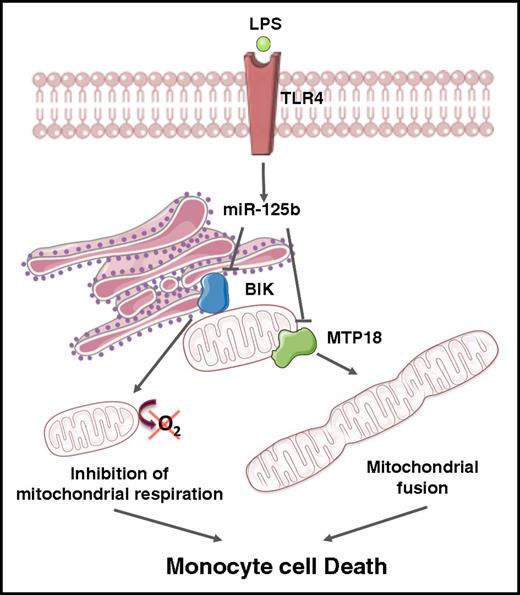
Our present work identifies the ER BH3-only protein BIK and the mitochondrial fission process 1 protein MTP18 as 2 novel cellular targets of miR-125b. In addition, we show that increased miR-125b through TLR4 affects mitochondrial respiration and dynamics through BIK and MTP18 silencing, respectively, promoting proinflammatory activation and apoptosis of monocytes (Figure 6). Our data suggest a novel role for miR-125b in regulating metabolic adaptation of monocytes to inflammation, thus extending its involvement in cell biology and disease pathogenesis, as well as providing potential candidates for therapeutic intervention.
The miR-125b/BIK/MTP18 axis controls monocyte mitochondrial metabolism. The ER BH3-only protein BIK and the mitochondrial fission process 1 protein (ie, MTP18) are 2 novel cellular targets of miR-125b. An increase of miR-125b expression through TLR4 engagement affects mitochondrial respiration and dynamics through BIK and MTP18 silencing, respectively. Mitochondrial fusion induction and decrease of mitochondrial respiration promote apoptosis of monocytes. The figure was produced using Servier Medical Art.
The miR-125b/BIK/MTP18 axis controls monocyte mitochondrial metabolism. The ER BH3-only protein BIK and the mitochondrial fission process 1 protein (ie, MTP18) are 2 novel cellular targets of miR-125b. An increase of miR-125b expression through TLR4 engagement affects mitochondrial respiration and dynamics through BIK and MTP18 silencing, respectively. Mitochondrial fusion induction and decrease of mitochondrial respiration promote apoptosis of monocytes. The figure was produced using Servier Medical Art.
Monocyte differentiation, polarization, and plasticity allow the tailoring of an immune response upon stimulation of a range of receptors, including TLRs.48 Changes in cell activation, cell fate, and cell functions are coupled to changes in cellular metabolism, and vice versa. As regulators of oxygen consumption and ATP production, mitochondria are instrumental to the metabolic adaptation of monocytes to environmental signals2,4 that drive effector functions such as migration, cytokine production, phagocytosis, antigen presentation, and polarization.49 Ligation of TLR4 induces metabolic changes characterized by a decrease in OXPHOS and an increase in glycolysis.50,51 This switch is observed when monocytes undergo differentiation into macrophages, and inhibition of this transition limits cell activation and life span. Profound metabolic reprogramming also occurs under macrophage polarizing conditions, with glycolysis being a hallmark of the inflamed state that occurs in M1 macrophages, and OXPHOS occurring in immunoregulatory M2 macrophages.44 M1 polarization uses anaerobic glycolysis to generate large quantities of ATP and to maintain a high mitochondrial membrane potential, despite the complete inhibition of respiration, allowing cells to adapt functions quickly and cope with the hypoxic tissue environment while providing a defense mechanism against cell death induced by endogenous reactive oxygen species. In contrast, M2 macrophages preferentially use OXPHOS metabolism to provide sustained ATP supply for tissue remodeling and repair and immunomodulation.52,53 The molecular events controlling the decrease in OXPHOS and survival response to LPS and to M1 polarization remain unclear. Here, we showed that one element controlling metabolic adaptation of monocytes/macrophages to these signals is miR-125b, through coordinated targeting of BIK and MTP18. Consistent with previous studies, we found that TLR4 ligation and M1 polarization reduced OXPHOS.44,50,54 We showed that response to TLR4 agonist is accompanied by a decrease in mitochondrial respiration, and that this effect is reversed when miR-125b is blocked. We also showed that miR-125b alone mimics metabolic adaptation of monocytes to TLR4 ligation, as evidenced by a decreased OCR. More specifically, miR-125b regulates the spare respiratory capacity, which represents the amount of extra ATP that can be produced by OXPHOS in the case of a sudden increase in energy demand, permitting rapid adaptation to metabolic changes. If the respiratory reserve is not sufficient to provide the cellular ATP levels required, cells are induced to apoptosis. This is consistent with our observation that miR-125b ectopic expression reduces cell proliferation and promotes apoptosis, whereas miR-125b blockade prevents cells from apoptosis. Although observed in a different cell context, miR-125b was recently reported to induce loss of mitochondrial membrane potential and sensitization to apoptosis in a human breast cancer cell line.20
In addition to identifying a miR-125b/BIK/MTP18 pathway controlling energy production and cellular survival in human monocytes, our work also substantially clarifies the nature of the complex cross talk between ER and mitochondria in human monocytes. The proapoptotic proteins BAK1, BMF, PUMA, and p53 and the anti-apoptotic proteins MCL-1, BCL-W, and BCL2 have been experimentally confirmed as cellular targets of miR-125b.18 Therefore, miR-125b can simultaneously modulate multiple genes playing opposite roles in apoptosis, acting as molecular hub that integrates various signals and takes different routes to modulate the mitochondrial apoptosis pathway. Our expression profiling evidenced that miR-125b redirects genetic programs of human monocytes toward metabolic cascades that promote apoptosis and inflammation, by modulating several genes implicated in mitochondrial functions. Here, we evidenced that, in human monocytes, miR-125b does not directly control BCL2, BAK1, and BMF expression, but promotes proinflammatory macrophage activation and apoptosis through direct regulation of BIK and MTP18 expression. Several miRNAs have been reported to regulate mitochondrial functions55 ; few of them are associated or localized in mitochondria. These so-called mitomiRs are different according to species and cell types, and their modulation is involved in inflammation and age-related diseases.56 Although miR-125b is considered a mitomiR in human skeletal muscular cells,57 we could not detect it in mitochondria isolated from human monocytes (data not shown).
BIK is an apoptosis-sensitizing protein anchored to the ER membrane33 that mediates release of ER Ca2+ and contributes to apoptosis through the mitochondria.40,41 Upon induction, BIK neutralizes prosurvival Bcl2 family members, relieving proapoptotic BAX and BAK proteins58 that are translocated to the mitochondrial membrane and induce mitochondrial fission, causing apoptotic cell death. BIK can also contribute to the regulation of autophagy by displacing BCL-2 from Beclin 1 and NAF-1.35,36 We showed that BIK silencing mimics miR-125b overexpression and TLR4 stimulation, decreasing the OCR and promoting cell death, with no impact on either mitochondrial network or autophagy. Our present work on human monocytes definitively reveals a totally new role for BIK in controlling mitochondrial respiration that fits with the reported role of BH3-only proteins in restraining inflammatory diseases.59 In addition, we observed an inverse correlation between low miR-125b expression levels and high mRNA levels of BIK and MTP18 in CD14+ blood monocytes from patients with RA, a systemic inflammatory condition in which CD14+ monocytes are abundant and abnormally activated in tissues and harbor a reduced propensity to undergo apoptosis.47
MTP18 is localized in the mitochondria and involved in shaping the mitochondrial network through the induction of fission.34 MTP18 loss-of-function promotes fusion of mitochondria, leading to increased susceptibility to apoptosis,42 as we observed in THP-1 cells using either siRNA against MTP18 or miR-125b mimics. Mitochondria fission facilitates the redistribution of mitochondria in response to local changes in the demand for ATP and for mitophagy, whereas mitochondria fusion promotes exchange of mitochondrial components to strengthen the mitochondrial network. When cells become committed to apoptosis, massive fragmentation of the mitochondrial network through fission induces the simultaneous release of cytochrome c from all mitochondria, promoting cell death. An alternative pathway termed stress-induced mitochondrial hyperfusion, however, promotes mitochondrial fusion to confer some resistance to low levels of stress but can also lead to apoptosis if further insults occur.60 Dynamic changes and balanced fusion/fission is crucial for cell survival, but how different forms of cellular stress converge on the mitochondria and are regulated remains unknown. Our data reveal a novel model regulating the mitochondrial network in monocytes, which is composed of miR-125b and MTP18.
MicroRNAs are fine-tuners of TLR signaling and some of them have been involved in macrophage plasticity and polarization.61,,,,-66 In THP-1, the expression of miR-125b is upregulated in macrophages, either after PMA-induced differentiation or M1 polarizing conditions, as well as in monocytes after LPS stimulation. Several reports showed that miR-125b is enriched in macrophages and is an LPS-regulated miRNA. Its expression is decreased shortly after LPS stimulation of macrophages27,67,68 but increased after sustained LPS exposure.25 Contrasting results from different studies show that miR-125b acts either as a negative regulator of the NF-κB pathway by stabilizing NKIRAS267 and repressing TNF-α24 or as a positive regulator that strengthens and prolongs NF-κB activity by targeting TNFAIP3.17 In mouse macrophages, miR-125b controls an activated phenotype through IRF4 silencing14 and regulates the differentiation/proliferation balance through Stat3 repression,13 2 genes that promote M2 polarization. In both THP-1 and primary CD14+ cells, we showed that the M1 phenotype was associated with decreased mitochondrial metabolism and increased mitochondrial fusion, as well as with repressed expression of BIK and MTP18, as observed when increasing miR-125b expression. The lack of autophagy induction upon miR-125b overexpression in human monocytes is consistent with early works showing that elongated mitochondria escape autophagy and that inhibition of autophagy switches macrophages from M2 to M1 polarization.44,45
Understanding how immune cells commit to particular microenvironments and metabolic fates and the immunologic consequences of reaching a metabolic end point is key. Here, we provide molecular clues for miR-125b involvement in the complex metabolic adaptation of human monocytes when activated by LPS and polarized toward the M1 phenotype. Our data suggest that miR-125b contributes to inflammation by controlling mitochondria integrity through BIK and MTP18 silencing in order to adapt oxygen consumption and mitochondrial dynamics of monocytes to environmental cues. These results identify important mechanistic connections between microRNAs and mitochondrial functions, with broad implications for cellular metabolism and adaptation to cellular stress, and deepen our understanding of the molecular events of human monocyte biology.
The chip data discussed in this article have been deposited in the Gene Expression Omnibus database (accession number GSE62693).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was funded by INSERM, University of Montpellier, and by grants from the ARTHRITIS Foundation COURTIN, the French Society of Rheumatology, UCB Pharma (Sirius 2010), and the European Community (AutoCure, contract No. LSHB-CT-2006-018661, and BeTheCure Innovative Medicines Initiative 2nd Call Topic 6, contract No. 115142).
Authorship
Contribution: I.D.-R. and F.A. designed the experiments, analyzed the data, and wrote the manuscript; I.D.-R. contributed to experiments in all figures; C.R., M.A., J.P., J.R.G., C.-H.L., T.H., P.C., V.B., J.E., C.J., Y.-M.P., and A.G. contributed to experiments and data analyses and helped to write the manuscript; and all authors read and approved the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Florence Apparailly, INSERM U 1183, Centre Hospitalier Universitaire Saint Eloi, 80 Rue Augustin Fliche, 34295 Montpellier cedex 5, France; e-mail: florence.apparailly@inserm.fr.