Key Points
RhoA-dependent differential activity of NMII isoforms determines its localization at the cleavage furrow of megakaryocytes.
Perturbation of the actin cytoskeleton was sufficient to induce the differential localization of NMII isoforms.
Megakaryocyte polyploidy is characterized by cytokinesis failure resulting from defects in contractile forces at the cleavage furrow. Although immature megakaryocytes express 2 nonmuscle myosin II isoforms (MYH9 [NMIIA] and MYH10 [NMIIB]), only NMIIB localizes at the cleavage furrow, and its subsequent absence contributes to polyploidy. In this study, we tried to understand why the abundant NMIIA does not localize at the furrow by focusing on the RhoA/ROCK pathway that has a low activity in polyploid megakaryocytes. We observed that under low RhoA activity, NMII isoforms presented different activity that determined their localization. Inhibition of RhoA/ROCK signaling abolished the localization of NMIIB, whereas constitutively active RhoA induced NMIIA at the cleavage furrow. Thus, although high RhoA activity favored the localization of both the isoforms, only NMIIB could localize at the furrow at low RhoA activity. This was further confirmed in erythroblasts that have a higher basal RhoA activity than megakaryocytes and express both NMIIA and NMIIB at the cleavage furrow. Decreased RhoA activity in erythroblasts abolished localization of NMIIA but not of NMIIB from the furrow. This differential localization was related to differences in actin turnover. Megakaryocytes had a higher actin turnover compared with erythroblasts. Strikingly, inhibition of actin polymerization was found to be sufficient to recapitulate the effects of inhibition of RhoA/ROCK pathway on NMII isoform localization; thus, cytokinesis failure in megakaryocytes is the consequence of both the absence of NMIIB and a low RhoA activity that impairs NMIIA localization at the cleavage furrow through increased actin turnover.
Introduction
Megakaryocytes are specialized cells of the bone marrow that naturally undergo polyploidization through endomitosis, a process characterized by defective cytokinesis.1 An absence of nonmuscle myosin II (NMII) and hence a defect in the cleavage furrow was observed during endomitosis.2 NMII is a constitutive component of the actomyosin ring formed at the cleavage furrow and provides the motor force required for cytokinesis and karyokinesis.3 A single unit of NMII consists of 2 heavy chains of NMII linked to 2 regulatory light chains that regulate NMII activity and 2 essential light chains that stabilize the heavy chain structure. The activity of NMII is regulated by the phosphorylation of key residues on the regulatory light chains that induces a conformational change in NMII molecules promoting their filament assembly and adenosine triphosphatase activity. NMII thus interacts with the actin filaments in an adenosine triphosphate–dependent manner to create tension and induce sliding of the actin filaments, thereby contributing to processes such as cytokinesis.4
In megakaryocytes, the activity of NMII correlated with ploidy and proplatelet formation. Blebbistatin, an inhibitor of NMII, increased both ploidy and proplatelet formation.2,5 Two isoforms of NMII—MYH9 (NMIIA) and MYH10 (NMIIB)—are expressed during megakaryocyte differentiation. The 2 isoforms are highly similar in their sequence, but vary considerably in their tail region; thus, although NMIIA and NMIIB can sometimes substitute for the absence of the other, they often have unique functions and subcellular localizations.6 The 2 isoforms also differ in their motor properties, with NMIIB showing a higher duty ratio when compared with NMIIA.7 During megakaryocyte maturation, the expression of NMIIA increases, whereas that of NMIIB decreases through RUNX1-mediated silencing of MYH10.2 Interestingly, NMIIB was the only isoform that could localize at the cleavage furrow of mitotic megakaryocytes.2 This localization was found to be dependent upon the C-terminal domain of NMIIB wherein a chimeric NMIIAhead-IIBtail could localize at the cleavage furrow.8 Given that both isoforms of NMII localize at the cleavage furrow of many cell types including erythroblasts,2 it remains unclear what leads to the selective abolition of NMIIA from the cleavage furrow during megakaryopoiesis.
In this study, we analyzed the effects of the RhoA/ROCK pathway on NMII isoform localization at the cleavage furrow/midzone of 4N megakaryocytes because it was demonstrated that RhoA activity is downregulated specifically at this stage.9 We found that the RhoA/ROCK pathway was a crucial determinant of NMII isoform activity and localization through regulation of the actin cytoskeleton.
Methods
In vitro culture of megakaryocytes and erythroblasts
Cord blood and leukapheresis samples were obtained after informed consent in accordance with the Declaration of Helsinki. Cord blood– and leukapheresis-derived CD34+ cells were isolated using immunomagnetic beads (AutoMacs; Miltenyi Biotec, Bergisch Gladbach, Germany) and cultured in vitro into erythroblast and megakaryocyte lineages, respectively. Details of culture conditions and treatments are provided in the supplemental Methods, available on the Blood Web site.
FRAP and immunofluorescence
Megakaryocytes and erythroblasts expressing GFP-NMIIA, GFP-NMIIB, and mCherry-LifeAct were used for fluorescence recovery after photobleaching (FRAP) experiments. Details of the experimental conditions are given in the supplemental Methods. For immunofluorescence, cells were plated on poly-l-lysine–coated slides (O. Kindler GmbH&Co, Freiburg, Germany) for 1 hour at 37°C. Immunofluorescence staining was performed using mouse anti-NMIIA (Sigma-Aldrich, France), rabbit anti-NMIIB antibodies (Cell Signaling Technology, Danvers, MA), and appropriate secondary antibodies conjugated with Alexa 488, Alexa 546, or Alexa 647 (Molecular Probes, Life Technology, Carlsbad, CA). A 4′,6-diamidino-2-phenylindole (DAPI; Molecular Probes, Life Technology) stain was applied for nuclear staining. Alexa 633-phalloidin (Molecular Probes, Life Technology) was used to stain filamentous actin. Cells were examined under Leica TCS SP8 MP (Leica Microsystemes SAS, Nanterre, France) with a 63× oil immersion objective under confocal mode. To measure the full width at half maxima (FWHM) of cortical actin, line scans were performed across the cell. Four diametric line scans were taken for each imaged cell. At least 17 cells were analyzed per sample. The resulting fluorescence intensities and the FWHM were obtained using Leica LAS X analysis software.
Cell fractionation and western blot analysis
Megakaryocytes and erythroblasts were fractionated according to a previously published protocol.10 Briefly, equal numbers of megakaryocytes/erythroblasts were pelleted for each condition in duplicate. Cells were lysed in cold radioimmunoprecipitation buffer (for whole-cell lysis) or Triton X-100–based fractionation buffer. After centrifugation according to the protocol,10 the supernatant was collected. Pellet fraction from Triton X-100 fractionation was resuspended in 2X Laemmli buffer. Western blots were performed as described previously.11 Anti-NMIIA and anti-NMIIB antibodies (Cell Signaling Technology) were used along with anti-HSC70 (Santa Cruz Biotechnologies Inc., Dallas, TX). RhoA-GTP assay was performed using the Rhotekin bead-based pulldown assay by following the manufacturer’s protocol (Cytoskeleton Inc., Denver, CO).
Statistical analysis
One-way analysis of variance testing was used to test the significance of the data.
Results
Differential activity of NMIIA and NMIIB in megakaryocytes under basal RhoA activity
Megakaryocytes were cultured in vitro from adult leukapheresis-derived CD34+ cells. The cultured megakaryocytes divided initially by mitosis until days 6 and 7 and thereafter switching to endomitosis. As a result, megakaryocytes were sorted on day 5 on the expression of CD41a and CD42; subsequent experiments were performed on days 6 or 7 of culture when they were mostly in 2N-4N ploidy state.
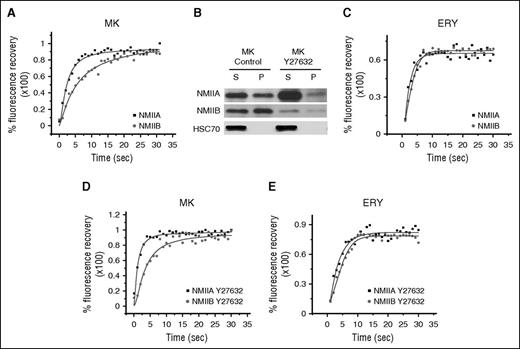
To address the selective abolition of NMIIA from the cleavage furrow of megakaryocytes, we hypothesized that the 2 isoforms (NMIIA and NMIIB) may have intrinsically different activity, contributing to different filament assembly properties and hence different mobility. Using FRAP, megakaryocytes expressing GFP-tagged NMIIA or NMIIB were studied to assess the mobility of the 2 isoforms. A faster and higher fluorescence recovery after photobleaching of NMIIA (half-life [t1/2], 2.10 ± 0.29 seconds; maximum percent recovery, 94 ± 3.6) was observed when compared with NMIIB (t1/2, 6.60 ± 1.41 seconds; maximum percent recovery, 88.0 ± 2.4; for t1/2P < .005) (Figure 1A; supplemental Figure 1). Further, an intrinsic difference in filament assembly and polymerization of NMIIA and NMIIB was observed by the analysis of Triton X-100 soluble and insoluble NMII complexes whereby a higher percentage of NMIIB was found in the pelleted insoluble fraction compared with NMIIA (Figure 1B; quantification in supplemental Figure 2). Together, the studies indicate that under basal RhoA activity, the 2 isoforms present different activity when NMIIB is the predominant active isoform in megakaryocytes. We also assessed the mobility of NMII isoforms in erythroblasts that are developmentally linked to megakaryocytes. In contrast to megakaryocytes however, erythroblasts express both NMIIA and NMIIB at the cleavage furrow and do not polyploidize; therefore, erythroblasts were used all along this study as control cells. We observed that NMIIA had a shorter recovery t1/2 (NMIIA t1/2, 4.01 ± 0.25; NMIIB t1/2, 5.59 ± 0.35; P < .001) without any significant change in the maximum percent recovery (NMIIA, 68.62 ± 0.57; NMIIB, 64.39 ± 1.08) when compared with NMIIB (Figure 1C; supplemental Figure 3). Importantly, compared with megakaryocytes, erythroblast NMII isoforms had reduced mobility as observed by decreased maximum percent recovery.
Differential activity of NMII isoforms. (A) Representative fluorescence recovery kinetics of NMIIA and NMIIB in CD41a+CD42+ megakaryocytes (MK) on day 7 of in vitro culture. (B) Western blot analysis of the expression of NMIIA and NMIIB in Triton X-100 fractionation-based supernatant (S) and pellet (P) fractions of in vitro cultured day 7 control megakaryocytes and megakaryocytes treated with Y27632 for 24 hours. Densitometric analysis is provided in supplemental Figure 2. (C) Representative fluorescence recovery kinetics of NMIIA-GFP and NMIIB-GFP in erythroblasts (ERY) on day 8 of culture. (D) Fluorescence recovery kinetics of NMIIA and NMIIB in CD41a+CD42+ megakaryocytes on day 7 of culture pretreated with Y27632 (ROCK inhibitor) for 24 hours. (E) Fluorescence recovery kinetics of NMIIA-GFP and NMIIB-GFP in erythroblasts on day 9 of culture after incubation in the presence of 10 μM Y27632 for 24 hours.
Differential activity of NMII isoforms. (A) Representative fluorescence recovery kinetics of NMIIA and NMIIB in CD41a+CD42+ megakaryocytes (MK) on day 7 of in vitro culture. (B) Western blot analysis of the expression of NMIIA and NMIIB in Triton X-100 fractionation-based supernatant (S) and pellet (P) fractions of in vitro cultured day 7 control megakaryocytes and megakaryocytes treated with Y27632 for 24 hours. Densitometric analysis is provided in supplemental Figure 2. (C) Representative fluorescence recovery kinetics of NMIIA-GFP and NMIIB-GFP in erythroblasts (ERY) on day 8 of culture. (D) Fluorescence recovery kinetics of NMIIA and NMIIB in CD41a+CD42+ megakaryocytes on day 7 of culture pretreated with Y27632 (ROCK inhibitor) for 24 hours. (E) Fluorescence recovery kinetics of NMIIA-GFP and NMIIB-GFP in erythroblasts on day 9 of culture after incubation in the presence of 10 μM Y27632 for 24 hours.
RhoA/ROCK pathway determines the activity of NMII isoforms
The RhoA/ROCK pathway directly contributes toward NMII activity by regulating phosphorylation of myosin light chain (MLC)12 ; furthermore, we observed that cultured erythroblasts had a higher basal RhoA activity when compared with megakaryocytes (supplemental Figure 4). We therefore analyzed whether the RhoA/ROCK pathway could differentially affect NMII isoform mobility and Triton X-100 solubility. Using FRAP, we observed that ROCK inhibition leads to an increased percentage of mobile fraction of both NMIIA and NMIIB (NMIIA, 98.0 ± 2.4; NMIIB, 98.0 ± 2.2) in megakaryocytes. Significantly, NMIIA still showed a shorter recovery time (t1/2, 0.96 ± 0.29 seconds) as compared with NMIIB (2.13 ± 0.86 seconds) (Figure 1D; supplemental Figure 5). Fractionation of megakaryocytes treated with ROCK inhibitor also revealed that both isoforms were present mostly in the supernatant fraction, indicating reduced filament assembly and hence activity (Figure 1B; quantification in supplemental Figure 2); thus, the activity of NMII isoforms was dependent on RhoA/ROCK pathway. We also tested the NMII isoform mobility in erythroblasts treated with ROCK inhibitor (Figure 1E; supplemental Figure 1). Treatment with Y27632 increased recovery of both the isoforms, with NMIIA showing a shorter recovery t1/2 as compared with NMIIB (NMIIA, 84.26 ± 2.14, t1/2, 4.81 ± 0.25; NMIIB, 74.90 ± 1.36, t1/2, 6.15 ± 0.46, for t1/2P < .019). Together, the results indicate that the activity of NMII isoforms is directly correlated with RhoA/ROCK activity.
RhoA/ROCK signaling determines the localization of NMII isoforms at the cleavage furrow
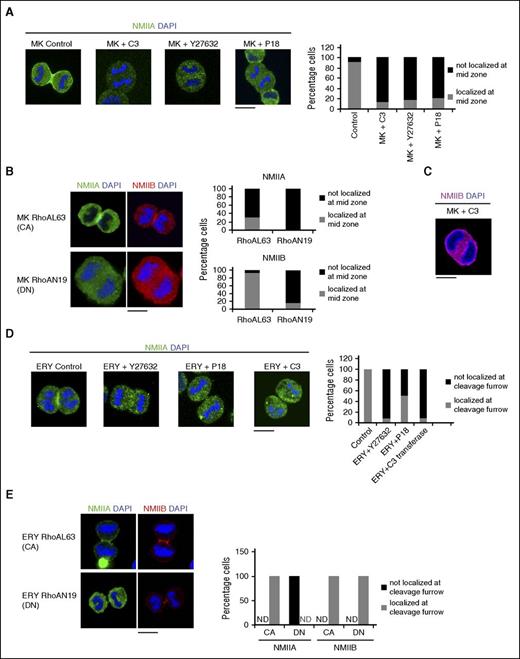
RhoA activity in mitotic cells is known to precisely localize the cytokinetic apparatus.13 Moreover, a specific decrease in RhoA activity at the cleavage furrow during megakaryocyte endomitosis was previously reported9 ; therefore, to understand whether RhoA/ROCK pathway–dependent changes in NMII isoform activity also translated to modifications in their localization at the cleavage furrow, we used specific inhibitors of RhoA/ROCK pathway including the MLC kinase inhibitor P18. As previously reported,2 control megakaryocytes showed distinct localization of NMIIB at the cleavage furrow with clear progression of mitosis (Figure 2A). Treatment of day 5 CD41a+CD42+ megakaryocytes with RhoA/ROCK inhibitors resulted in the absence of NMIIB from the midzone (Figure 2A). NMIIB was similarly abolished from the midzone of megakaryocytes expressing dominant negative RhoAN19 (Figure 2B). On the other hand, constitutively active RhoAL63 induced the expression of both the isoforms at the cleavage furrow of megakaryocytes (Figure 2B). Significantly, endomitosis was not completely hindered in megakaryocytes expressing constitutively active RhoAL63 as evidenced by binuclear megakaryocytes (supplemental Figure 7). Absence of NMIIB from the midzone of megakaryocytes also resulted in the formation of elongated or sometimes nearly spherical cells that failed to form the cleavage furrow. Furthermore, many elongated megakaryocytes were observed, with 2 distinct nuclei at the poles, indicating failed cytokinesis (Figure 2C). Remarkably, identical treatment of primary erythroblasts did not change the localization of NMIIB (supplemental Figure 8), but abolished the localization of NMIIA from their cleavage furrow (Figure 2D). This was further demonstrated by the expression of dominant negative RhoAN19 (Figure 2E). Finally, inhibition of Rac or Cdc42 had no effect on the localization of NMIIB at the megakaryocyte cleavage furrow (supplemental Figure 9); thus, RhoA/ROCK signaling determined the differential localization of NMII isoforms at the cleavage furrow of both megakaryocytes and erythroblasts with high RhoA/ROCK activity required for the localization of NMIIA.
RhoA-dependent NMII isoform localization. (A) Representative confocal microscopy images of in vitro cultured control and inhibitor-treated MKs sorted on day 5 of culture on the expression of CD41a+CD42+ followed by treatment with the indicated inhibitors: (from left) control, C3 transferase (RHO inhibitor), Y27632 (ROCK inhibitor), or P18 (MLC kinase inhibitor). Cells were stained for NMIIB and costained with DAPI to indicate the mitotic cells. At least 3 independent samples were analyzed and the number of mitotic cells was counted for NMII localization. The corresponding histogram plot indicates the percentage of cells with NMIIB localization at the cleavage furrow. (B) A representative confocal image of MKs expressing constitutively active (CA) RhoA (RhoAL63) or dominant negative (DN) RhoA (RhoAN19). Cells were fixed and stained for NMIIA, NMIIB, and DAPI. At least 3 independent samples were analyzed and the number of mitotic cells was counted for NMII localization. The corresponding histogram plot indicates the percentage of cells with NMIIA and NMIIB localization at the cleavage furrow. (C) Representative confocal microscopy image of in vitro cultured MKs treated with exozyme C3 transferase. Cells were stained for NMIIB and costained with DAPI to indicate the nucleus. (D) Representative confocal microscopy images of day 9 in vitro cultured control and inhibitor treated ERYs: (from left) control, Y27632, P18, and C3 transferase. Cells were stained for NMIIA and costained with DAPI to indicate the mitotic cells. At least 3 independent samples were analyzed and the number of mitotic cells was counted for NMIIA localization. The corresponding histogram plot indicates the percentage of cells with NMIIA localizing at the cleavage furrow. (E) A representative confocal image of erythroblasts expressing CA RhoA (RhoAL63) or DN RhoA (RhoAN19). Cells were fixed and stained for NMIIA, NMIIB, and DAPI. At least 3 independent samples were analyzed and the number of mitotic cells was counted for NMII localization. The corresponding histogram plot indicates the percentage of cells with NMIIA and NMIIB localization at the cleavage furrow. ND, not detected. All bars represent 10 μm.
RhoA-dependent NMII isoform localization. (A) Representative confocal microscopy images of in vitro cultured control and inhibitor-treated MKs sorted on day 5 of culture on the expression of CD41a+CD42+ followed by treatment with the indicated inhibitors: (from left) control, C3 transferase (RHO inhibitor), Y27632 (ROCK inhibitor), or P18 (MLC kinase inhibitor). Cells were stained for NMIIB and costained with DAPI to indicate the mitotic cells. At least 3 independent samples were analyzed and the number of mitotic cells was counted for NMII localization. The corresponding histogram plot indicates the percentage of cells with NMIIB localization at the cleavage furrow. (B) A representative confocal image of MKs expressing constitutively active (CA) RhoA (RhoAL63) or dominant negative (DN) RhoA (RhoAN19). Cells were fixed and stained for NMIIA, NMIIB, and DAPI. At least 3 independent samples were analyzed and the number of mitotic cells was counted for NMII localization. The corresponding histogram plot indicates the percentage of cells with NMIIA and NMIIB localization at the cleavage furrow. (C) Representative confocal microscopy image of in vitro cultured MKs treated with exozyme C3 transferase. Cells were stained for NMIIB and costained with DAPI to indicate the nucleus. (D) Representative confocal microscopy images of day 9 in vitro cultured control and inhibitor treated ERYs: (from left) control, Y27632, P18, and C3 transferase. Cells were stained for NMIIA and costained with DAPI to indicate the mitotic cells. At least 3 independent samples were analyzed and the number of mitotic cells was counted for NMIIA localization. The corresponding histogram plot indicates the percentage of cells with NMIIA localizing at the cleavage furrow. (E) A representative confocal image of erythroblasts expressing CA RhoA (RhoAL63) or DN RhoA (RhoAN19). Cells were fixed and stained for NMIIA, NMIIB, and DAPI. At least 3 independent samples were analyzed and the number of mitotic cells was counted for NMII localization. The corresponding histogram plot indicates the percentage of cells with NMIIA and NMIIB localization at the cleavage furrow. ND, not detected. All bars represent 10 μm.
Differential localization of NMII isoforms cannot be explained by MLC phosphorylation levels
RhoA/ROCK pathway induces MLC phosphorylation and hence NMII activation.12 By inducing both NMII activation and actin polymerization, RhoA/ROCK signaling leads to assembly and constriction of the actomyosin ring at the cleavage furrow. Accordingly, RhoA/ROCK inhibitors decreased MLC phosphorylation (supplemental Figure 10A). Although, MLC2 phosphorylation increased during megakaryopoiesis11 and phosphorylated MLC2 is recruited at the cleavage furrow of mitotic megakaryocytes, NMIIA was unable to localize at the cleavage furrow (supplemental Figure 10B-C). To go further, we expressed the double phospho-mimetic MLC-DD in megakaryocytes, but it could not recruit NMIIA at the cleavage furrow (supplemental Figure 10D); thus, the differential localization of NMII isoforms cannot be explained by a decreased level of MLC2 phosphorylation induced by the low RhoA/ROCK activity in megakaryocytes. As a result, other mechanisms must exist that correlate RhoA/ROCK activity with the localization of NMII isoforms.
RhoA/ROCK signaling determines actin turnover in megakaryocytes and erythroblasts
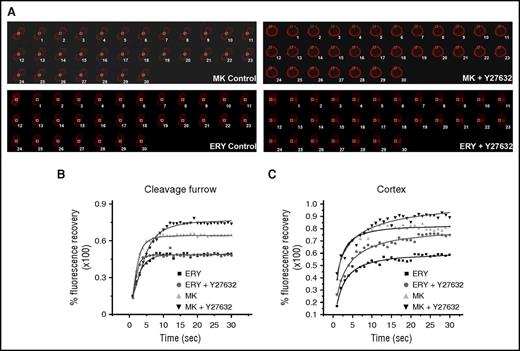
To understand how RhoA/ROCK signaling differentially regulated NMII isoforms, we analyzed the actin cytoskeleton at the cleavage furrow in megakaryocytes and erythroblasts. Using mCherry-tagged LifeAct, which allowed us to monitor changes in actin cytoskeleton, we analyzed the fluorescence recovery of mCherry upon photobleaching specifically within the cleavage furrow of megakaryocytes and erythroblasts (Figure 3A-B). Our results show that erythroblasts have a lower percent recovery (49.037 ± 0.037; t1/2, 6.48 ± 0.02 seconds) as compared with megakaryocytes (61.79 ± 2.52; t1/2, 4.14 ± 0.28 seconds). Treatment of erythroblasts with ROCK inhibitor decreased the recovery t1/2 without significantly altering the maximum percent recovery of fluorescence (48.19 ± 1.16; t1/2, 4.25 ± 0.12 seconds, for t1/2P < .001). On the other hand, megakaryocytes treated with ROCK inhibitor showed loss of NMIIB from the cleavage furrow and consequently increased dynamic actin at the midzone (maximum recovery, 73.57 ± 2.48; t1/2, 5.11 ± 0.7). Because the effect of ρ/ROCK inhibition on actin cytoskeleton is not limited only to the cleavage furrow, we also explored the actin turnover at the cell cortex (Figure 3C; supplemental Figures 11-12). A prominent cortical actin bundle was observed in both erythroblasts and megakaryocytes. Megakaryocytes showed increased fluorescence recovery percent (maximum recovery, 80.15 ± 1.91; t1/2, 5.43 ± 0.17 seconds) when compared with erythroblasts (maximum percent recovery, 60.39 ± 1.29; t1/2, 5.13 ± 0.21 seconds). Further, treatment with a ROCK inhibitor increased the recovery in both cell types (megakaryocyte maximum percent recovery, 90.39 ± 0.40, t1/2, 5.49 ± 0.34 seconds; erythroblast maximum recovery, 75.15 ± 1.73%, t1/2, 4.97 ± 0.16 seconds). However, megakaryocytes still showed a faster and higher recovery when compared with erythroblasts (Figure 3C). Together, these results indicate that megakaryocytes have a higher actin turnover when compared with erythroblasts. It also indicates that the ρ/ROCK pathway affects actin turnover both at the cleavage furrow/midzone as well as at the cortex of megakaryocytes and erythroblasts.
RhoA-dependent actin turnover in megakaryocytes and erythroblasts. MKs and ERYs were transduced with lentiviral vector encoding mCherry-tagged LifeAct on days 3 and 4 of in vitro culture, respectively. MKs were sorted on day 5 on the expression of CD41a CD42 and mCherry. ERYs were sorted on mCherry expression on day 7 of culture. (A) MKs and ERYs either control or pretreated for 24 hours with Y27632 were assessed by FRAP. MKs were analyzed on day 6 of culture and ERYs on day 8 of culture under a confocal microscope for the fluorescence recovery of mCherry at the cleavage furrow/midzone. Representative montage images of fluorescence recovery are shown. A white box marks the bleached area. (B) At least 2 FRAP curves were obtained from each cell analyzed. At least 5 individual cells were analyzed and the resulting FRAP curves were fitted. An example of the FRAP recovery curve is presented. (C) Fluorescence recovery kinetics of mCherry-tagged LifeAct in control and Y27632-treated MKs and ERYs at the cell cortex. A representative FRAP recovery kinetics is shown here.
RhoA-dependent actin turnover in megakaryocytes and erythroblasts. MKs and ERYs were transduced with lentiviral vector encoding mCherry-tagged LifeAct on days 3 and 4 of in vitro culture, respectively. MKs were sorted on day 5 on the expression of CD41a CD42 and mCherry. ERYs were sorted on mCherry expression on day 7 of culture. (A) MKs and ERYs either control or pretreated for 24 hours with Y27632 were assessed by FRAP. MKs were analyzed on day 6 of culture and ERYs on day 8 of culture under a confocal microscope for the fluorescence recovery of mCherry at the cleavage furrow/midzone. Representative montage images of fluorescence recovery are shown. A white box marks the bleached area. (B) At least 2 FRAP curves were obtained from each cell analyzed. At least 5 individual cells were analyzed and the resulting FRAP curves were fitted. An example of the FRAP recovery curve is presented. (C) Fluorescence recovery kinetics of mCherry-tagged LifeAct in control and Y27632-treated MKs and ERYs at the cell cortex. A representative FRAP recovery kinetics is shown here.
RhoA/ROCK signaling determines the cortical localization of NMII isoforms
Because RhoA/ROCK activity was found to alter the actin turnover at the cell cortex, we next analyzed whether RhoA/ROCK activity may also differentially affect cortical localization of NMII isoforms. We stained megakaryocytes and erythroblasts with fluorescent-tagged phalloidin or anti-NMIIA or anti-NMIIB antibodies (Figure 4A). Analysis of the thickness of the cortical actin bundle by measuring the FWHM of the fluorescence intensity of tagged phalloidin at the cell cortex revealed a thicker cortical actin in erythroblasts when compared with megakaryocytes (Figure 4A-B). Inhibition of the RhoA/ROCK pathway decreased the thickness of the cortical actin in erythroblasts and megakaryocytes (Figure 4A-B). Both megakaryocytes and erythroblasts showed prominent cortical NMIIB localization. No significant change in the thickness of cortical NMIIB (by measuring FWHM of the cortical NMIIB signal) was observed between the 2 cell types (megakaryocyte NMIIB FWHM, 280.5 ± 8.6 nm; erythroblast NMIIB FWHM, 293.8 ± 9.5 nm). Treatment with ROCK inhibitor caused a significant decrease in the thickness of cortical NMIIB in erythroblasts (erythroblast NMIIB FWHM, 293.8 ± 9.5 nm; erythroblast+Y27632 NMIIB FWHM, 260 ± 8.4 nm; P < .05). Interestingly, ROCK inhibition in megakaryocytes abolished cortical NMIIB in most cells. Cortical NMIIA was detected in erythroblasts, but we failed to detect a continuous cortical NMIIA in megakaryocytes. Further, ROCK inhibition completely abolished NMIIA from the cell cortex of erythroblasts (Figure 4A). Strikingly, expression of RhoAL63 but not MLCDD could restore the localization of NMIIA at megakaryocyte cortex (supplemental Figure 13); thus, NMIIA localization at the cell cortex as well as at the cleavage furrow requires high RhoA/ROCK activity. On the other hand, NMIIB could localize even at low RhoA/ROCK activity; therefore, a robust decrease in RhoA/ROCK activity was required to manifest a change in the localization of NMIIB. Taken together, our experiments clearly indicate a correlation between RhoA/ROCK activity, actin turnover, and differential localization of NMII isoforms. It further raises the possibility that the actin cytoskeleton may be the crucial mediator that differentially relays the effects of RhoA/ROCK signaling to the 2 NMII isoforms.
RhoA-dependent cortical actin and NMII isoform localization. (A) Representative confocal microscopy images of in vitro cultured control MKs and ERYs and those treated with Y27632. Cells were stained for actin (phalloidin), NMIIA, or NMIIB and their cortical localization was observed. Enlarged views of the areas within the white boxes in each panel are also shown (insets). In 3 independent samples, Y27632 decreased the localization of NMIIB at the cortex in erythroblasts. At least 4 measurements were taken in each cell and at least 17 cells per sample were assessed. (B) A representative image of cortical actin cytoskeleton (phalloidin-stained) in MKs and the fluorescence intensity across the cell obtained by a line scan. The cortical actin intensity is further analyzed to obtain the FWHM, which is indicated by a black line on the fluorescence intensity plot. The corresponding histogram plot represents the FWHM of cortical actin in control and Y27632-treated megakaryocytes and erythroblasts. The data represent the mean ± standard error of the mean of each condition (P < .01). At least 4 measurements were taken in each cell and at least 17 cells per sample per slide were assessed.
RhoA-dependent cortical actin and NMII isoform localization. (A) Representative confocal microscopy images of in vitro cultured control MKs and ERYs and those treated with Y27632. Cells were stained for actin (phalloidin), NMIIA, or NMIIB and their cortical localization was observed. Enlarged views of the areas within the white boxes in each panel are also shown (insets). In 3 independent samples, Y27632 decreased the localization of NMIIB at the cortex in erythroblasts. At least 4 measurements were taken in each cell and at least 17 cells per sample were assessed. (B) A representative image of cortical actin cytoskeleton (phalloidin-stained) in MKs and the fluorescence intensity across the cell obtained by a line scan. The cortical actin intensity is further analyzed to obtain the FWHM, which is indicated by a black line on the fluorescence intensity plot. The corresponding histogram plot represents the FWHM of cortical actin in control and Y27632-treated megakaryocytes and erythroblasts. The data represent the mean ± standard error of the mean of each condition (P < .01). At least 4 measurements were taken in each cell and at least 17 cells per sample per slide were assessed.
Perturbation of actin polymerization is sufficient for the differential localization of NMII isoforms
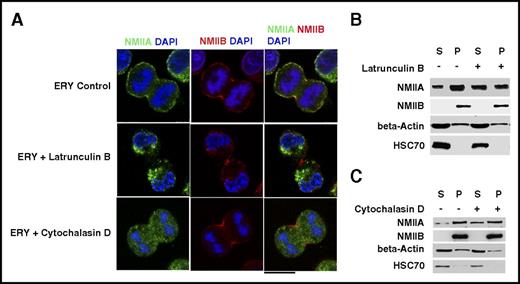
Because RhoA/ROCK pathway primarily acts on the actin cytoskeleton network and because the 2 isoforms are known to have significantly different duty ratio (a measure of actin binding),7 we hypothesized that changes in the actin cytoskeleton may be differentially sensed by the NMII isoforms. To support this hypothesis, we studies erythroblasts that express both isoforms at the cleavage furrow. Erythroblasts were treated with 40 nM latrunculin B or 20 nM cytochalasin D (actin depolymerizing agents) for 30 minutes and fixed and stained for NMII isoforms. At these conditions, total F-actin content decreased in erythroblasts while keeping its localization at the cleavage furrow unchanged (data not shown). We observed that nearly 40% of mitotic cells treated with latrunculin B and 70% of cells treated with cytochalasin D did not show the presence of NMIIA at the cleavage furrow (Figure 5A). Additionally, we observed many mitotic cells that showed prominent NMIIA staining that failed to localize to the cytokinetic ring; however, NMIIB continued to be expressed at the cleavage furrow (supplemental Videos 1-2). As a result, perturbation of the actin cytoskeleton could induce the differential localization of the 2 NMII isoforms in erythroblasts. Similar to erythroblasts, identical treatment of megakaryocytes did not cause any change in the localization of NMIIB (supplemental Figure 14). Finally, we studied the Triton X-100 solubility of NMII isoforms in day 10 erythroblasts with or without treatment with cytochalasin D or latrunculin B. Treatment with cytochalasin D or latrunculin B for 30 minutes increased the soluble fraction of NMIIA (Figure 5 B-C; quantification in supplemental Figure 15). No such change was observed for NMIIB; thus, our results indicate that changes in the actin cytoskeleton are sufficient to induce the differential localization of NMII isoforms at the cleavage furrow of erythroblasts.
Actin network and NMII isoform localization. (A) Representative confocal microscopy images of in vitro–cultured day 9 control ERYs and ERYs treated with the indicated inhibitors. Cells were costained for NMIIA and NMIIB. DAPI was used to indicate the mitoticcells. Bar represents 10 μm. (B,C) Representative western blot of day 10 ERYs treated with/without cytochalasin D and latrunculin B. Cells were subjected to Triton X-100 fractionation and the soluble S and insoluble P fractions were probed for NMIIA, NMIIB, and β-actin. HSC70 was used as a loading control. The densitometric analysis of the blots is provided in supplemental Figure 16.
Actin network and NMII isoform localization. (A) Representative confocal microscopy images of in vitro–cultured day 9 control ERYs and ERYs treated with the indicated inhibitors. Cells were costained for NMIIA and NMIIB. DAPI was used to indicate the mitoticcells. Bar represents 10 μm. (B,C) Representative western blot of day 10 ERYs treated with/without cytochalasin D and latrunculin B. Cells were subjected to Triton X-100 fractionation and the soluble S and insoluble P fractions were probed for NMIIA, NMIIB, and β-actin. HSC70 was used as a loading control. The densitometric analysis of the blots is provided in supplemental Figure 16.
Discussion
Megakaryocyte endomitosis corresponds to a failure of cytokinesis resulting from the absence of NMII accumulation at the cleavage furrow/midzone.1 We have previously demonstrated that RUNX1-mediated silencing of MYH10 plays a major role in the switch from mitosis to endomitosis.2 However, it remained unknown why the other highly abundant isoform (NMIIA) could not localize to the megakaryocyte cleavage furrow,2 in contrast to many other cell types, including T cells and erythroblasts.2,14
The formation of myosin II filaments is dependent upon the phosphorylation of its regulatory domain. This process requires a conformational change that stretches the globular myosin II structure into an elongated conformation that, at physiological conditions, forms multimeric filaments.4 We hypothesized that the differences in the localization of the 2 NMII isoforms were related to their basal activity. Using the Triton X-100 solubility assay and the FRAP kinetics of NMII isoforms as indirect methods to determine their relative activities, we observed that NMIIB was the predominant active isoform in megakaryocytes. This result was in agreement with a previous report showing that NMIIA was inactive during megakaryocyte differentiation and activated only during pre-/proplatelet fission.15 Importantly, differential activity of NMII isoforms was also detected in erythroblasts. Differences in basal NMII isoform activity were also evident in migrating fibroblasts where NMII isoform activity correlated with their localization as well as RhoA/ROCK activity.16 We observed that inhibition of RhoA/ROCK pathway abolished the localization of NMIIB from the midzone of megakaryocytes, whereas expression of a constitutively active RhoAL63 induced localization of NMIIA at the cleavage furrow. It is noteworthy that inhibition of the RhoA/ROCK pathway using the same set of inhibitors was previously reported to increase the ploidy of megakaryocytes.1,17 A similar inhibition in erythroblasts abolished the localization of NMIIA, but not of NMIIB at the cleavage furrow. Additionally, inhibition of Rac or Cdc42 had no effect on the localization of NMIIB at the megakaryocyte cleavage furrow. That RhoA and ROCK inhibition had similar effects suggests that ROCK is the main RhoA effector in determining NMII isoform localization and the consequent ploidization; however, this does not exclude that other RhoA effectors such as DIAPH1, DIAPH2, and DIAPH3 may also be involved in the regulation of ploidization.18 Interestingly, a recent report in mice demonstrated that RhoA knockout in erythroblasts induces a cytokinesis failure and polyploidization.19
The RhoA/ROCK pathway affects the activity of NMII isoforms by regulating MLC phosphorylation.20,21 However, the differential localization of NMII isoforms could not be explained by RhoA/ROCK-dependent changes in MLC phosphorylation. More particularly, the double phospho-mimetic MLC-DD could not recruit NMIIA at the cleavage furrow of megakaryocytes. We therefore investigated other pathways implicated in RhoA/ROCK signaling that may also affect NMII localization. The actin cytoskeleton being the most important effector of the RhoA/ROCK pathway,22 we investigated actin dynamics at the cleavage furrow of megakaryocytes and erythroblasts. Increased actin turnover was observed at the megakaryocyte cleavage furrow when compared with erythroblasts. Further, ROCK inhibitor increased actin turnover at the midzone of megakaryocytes, which correlated with the absence of NMIIB at the midzone. We also investigated the localization of actin and NMII isoforms at the cell cortex of megakaryocytes and erythroblasts and observed a correlation between ROCK activity and the thickness of the cortical actin and cortical localization of NMII isoforms. Whereas in erythroblasts both NMIIA and NMIIB localized at the cell cortex, only NMIIB was observed to have a distinct cortical localization in megakaryocytes. NMIIB could be abolished by treatment with ROCK inhibitor, recapitulating the effects seen at the cleavage furrow of megakaryocytes. Moreover, cortical localization of NMIIA was abolished in erythroblasts treated with ROCK inhibitor, an effect already seen at the erythroblast cleavage furrow. Interestingly, a previous report also showed that NMIIA localization at the cell cortex of mitotic round cells is abolished by treatment with RhoA/ROCK inhibitors.23
Given the correlation observed among RhoA/ROCK activity, actin dynamics, and differential localization of NMII isoforms, we hypothesized that the actin cytoskeleton may be involved in the differential localization of the NMII isoforms. Such a possibility was also raised in a recent report in which in silico simulation of actomyosin complexes on an elastic matrix suggested that NMIIA could efficiently generate force only at higher matrix stiffness.24 Furthermore, inhibition of actin polymerization by cytochalasin B was reported to increase the ploidy of several megakaryocytic cell lines, which was also suggested to be related to a defect in the cleavage furrow25,26 ; therefore, we tested whether perturbation of the actin cytoskeleton was sufficient to induce the differential localization of the NMII isoforms using conventional actin filament destabilizing agents. Strikingly, erythroblasts treated with these agents showed an absence of NMIIA but not NMIIB from the cleavage furrow of mitotic cells. Our study thus implicates the actin cytoskeleton as a mediator of RhoA/ROCK-dependent changes in NMII isoform-specific localization.
It was previously shown that the transition from mitotic division to endomitosis is associated with a decrease in RhoA activity at the midzone of megakaryocytes because of a downregulation of the Rho-GEFs GEF-H1 and ECT2.9 Our results underscore that this decrease leads to the absence of NMIIA from the cleavage furrow; thus, the mechanism of cytokinesis failure in megakaryocytes is due to both the silencing of MYH10 gene and the absence of NMIIA accumulation in the cleavage furrow because of low RhoA activity. Importantly, the results have been obtained using megakaryocytes derived from adult CD34+ cells; it remains unknown if a different regulation of NMIIA and NMIIB accumulation at the cleavage furrow explains the lower ploidy of fetal and neonatal megakaryocytes. Together, the combination of these 2 mechanisms explains the switch from mitosis to endomitosis.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Anthony Means and Robert Adelstein for providing GFP tagged NMII isoform constructs. The authors also thank the Cytometry and Imaging platform of Gustave Roussy.
This work was supported by grants from the Ligue Nationale Contre le Cancer (“Equipe labellisée HR 2013 and 2016”) (A.R., N.D., W.V., and H.R.) and INSERM, Fondation de la Recherche médicale (SPF20140129106) (A.R.), and the Laboratory of Excellence Globule Rouge-Excellence “Investissements d’avenir” (W.V.).
Authorship
Contribution: A.R., H.R., and W.V. conceived and designed the study, interpreted the data, and wrote the paper. A.R. designed and performed experiments of cell and molecular biology. L.L. and S.M. performed experiments of cell and molecular biology. N.D. and Y.C. analyzed the data and provided essential reagents. V.L. and J.L. provided essential reagents.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: William Vainchenker, INSERM UMR1170, Gustave Roussy, Université Paris-Saclay, 114 rue Edouard Vaillant, 94805 Villejuif, France; e-mail: william.vainchenker@gustaveroussy.fr.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal