Key Points
The expression level of ROR1 on CLL cells varies between patients.
High-level CLL-cell expression of ROR1 associates with more aggressive disease.
Abstract
ROR1 is an oncoembryonic orphan receptor found on chronic lymphocytic leukemia (CLL) B cells, but not on normal postpartum tissues. ROR1 is a receptor for Wnt5a that may complex with TCL1, a coactivator of AKT that is able to promote development of CLL. We found the CLL cells of a few patients expressed negligible ROR1 (ROR1Neg), but expressed TCL1A at levels comparable to those of samples that expressed ROR1 (ROR1Pos). Transcriptome analyses revealed that ROR1Neg cases generally could be distinguished from those that were ROR1Pos in unsupervised gene-expression clustering analysis. Gene-set enrichment analyses demonstrated that ROR1Neg CLL had lower expression and activation of AKT signaling pathways relative to ROR1Pos CLL, similar to what was noted for leukemia that respectively developed in TCL1 vs ROR1xTCL1 transgenic mice. In contrast to its effect on ROR1Pos CLL, Wnt5a did not enhance the proliferation, chemotaxis, or survival of ROR1Neg CLL. We examined the CLL cells from 1568 patients, which we randomly assigned to a training or validation set of 797 or 771 cases, respectively. Using recursive partitioning, we defined a threshold for ROR1 surface expression that could segregate samples of the training set into ROR1-Hi vs ROR1-Lo subgroups that differed significantly in their median treatment-free survival (TFS). Using this threshold, we found that ROR1-Hi cases had a significantly shorter median TFS and overall survival than ROR1-Lo cases in the validation set. These data demonstrate that expression of ROR1 may promote leukemia-cell activation and survival and enhance disease progression in patients with CLL.
Introduction
ROR1 is a developmentally restricted, type I tyrosine kinase–like orphan receptor expressed on the neoplastic B cells of patients with chronic lymphocytic leukemia (CLL), but not on the presumed normal counterpart to CLL, the CD5 B cell.1-3 ROR1 is a receptor for Wnt5a,1 which recent studies show can promote CLL cell survival, proliferation, and migration in a ROR1-dependent manner.4 On the other hand, reducing ROR1 expression on CLL cells via small interfering RNA can decrease leukemia-cell survival.5
Studies indicate that ROR1 can accelerate the development and progression of leukemia in mouse models of human CLL. ROR1 may complex with a known coactivator of AKT, namely, TCL1, which, when expressed under the control of an immunoglobulin promoter/enhancer, promotes development of leukemia similar to that of patients with CLL in TCL1 transgenic mice.6 However, in contrast to human CLL, the leukemia that develops in TCL1 transgenic mice does not express ROR1.7 On the other hand, mice that have both human TCL1 and ROR1 transgenes under the control of an immunoglobulin promoter/enhancer develop leukemia that is ROR1-positive. Such double transgenic mice develop leukemia at a significantly younger median age and have a shorter median survival due to aggressive leukemia than do otherwise syngeneic TCL1 transgenic mice.7 Furthermore, the ROR1-positive leukemia that develops in ROR1xTCL1 transgenic mice have higher-level expression of pathways implicated in embryonic and/or tumor-cell proliferation, but lower expression of pathways involved in cell-cell adhesion or cell death, than the ROR1-negative leukemia that develops in TCL1 mice. ROR1xTCL1 leukemia cells also had higher levels of phospho-AKT, higher proportions of Ki-67–positive cells, lower proportions of cells undergoing spontaneous apoptosis, and produced more aggressive disease upon adoptive transfer than TCL1 leukemia cells that lacked ROR1. Collectively, these studies demonstrated that ROR1 could accelerate the development and progression of leukemia in this transgenic mouse model.
Nevertheless, prior studies found ROR1 expressed by virtually all cases of CLL, independent of established prognostic markers, such as the mutation status of immunoglobulin heavy-chain variable region genes (IGHV) or the expression of ZAP-70.1-3 However, we reinterrogated the published database of the Microarray Innovations in Leukemia study and noted that a few of the 448 cases examined in this study expressed low to negligible amounts of ROR1.8 Such cases expressed the gene encoding TCL1, TCL1A, at levels comparable to those observed for CLL samples that expressed high levels of ROR1 (supplemental Figure 1, available on the Blood Web site). We hypothesized that low-level leukemia-cell expression of ROR1 might be associated with more indolent disease.
In the present study, we examined the transcriptomes of 12 CLL cases with low to negligible ROR1 (designated as ROR1Neg) and 12 CLL cases that expressed levels of ROR1 more commonly found on CLL (designated as ROR1Pos). We then examined the relationship between leukemia-cell expression of ROR1 and clinical progression in 1568 patients followed by clinical investigators in the CLL Research Consortium (CRC).
Materials and methods
Sample collection and preparation
This study was conducted in accordance with the Declaration of Helsinki for the protection of human subjects. Blood samples were collected from consenting patients who satisfied diagnostic criteria for CLL and who enrolled in the CRC Tissue Core. Blood samples were obtained from CLL patients examined at CRC sites: UC San Diego Moores Cancer Center (n = 353), MD Anderson Cancer Center (n = 382), North Shore-Long Island Jewish Cancer Institute (n = 264), Mayo Clinic (n = 192), Dana-Farber Cancer Institute (n = 166), The Ohio State University (n = 155), and Barts Cancer Institute (n = 56). At the time of sample collection, 1331 patients (85%) had not received prior therapy; 237 patients (15%) had received prior therapy; 163 patients (10%) had samples obtained at diagnosis, and the median time between diagnosis and sample collection (delayed entry time) was 1.3 years (the lower quartile time and upper quartile time are 0.2 years and 4.8 years, respectively). We used Ficoll-Hypaque density-gradient centrifugation to isolate mononuclear cells, which were cryopreserved in 10% dimethyl sulfoxide for storage in liquid nitrogen until analysis. Samples with >90% CD19+CD5+ CLL cells were used without further purification throughout this study. IGHV mutation status was assessed as per established criteria.9
Patient cohorts and ROR1 expression
Cohort 1: Previously published gene expression data from Microarray Innovations in Leukemia study datasets (GSE 13204) of 448 CLL patients deposited in PubMed GEO database were downloaded to analyze for expression of ROR1 and TCL1A8 ; Cohort 2: Next-generation sequencing on the transcriptome of 12 patient samples that had low to negligible expression of ROR1 (designated as ROR1Neg) and 12 samples that expressed ROR1 (designated as ROR1Pos). We used flow cytometry to determine the “ROR1 ΔMFI,” which is the mean fluorescence intensity (MFI) of CD19+ cells stained with an Alexa-647–conjugated anti-ROR1 monoclonal antibody (mAb; 4A5) minus the MFI of the CD19+ cells stained with an Alexa-647-conjugated nonspecific mAb of the same isotype; Cohort 3: We examined for expression of ROR1 on CLL cells in serial samples collected at different times from 11 untreated patients and 10 patients who had received therapy for CLL. The median time that lapsed between the first and last sample collected for patients in this longitudinal survey was 8.3 years; Cohort 4: 1568 patient samples from CRC Tissue Core were randomly segregated into either of 2 cohorts: a training set of 797 and a validation set of 771 cases. Training and validation cohorts could be segregated into 2 disparate subgroups based upon whether the CLL cells had a ROR1 ΔMFI less than optimal threshold 32 (“ROR1-Lo”) or greater than or equal to optimal threshold 32 (“ROR1-Hi”).
Descriptions of flow cytometry analyses, library preparation, sequencing, gene differential expression, unsupervised clustering, subnetwork analyses, gene set enrichment analysis, immunoblot analysis, viability, proliferation and migration assays, and statistical analyses are provided in the supplemental Methods.
Results
Transcriptome analyses of ROR1Neg and ROR1Pos CLL
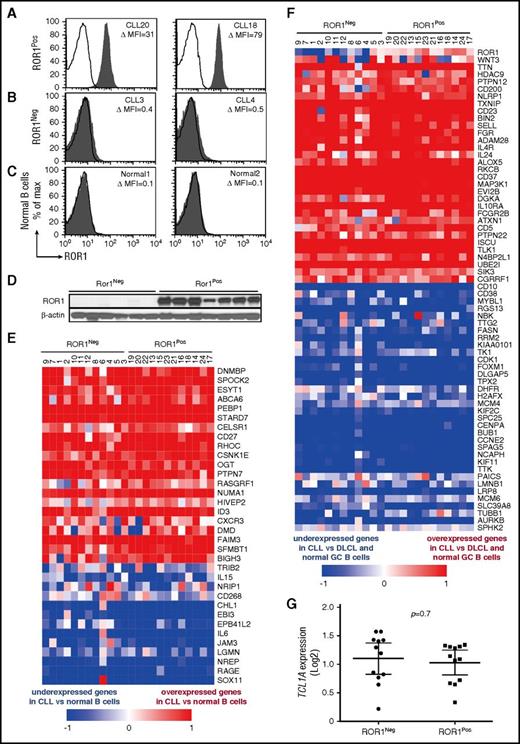
We examined the leukemia cells of 12 ROR1Neg cases with low to negligible ROR1 and 12 ROR1Pos cases, which expressed levels of ROR1 comparable with that commonly found in CLL (Figure 1A-C). Six of the 12 ROR1Neg cases used unmutated immunoglobulin heavy-chain variable region genes (U-IGHV), whereas 8 of the 12 ROR1Pos cases used U-IGHV (supplemental Table 1). The average ROR1 ΔMFI for the ROR1Neg samples was 2 ± 2 (± SD), whereas the average ROR1 ΔMFI for the ROR1Pos samples was 44 ± 15. For comparison, the average ROR1 ΔMFI for representative normal blood B cells of adults was 1.0 ± 1.1, n = 14. Immunoblot analyses of lysates from representative cases demonstrated the ROR1Neg samples had negligible ROR1, in contrast to the ROR1Pos cases, indicating that the CLL cells of ROR1Neg cases also lacked detectable cytoplasmic ROR1 (Figure 1D).
Expression of CLL signature genes by ROR1Negor ROR1PosCLL cells. Representative histograms depicting the fluorescence of CD19+ cells labeled with Alexa-647–conjugated 4A5 (anti-ROR1 mAb) (shaded histograms) or Alexa-647-conjugated nonspecific immunoglobulin G (IgG) of the same isotype (open histograms) for (A) ROR1Pos CLL B cells, (B) ROR1Neg CLL cells, or (C) representative normal blood B cells. The case identifier and the ROR1 ΔMFI are indicated at the top right corner of each panel. (D) Immunoblot analyses of whole cell lysates of ROR1Neg CLL or ROR1Pos CLL, as indicated at the top. Each lane represents a separate case. The membranes were probed with mAbs specific for ROR1 (top row) or β-actin (bottom row), as indicated on the left. (E-F) Each column represents a separate case of ROR1Neg or ROR1Pos CLL, as indicated at the top. The relative expression of the genes indicated on the right margin are provided in rows, using the color coding for log2 normalized effective count z scores as per the scale provided at the bottom of each heat map. (E) Heat map for 34 genes previously found expressed differentially by CLL cells vs normal B cells. (F) Heat map for 65 genes previously found expressed differentially by CLL cells vs diffuse large cell lymphoma (DLCL) cells or normal germinal center (GC) B cells. (G) Relative amount of TCL1A transcripts found in ROR1Neg or ROR1Pos CLL cases, as indicated at the bottom of the graph. Each dot represents the relative TCL1A of a separate case. The large horizontal bar indicates the mean level of TCL1A transcripts, and the 2 smaller horizontal bars show the 95% confidence interval.
Expression of CLL signature genes by ROR1Negor ROR1PosCLL cells. Representative histograms depicting the fluorescence of CD19+ cells labeled with Alexa-647–conjugated 4A5 (anti-ROR1 mAb) (shaded histograms) or Alexa-647-conjugated nonspecific immunoglobulin G (IgG) of the same isotype (open histograms) for (A) ROR1Pos CLL B cells, (B) ROR1Neg CLL cells, or (C) representative normal blood B cells. The case identifier and the ROR1 ΔMFI are indicated at the top right corner of each panel. (D) Immunoblot analyses of whole cell lysates of ROR1Neg CLL or ROR1Pos CLL, as indicated at the top. Each lane represents a separate case. The membranes were probed with mAbs specific for ROR1 (top row) or β-actin (bottom row), as indicated on the left. (E-F) Each column represents a separate case of ROR1Neg or ROR1Pos CLL, as indicated at the top. The relative expression of the genes indicated on the right margin are provided in rows, using the color coding for log2 normalized effective count z scores as per the scale provided at the bottom of each heat map. (E) Heat map for 34 genes previously found expressed differentially by CLL cells vs normal B cells. (F) Heat map for 65 genes previously found expressed differentially by CLL cells vs diffuse large cell lymphoma (DLCL) cells or normal germinal center (GC) B cells. (G) Relative amount of TCL1A transcripts found in ROR1Neg or ROR1Pos CLL cases, as indicated at the bottom of the graph. Each dot represents the relative TCL1A of a separate case. The large horizontal bar indicates the mean level of TCL1A transcripts, and the 2 smaller horizontal bars show the 95% confidence interval.
We performed massive parallel sequencing to examine the transcriptomes of these 24 leukemia samples. Each had a CLL gene-expression signature (Figure 1E-F), which was distinct from those identified in earlier studies of normal B cells or other B-cell malignancies.10-13 Between ROR1Pos and ROR1Neg cases, we found differential expression in only 1 of the 20 genes found in prior studies to be differentially expressed between CLL with U-IGHV vs CLL with mutated-IGHV (M-IGHV), namely CTLA4, P = .0006, Bonferroni’s multiple comparisons test (supplemental Figure 2A-B).12 Furthermore, we did not find significant differences between ROR1Pos and ROR1Neg cases in their relative expression of the genes that reportedly were upregulated in monoclonal B-cell lymphocytosis relative to CLL (supplemental Figure 2C).14 Finally, ROR1Neg and ROR1Pos cases expressed comparable levels of TCL1A (P = .7; Figure 1G).
We did find that 3094 of 14 761 protein-coding genes were differentially expressed in ROR1Neg vs ROR1Pos cases, 1 of which was ROR1 (DESeq2, BH-adjusted; P < .05). We performed unsupervised clustering on the log2 effective count z scores for the 2000 genes expressed in all samples that had the highest coefficients of variation. Unsupervised clustering distinguished the ROR1Neg and ROR1Pos cases, except for 1 outlier (Figure 2A).
Differentially expressed genes and subnetworks in ROR1Posvs ROR1NegCLL cells. (A) Unsupervised clustering of ROR1Pos (n = 12, red) and ROR1Neg (n = 12, blue) CLL cases using log2 effective count z scores for 2000 genes expressed in all samples (effective count >0) with the largest coefficients of variation. Each column represents a separate case. (B) z scores from Ingenuity Pathway Analysis depicting the subnetwork–gene expression differences between ROR1Pos vs ROR1Neg CLL cases. The z score heat map depicts the top 10 most upregulated and top 10 most downregulated subnetworks. Each row represents the data for the subnetwork indicated at the right margin. (C) The z score of each of the 14 subnetworks that are expressed differentially by ROR1Pos (n = 12) vs ROR1Neg (n = 12) CLL cases that also were expressed differentially by the leukemia cells of ROR1xTCL1 (n = 4) vs TCL1 transgenic mice (n = 4). The color of the bar indicates whether the subnetwork is expressed at higher or lower level by ROR1Pos CLL relative to ROR1Neg CLL, or by ROR1xTCL1 leukemia cells relative to TCL1 leukemia cells, as indicated in the legend at the bottom of the figure. (D) Immunoblot analyses of whole cell lysates of ROR1Neg and ROR1Pos CLL as indicated at the top of the panel. Each lane represents a separate case. The membranes were probed with mAb-specific phospho-AKT (p-AKT Ser473) or total AKT (t-AKT) as indicated on the left margin. The ratios of the band densities for each case of p-AKT/t-AKT are provided in the dot plot on the right for ROR1Neg CLL cases vs ROR1Pos CLL cases, as indicated at the bottom. The horizontal bar provides the median ratio observed for each group. The Mann-Whitney U test was used to calculate the P value, indicating the significance of the difference in median values between the 2 groups.
Differentially expressed genes and subnetworks in ROR1Posvs ROR1NegCLL cells. (A) Unsupervised clustering of ROR1Pos (n = 12, red) and ROR1Neg (n = 12, blue) CLL cases using log2 effective count z scores for 2000 genes expressed in all samples (effective count >0) with the largest coefficients of variation. Each column represents a separate case. (B) z scores from Ingenuity Pathway Analysis depicting the subnetwork–gene expression differences between ROR1Pos vs ROR1Neg CLL cases. The z score heat map depicts the top 10 most upregulated and top 10 most downregulated subnetworks. Each row represents the data for the subnetwork indicated at the right margin. (C) The z score of each of the 14 subnetworks that are expressed differentially by ROR1Pos (n = 12) vs ROR1Neg (n = 12) CLL cases that also were expressed differentially by the leukemia cells of ROR1xTCL1 (n = 4) vs TCL1 transgenic mice (n = 4). The color of the bar indicates whether the subnetwork is expressed at higher or lower level by ROR1Pos CLL relative to ROR1Neg CLL, or by ROR1xTCL1 leukemia cells relative to TCL1 leukemia cells, as indicated in the legend at the bottom of the figure. (D) Immunoblot analyses of whole cell lysates of ROR1Neg and ROR1Pos CLL as indicated at the top of the panel. Each lane represents a separate case. The membranes were probed with mAb-specific phospho-AKT (p-AKT Ser473) or total AKT (t-AKT) as indicated on the left margin. The ratios of the band densities for each case of p-AKT/t-AKT are provided in the dot plot on the right for ROR1Neg CLL cases vs ROR1Pos CLL cases, as indicated at the bottom. The horizontal bar provides the median ratio observed for each group. The Mann-Whitney U test was used to calculate the P value, indicating the significance of the difference in median values between the 2 groups.
We used the Ingenuity Pathway Analysis based on Ingenuity Knowledge Base to define gene expression subnetworks, which were derived from integrating the expression levels of genes encoding proteins known to interact with 1 another.7,15 We identified 55 subnetworks that were differentially expressed between ROR1Pos vs ROR1Neg cases, with a false discovery rate of <0.05 (supplemental Figure 3A). The ROR1Pos leukemia cells had higher levels of subnetworks associated with protein kinase activation and proliferation of tumor cells, but lower levels of subnetworks associated with apoptosis and RNA decay and processing, than did ROR1Neg leukemia cells (Figure 2B; supplemental Figure 3B). The gene encoding AKT1 contributed to many of these subnetworks (supplemental Figure 3B). ROR1 and AKT1 were found included in 7 subnetworks associated with cell viability, proliferation, hematologic cancer, or inhibition of cell death. Fourteen (25%) of these 55 subnetworks also were differentially expressed between the ROR1-positive leukemia cells of ROR1xTCL1 transgenic mice vs the ROR1-negative leukemia cells of TCL1 transgenic mice (Figure 2C), only 2 of which had discordant activation z scores.7
We performed gene-set enrichment analysis, focusing on selected signaling pathways in the BIOCARTA and Reactome databases. We did not observe significant differences in the expression levels of genes encoding proteins upregulated by or involved in NF-κB signaling, Src activation, B-cell receptor signaling, CREB signaling, or mTOR signaling (supplemental Table 2). On the other hand, ROR1Pos CLL cases had higher expression levels of genes encoding proteins in AKT-signaling pathway (Reactome), which were the 2 most significantly enriched gene sets (false discovery rate <25%) (supplemental Figure 4; supplemental Table 2). Consistent with the notion that ROR1Pos CLL cells have enhanced activation of AKT-signaling pathways, we found that the average normalized ratio of phosphorylated AKT to total AKT in ROR1Pos CLL cases (8 ± 9 [± SD], n = 6) was significantly higher than that noted in ROR1Neg CLL samples (1 ± 1, n = 6, P = .003, Mann-Whitney U test), as assessed via immunoblot analyses (Figure 2D).
ROR1Pos CLL cells have enhanced survival, migration, and proliferation
Prior studies found that Wnt5a could enhance CLL-cell viability, migration, and proliferation in a ROR1-dependent manner.4,16,17 We found that ROR1Neg CLL cells had a poorer viability than ROR1Pos CLL cells after 48 hours in vitro (Figure 3A), possibly due the inability of ROR1Neg cells to respond to endogenous Wnt5a, which might act as an autocrine.16 Furthermore, we found exogenous Wnt5a could enhance the viability (Figure 3B), migration (Figure 3C), and CXCL12- or CCL19-directed chemotaxis of ROR1Pos CLL cells (Figure 3D-E), but had no such effects on ROR1Neg CLL cells (Figure 3B-E). Exogenous Wnt5a also enhanced the proliferation of ROR1Pos CLL cells induced by CD154 in media supplemented with interleukin-4 (IL-4) and IL-10, an effect that could be inhibited by the anti-ROR1 mAb, UC-961 (cirmtuzumab) (Figure 3F-H). On the other hand, Wnt5a did not enhance the proliferation of ROR1Neg CLL cells treated under similar conditions (Figure 3F-H).
ROR1PosCLL cells have enhanced survival, migration, and proliferation. (A) CLL-cell viability was assessed after 0, 3, 6, 24, or 48 hours and normalized to that of the cells at 0 hour (mean ± standard error of the mean [SEM]; n = 10 for each group). The initial absolute viability of all samples at 0 hour was of 81% ± 3% (mean ± SEM). Significant difference of the percentage live CLL cells between ROR1Pos (n = 10) and ROR1Neg (n = 10) subgroup is indicated by asterisks (Student t test; **P < .01). (B) Percentage of viability after 24 hours (left) and 48 hours (right) incubation. P values were determined by Wilcoxon matched paired nonparametric test. (C-E) After 3 hours of incubation, relative percentage of basal migration (C) and chemotaxis toward CXCL12 (D) or CCL19 (E) of ROR1Neg or ROR1Pos CLL cells, with or without Wnt5a (200 ng/mL), as indicated at the bottom of the graph. P values were determined by Wilcoxon matched paired nonparametric test. (F-H) CD154-induced proliferation of carboxyfluorescein diacetate succinimidyl ester (CFSE)-labeled ROR1Neg (F) or ROR1Pos (G) CLL cells (n = 3 per group) with (right) or without (left) exogenous Wnt5a in the presence of control IgG antibody or UC-961, as indicated in the upper left of each histogram. The results of assays on 1 representative CLL sample are shown with the percent of dividing cells. (H) The bars indicate the mean proportions of ROR1Pos or ROR1Neg CLL cells with diminished CFSE fluorescence from each of 3 different patients for each culture condition indicated at the bottom of the graph. Data are shown as mean ± SEM; **P < .01, *P < .05.
ROR1PosCLL cells have enhanced survival, migration, and proliferation. (A) CLL-cell viability was assessed after 0, 3, 6, 24, or 48 hours and normalized to that of the cells at 0 hour (mean ± standard error of the mean [SEM]; n = 10 for each group). The initial absolute viability of all samples at 0 hour was of 81% ± 3% (mean ± SEM). Significant difference of the percentage live CLL cells between ROR1Pos (n = 10) and ROR1Neg (n = 10) subgroup is indicated by asterisks (Student t test; **P < .01). (B) Percentage of viability after 24 hours (left) and 48 hours (right) incubation. P values were determined by Wilcoxon matched paired nonparametric test. (C-E) After 3 hours of incubation, relative percentage of basal migration (C) and chemotaxis toward CXCL12 (D) or CCL19 (E) of ROR1Neg or ROR1Pos CLL cells, with or without Wnt5a (200 ng/mL), as indicated at the bottom of the graph. P values were determined by Wilcoxon matched paired nonparametric test. (F-H) CD154-induced proliferation of carboxyfluorescein diacetate succinimidyl ester (CFSE)-labeled ROR1Neg (F) or ROR1Pos (G) CLL cells (n = 3 per group) with (right) or without (left) exogenous Wnt5a in the presence of control IgG antibody or UC-961, as indicated in the upper left of each histogram. The results of assays on 1 representative CLL sample are shown with the percent of dividing cells. (H) The bars indicate the mean proportions of ROR1Pos or ROR1Neg CLL cells with diminished CFSE fluorescence from each of 3 different patients for each culture condition indicated at the bottom of the graph. Data are shown as mean ± SEM; **P < .01, *P < .05.
Relationship between expression of ROR1 and disease progression
To study the stability of ROR1 expression on the leukemia cells over time, we examined serial CLL samples collected at different times from 11 untreated patients (supplemental Figure 5) and 10 patients who had received therapy for CLL (supplemental Figure 6). The median time that lapsed between the first and last sample collected from patients in this longitudinal survey was 8.3 years. Although the expression levels of ROR1 varied between the different patients, we did not observe substantial variation of the relative expression of ROR1 on the CLL cells of any 1 patient over time or with therapy (supplemental Figures 5 and 6). Collectively, the mean serial-sample variation in ROR1 ΔMFI was 1.5 ± 3.9 (mean ± SD, n = 21; supplemental Figures 5 and 6).
We next examined the data on 1568 CLL patients assessed for ROR1 by the CRC Tissue Core (supplemental Table 3). The median ROR1 ΔMFI for all cases was 35.2 and, as expected, the vast majority expressed ROR1. However, there was considerable heterogeneity in the expression levels of ROR1 between cases, with ΔMFI ranging from 1 to above 400. Five percent of all patients had leukemia cells with a median ΔMFI of 1.6 ± 1.6 (± SD, n = 80), which was not significantly different from the ROR1 ΔMFI of blood B cells obtained from healthy adults (1.0 ± 1.1, n = 14; P = .18; Figure 1B-C).
We randomly segregated these cases into a training set of 797 cases and a validation set of 771 cases. In the training dataset, 40% of patients had received treatment (n = 320), with median treatment-free survival (TFS) of 7.8 years, and 8% were deceased (n = 65). On the other hand, the median TFS for patients in the validation cohort was 6.7 years; 40% (n = 306) of the patients in this subset had received therapy and 9% (n = 69) were deceased. We obtained samples at diagnosis for 10% (n = 69) of the 797 cases in the training set and 12% of 771 samples in the validation set.
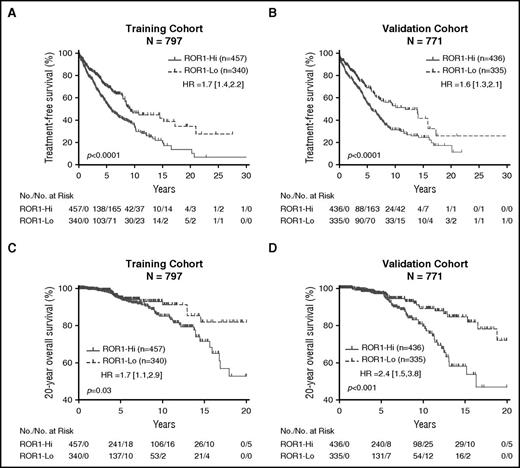
We employed the profile-likelihood method in a Cox regression model of TFS by R ‘rpart’ package from diagnosis to determine whether we could assign an optimal ROR1 ΔMFI threshold that could segregate patients in the training set into subgroups with disparate TFS. We found this cohort could be segregated into 2 disparate subgroups based upon whether the CLL cells had a ROR1 ΔMFI <32 (defined as “ROR1-Lo”), or ≥32 (defined as “ROR1-Hi”) (supplemental Figure 7). Three-hundred forty patients (43% of the total) were classified as “ROR1-Lo”; the median ROR1 ΔMFI for this subgroup was 20 ± 10 (± SD) (interquartile range 15; supplemental Figure 7). On the other hand, 457 patients were classified as being “ROR1-Hi”; the median ROR1 ΔMFI for this subgroup was 49 ± 34 (interquartile range 21; supplemental Figure 7). The patients in the ROR1-Hi subgroup had a median TFS of 5.7 years, whereas those in the ROR1-Lo subgroup had a median TFS of 9.3 years (Figure 4A). Although not factored in the analysis to define the ΔMFI threshold for ROR1-Hi vs ROR1-Lo, we noted that patients in the training cohort who were in the ROR1-Hi subgroup had shorter median overall survival (OS) than those in the ROR1-Lo subgroup (P < .05, hazard ratio [HR] 1.7; 95% confidence interval [CI] 1.1, 2.9; Figure 4C).
Relationship between expression levels of ROR1, TFS, and OS. Patients in the training cohort were segregated into ROR1-Lo vs ROR1-Hi subgroups using the ROR1 ΔMFI threshold found optimal for defining 2 groups that had the largest difference in TFS, whereas patients in the validation cohort were segregated into 2 subgroups using the ROR1 ΔMFI threshold found optimal for cases in the training cohort. (A-B) Kaplan-Meier curves depict the TFS probability over time for ROR1-Lo (dashed line) vs ROR1-Hi (continuous line) subgroups in the training cohort (A) or validation cohort (B). (C-D) Kaplan-Meier curves depict the OS probability over time for ROR1-Lo (dashed line) vs ROR1-Hi (continuous line) subgroups in the training cohort (C) or validation cohort (D). The number of patients in each category and the number of treatment events are shown in the tables under each figure. Statistical significance was determined by log-rank test (P < .05). The P values for the comparisons between subgroups are indicated below each graph.
Relationship between expression levels of ROR1, TFS, and OS. Patients in the training cohort were segregated into ROR1-Lo vs ROR1-Hi subgroups using the ROR1 ΔMFI threshold found optimal for defining 2 groups that had the largest difference in TFS, whereas patients in the validation cohort were segregated into 2 subgroups using the ROR1 ΔMFI threshold found optimal for cases in the training cohort. (A-B) Kaplan-Meier curves depict the TFS probability over time for ROR1-Lo (dashed line) vs ROR1-Hi (continuous line) subgroups in the training cohort (A) or validation cohort (B). (C-D) Kaplan-Meier curves depict the OS probability over time for ROR1-Lo (dashed line) vs ROR1-Hi (continuous line) subgroups in the training cohort (C) or validation cohort (D). The number of patients in each category and the number of treatment events are shown in the tables under each figure. Statistical significance was determined by log-rank test (P < .05). The P values for the comparisons between subgroups are indicated below each graph.
We applied this same threshold to stratify the 771 patients in the validation cohort. Three hundred thirty-five patients (43% of 771) fell into the ROR1-Lo subgroup; the median ROR1 ΔMFI for this subgroup was 18 ± 9 (± SD) (interquartile range 14). The remaining 436 cases fell into the ROR1-Hi subgroup; the median ROR1 ΔMFI for this subgroup was 46 ± 28 (interquartile range 18). Similar to our observations with the training cohort, the patients in the ROR1-Hi subgroup had a median TFS of 5.7 years, which was significantly shorter than that of the patients in the ROR1-Lo subgroup, who had a median TFS of 11.8 years (Figure 4B). The patients in the ROR1-Hi subgroup of the validation cohort had a median OS of 16.4 years, which was significantly shorter than that of patients in the ROR1-Lo subgroup (>20 years) (P < .001, HR 2.4; 95% CI 1.5, 3.8; Figure 4D).
Relationship between expression of ROR1 and other leukemia characteristics
The ROR1-Hi and ROR1-Lo cases in either the training set or the validation set expressed comparable levels of surface antigens typically found on CLL cells (eg, CD5, CD20, and CD23) (supplemental Figure 8). Although largely overlapping, the median ΔMFI for CD5 was significantly higher for ROR1-Hi cases than in ROR1-Lo cases (P < .001; supplemental Table 4). On the other hand, the ROR1-Hi cases had a lower median ΔMFI for CD20 than the ROR1-Lo cases, in both the training and the validation cohorts (supplemental Table 4).
The IGHV mutation status was known for 593 of the 797 patients in the training set (74%) and for 586 of the 771 patients in the validation set (76%) (supplemental Table 3). CLL samples with U-IGHV had higher average ROR1 ΔMFI in the training cohort (45 ± 25 [± SD]) or validation cohort (42 ± 20) than CLL samples with M-IGHV in either the training cohort (37 ± 32, P < .001) or validation cohort (36 ± 26; P < .001), respectively (supplemental Figure 9A-B). CLL samples with U-IGHV also comprised a higher proportion of the ROR1-Hi subgroup in either the training cohort (54%) or the validation cohort (59%) than CLL samples with M-IGHV (P < .0001, Pearson χ2 test; Table 1). Conversely, the CLL samples that used M-IGHV comprised a higher proportion of the ROR1-Lo subgroup in either the training cohort (67%) or the validation cohort (61%) than samples with U-IGHV. Nevertheless, half of all 608 cases that used M-IGHV were in the ROR1-Hi subgroup and approximately one-third (30%, n = 171) of the 571 cases that used U-IGHV were in the ROR1-Lo subgroup, indicating that the association between ROR1-Hi and use of U-IGHV was not absolute.
Association between high- vs low-level expression of ROR1 and IGHV mutation status
| . | Training dataset . | Validation dataset . | ||
|---|---|---|---|---|
| . | ROR1-Lo (%) (N = 236) . | ROR1-Hi (%) (N = 357) . | ROR1-Lo (%) (N = 236) . | ROR1-Hi (%) (N = 350) . |
| M-IGHV | 157 (67) | 163 (46) | 144 (61) | 144 (41) |
| U-IGHV | 79 (33) | 194 (54) | 92 (39) | 206 (59) |
| P < .0001 (Pearson χ2 test) | P < .0001 (Pearson χ2 test) | |||
| . | Training dataset . | Validation dataset . | ||
|---|---|---|---|---|
| . | ROR1-Lo (%) (N = 236) . | ROR1-Hi (%) (N = 357) . | ROR1-Lo (%) (N = 236) . | ROR1-Hi (%) (N = 350) . |
| M-IGHV | 157 (67) | 163 (46) | 144 (61) | 144 (41) |
| U-IGHV | 79 (33) | 194 (54) | 92 (39) | 206 (59) |
| P < .0001 (Pearson χ2 test) | P < .0001 (Pearson χ2 test) | |||
High-level expression of ROR1 also appeared associated with other adverse prognostic markers (supplemental Figure 9C-D; supplemental Table 5). We noted a significant association between high-level ROR1 and expression of ZAP-70 in the training cohort (P < .05) and the validation cohort (P < .01; supplemental Table 5); however, this association was not as strong as that noted with the use of U-IGHV genes (P < .0001 in both cohorts; Table 1). On the other hand, we did not discern a significant difference in the expression level of CD38 on ROR1-Hi CLL vs ROR1-Lo CLL in either the training or the validation cohort (supplemental Table 4).
Fluorescence in situ hybridization data were available for 455 of the 797 patients in the training set (57%) and for 431 of the 771 patients in the validation set (56%) (supplemental Table 6). Training and validation cohorts showed similar distribution of cytogenetic abnormalities (supplemental Table 7).
Multivariate analyses of ROR1 with other prognostic parameters
Using a multivariate Cox regression model with either a nondelayed entry or a delayed entry to account for a lag between diagnosis and time of collecting the samples in the validation cohort, we observed that high-level expression of ROR1 had predictive value for short TFS (nondelayed entry HR 1.4, P < .01; delayed entry HR 1.7, P < .01), even when we factored in the model IGHV mutation status (nondelayed entry HR 2.7, P < .001; delayed entry HR 2.7, P < .001; supplemental Table 8) or adverse cytogenetic features, such as del(17p) or del(11q) (supplemental Table 9). In particular, patients in the ROR1-Hi subgroup of the validation cohort who had CLL cells with M-IGHV (n = 144) had a significantly shorter median TFS (10.7 years) than patients in the ROR1-Lo subset who had CLL cells that used M-IGHV (14.1 years, n = 144) (supplemental Figure 10B). On the other hand, the apparent difference in the TFS for ROR1-Hi vs ROR1-Lo patients who had CLL cells that used U-IGHV genes did not reach statistical significance. Similar results were noted for patients in the training cohort (supplemental Figure 10A).
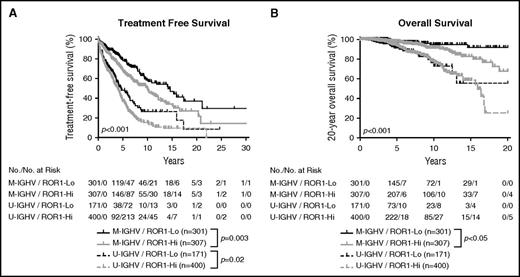
We assimilated the data from both training and validation cohorts, allowing us to examine the relationship between expression level of ROR1, IGHV mutation status, and outcome for the entire group of patients for whom we have IGHV mutation status. As expected, among patients who had CLL cells with M-IGHV in the combined cohort, those in the ROR1-Hi subgroup had a significantly shorter median TFS (10 years, n = 307) and OS than patients who were in the ROR1-Lo subset (median TFS = 14.1 years, n = 301; P < .01; Figure 5). Among patients who had CLL cells with U-IGHV, those within the ROR1-Hi subgroup had a shorter median TFS (4.0 years, n = 400) than those in the ROR1-Lo subset (4.6 years, n = 171; Figure 5A) that now reached statistical significance (P < .02); however, we did not observe a significant difference between the median OS of those in ROR1-Hi subgroup (15.9 years, n = 400) vs the median OS of those in the ROR1-Lo subgroup (undefined, n = 171; Figure 5B), despite the larger numbers of patients included in these analyses.
Relationship between expression levels of ROR1 and TFS, OS on CLL cells segregated on the basis of IGHV mutation status. Kaplan-Meier curves depict the TFS (A) or OS (B) probability over time for ROR1-Lo with M-IGHV (black solid line), ROR1-Lo with U-IGHV (black dashed line), ROR1-Hi with M-IGHV (gray solid line), or ROR1-Hi with U-IGHV (gray dashed line) subgroups of all CLL patients for whom the IGHV mutation status is known.
Relationship between expression levels of ROR1 and TFS, OS on CLL cells segregated on the basis of IGHV mutation status. Kaplan-Meier curves depict the TFS (A) or OS (B) probability over time for ROR1-Lo with M-IGHV (black solid line), ROR1-Lo with U-IGHV (black dashed line), ROR1-Hi with M-IGHV (gray solid line), or ROR1-Hi with U-IGHV (gray dashed line) subgroups of all CLL patients for whom the IGHV mutation status is known.
The IGHV mutational status and fluorescence in situ hybridization data were available for a total of 680 patients in the combined cohorts (supplemental Table 6). Although there was a higher frequency of cases with adverse cytogenetic features, (eg, del(17p) or del(11q)) among patients in the ROR1-Hi subgroup (supplemental Table 7), this apparently reflected the higher prevalence of such cytogenetic abnormalities among cases with U-IGHV (supplemental Table 10), as noted in prior studies.18 For example, among patients with U-IGHV, the proportion of cases with del(17p) (20%, or 49 of 250 cases) or del(11q) (20%, or 49 of 250 cases) in the ROR1-Hi subset was comparable to that for del(17p) (16%, or 13 of the 80 cases) or del(11q) (13%, or 10 of the 80 cases) in the ROR1-Lo subset. On the other hand, the proportions of patients with M-IGHV who had such cytogenetic abnormalities were lower regardless of the level of ROR1. Moreover, among patients with M-IGHV, the proportion of cases with del(17p) (9%, or 18 of 192 cases) or del(11q) (3%, or 5 of 192 cases) in the ROR1-Hi subset was comparable to that for del(17p) (6%, or 9 of the 158 cases) or del(11q) (1%, or 1 of the 158 cases) in the ROR1-Lo subset.
Discussion
In this study, we found that the levels of ROR1 on CLL cells varied between patients. Whereas the vast majority of cases expressed detectable ROR1, ∼5% of cases had negligible levels of ROR1 that were comparable to that of B cells of healthy adults. In addition to satisfying diagnostic criteria for CLL, such cases also had CLL gene-expression signatures, indicating that they indeed represented CLL and not some other B-cell malignancy masquerading as CLL. Among those cases that did express detectable ROR1, we also noted considerable heterogeneity in the levels of this orphan receptor on the CLL cells of different patients. Evaluation of serial samples revealed that the expression levels of ROR1 did not vary substantially over time, as noted in a prior study.19 However, this earlier study also noted an increase in the frequency of ROR1+ cells upon development of more progressive disease. Although we did not discern an increase in the expression levels of ROR1 on the CLL cells of any 1 patient over time, we cannot exclude the possibility the leukemia cells of some patients might change their relative expression of ROR1 in certain clinical settings.
Transcriptome analyses revealed that cases with negligible ROR1 had gene expression signatures that were distinct from those of ROR1Pos cases. Except for 1 outlier, unsupervised gene-expression-clustering analysis could distinguish ROR1Pos cases from ROR1Neg samples. Subnetwork analysis showed that RORNeg CLL cases had relatively low-level expression of protein kinase activation and proliferation subnetworks, particularly those involving activation of AKT. Consistent with the notion that ROR1Pos CLL cells had enhanced activation of AKT signaling pathways, we found that ratios of activated AKT (phosphorylated AKT) to total AKT in ROR1Pos CLL were higher than those of ROR1Neg CLL samples. Such differences were comparable to those noted between the ROR1Pos leukemia of ROR1xTCL1 transgenic mice relative to the ROR1Neg leukemia that develops in otherwise syngeneic TCL1 transgenic mice.7 Moreover, the ROR1Pos leukemia of ROR1xTCL1 transgenic mice had higher levels of activated AKT than the ROR1Neg leukemia of TCL1 transgenic mice; this was associated with higher rates of leukemia-cell proliferation and lower levels of spontaneous apoptosis, resulting in a more aggressive disease and shorter survival of ROR1xTCL1 transgenic mice compared with TCL1 transgenic mice.
Wnt5a influences a variety of cellular functions, such as proliferation, differentiation, migration, adhesion, and planar-cell polarity. As noted in this and prior studies,4,16,17 Wnt5a could enhance leukemia-cell migration, proliferation, and survival of ROR1Pos CLL cells, but had no such activity on ROR1Neg CLL cells. Moreover, we used an in vitro system of coculture of CLL cells with HelaCD154 in media containing IL-4/IL-10 and confirmed that Wnt5A could enhance the proliferation CLL cells that express ROR1; this effect could be blocked by treatment of the CLL cells with an anti-ROR1 mAb (UC-961, cirmtuzumab).4 On the other hand, Wnt5a did not enhance the proliferation of ROR1Neg samples, consistent with this effect being dependent upon ROR1. The ROR1-dependent effects that Wnt5a has on CLL cells in vitro infer that expression of ROR1 serves to stimulate leukemia-cell survival, migration, and proliferation in vivo and thus might influence disease progression.
That a high level of ROR1 expression may drive more aggressive disease was suggested by the results of a prior study of 20 CLL patients that found cases with progressive disease had higher frequencies of ROR1+ CLL cells than a matched number of CLL patients with nonprogressive disease.20 Patients were considered to have progressive disease if they had worsening disease-related anemia (hemoglobin <10 g/dL), thrombocytopenia (<100 × 109/L), lymphadenopathy, or doubling of the blood lymphocyte count in the preceding 3-month period; patients were considered nonprogressive if they lacked these criteria. However, 8 of the 10 CLL patients in the progressive disease group had previously received treatment, whereas none of the patients in the nonprogressive group had received therapy. As such, the authors of this study acknowledged that the differences in the expression of ROR1 between the 2 groups could have been the result of prior therapy or disease duration. In the current study, we found that the relative expression levels of ROR1 were not influenced by prior therapy (supplemental Figure 6), suggesting that high levels of ROR1 per se influence disease progression.
We thus examined whether high-level expression of ROR1 was associated with accelerated disease progression among patients with CLL. Randomly assigning cases to a training set or a validation set allowed us to define a threshold for the ROR1 ΔMFI in the training set that could then be applied to cases in the validation cohort to determine whether it could be used to “predict” differences in the clinical outcome. Consistent with the random assignment of cases to either subset, we noted that distribution of the expression levels of ROR1 were comparable between the training and validation cohorts (supplemental Figure S7). Because there was no clear threshold with which to segregate cases into subgroups with high vs low levels of ROR1, we used the profile-likelihood method in a Cox regression model based on TFS to define a ROR1 ΔMFI threshold that best could segregate patients in the training set into ROR1-Hi vs ROR1-Lo subsets that had different median TFS. This analysis defined an optimal ROR1 ΔMFI threshold of 32, which was considerably higher than the ROR1 ΔMFI of CLL cases considered ROR1Neg. As such, the ROR1-Lo subgroup, which included 43% of the patients in the training cohort, largely comprised samples that expressed some detectable, albeit low, level of ROR1. Nevertheless, when this threshold was applied to segregate samples in the validation cohort, we found that patients in the ROR1-Hi subgroup had a significantly shorter median TFS and OS than patients in the ROR1-Lo subgroup, even though OS was not used to define the ROR1 ΔMFI threshold used to segregate cases into ROR1-Hi or ROR1-Lo subgroups. This analysis indicates that quantitative differences in ROR1 may influence clinical outcome and that, on average, cases with low-level expression of ROR1 have a more indolent clinical course than patients with high-level expression of this orphan receptor.
Although high-level expression of ROR1 more often was associated with use of U-IGHV, this association was not absolute. Indeed, more than half of the cases with M-IGHV were classified as ROR1-Hi. Furthermore, high-level expression of ROR1 was associated with shorter TFS even when the IGHV mutation status was factored into a multivariate model. The association of high-level expression of ROR1 with adverse outcome was particularly apparent for patients with CLL cells that use M-IGHV. Moreover, high-level expression of ROR1 was associated with a significantly shorter TFS and OS in patients with M-IGHV. However, for patients with CLL that used U-IGHV, the differences in TFS did not reach statistical significance, unless we performed the analysis on the entire group of patients for whom we had IGHV sequence data. Furthermore, we find that patients with CLL cells that use U-IGHV and have low-level expression of ROR1 apparently have a shorter TFS and OS than patients with CLL cells that use M-IGHV and have high-level ROR1, indicating that high-level expression of ROR1 cannot account for the relatively aggressive clinical course already noted for patients with CLL cells that use U-IGHV.21,22 In any case, we conclude that high-level expression of ROR1 is associated with more aggressive disease, indicating that ROR1 may act as a driver of leukemia progression, as noted for ROR1xTCL1 transgenic mice.
The relationship between high-level expression of ROR1 and adverse outcome also has been noted for malignancies other than CLL. A variety of different cancers have been found to express ROR1.23 To date, high-level expression of ROR1 has been found to have adverse prognostic significance for patients with breast cancer,24-27 ovarian cancer,28,29 melanoma,30 or Ewing sarcoma.31 Moreover, high-level expression of ROR1 also has been associated with higher levels of AKT activation,24,30,32,33 consistent with the notion that ROR1 signaling leading to activation of AKT may serve as a driver, not just in CLL, but also in other malignancies. Because of this and its restricted postpartum expression, ROR1 may be an attractive target for therapy with either mAbs,7,34 or other agents that specifically inhibit this orphan receptor and/or its downstream signaling pathways.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Erin N. Smith for insightful comments and Kristen Jepsen for sample preparation. The authors acknowledge Ling Zhang for technical assistance and Andrew W. Greaves for database analysis. Thanks to the CRC for its participation in this study.
This work was supported in part by the National Institutes of Health, National Cancer Institute (grant R37-CA049870) (T.J.K.), CRC (grant PO1-CA81534) (L.Z.R. and T.J.K.), and the Blood Cancer Research Fund. C.D. is supported in part by the University of California, San Diego (UCSD), Genetics Training Program through an institutional training grant from the National Institute of General Medical Sciences (T32GM008666) and the California Institute for Regenerative Medicine Interdisciplinary Stem Cell Training Program at UCSD II (TG2-01154).
Authorship
Contribution: B.C., E.M.G., and L.C. designed and performed research, analyzed data, and wrote the paper; L.Z.R., G.F.W., and J.Y. performed research and contributed to scientific discussion, data interpretation, and paper revision; C.D., K.A.F., and D.S.N. analyzed the data, performed statistical analysis, and revised the paper; W.G.W., K.R.R., N.E.K., J.R.B., J.A.J., and J.G.G. contributed patient samples; and T.J.K. supervised the study, designed research, analyzed data, provided patient samples, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
A complete list of the members of the Chronic Lymphocytic Leukemia Research Consortium appears in the online appendix.
Correspondence: Thomas J. Kipps, 3855 Health Sciences Dr, Room 4307, San Diego, CA 92093-0820; e-mail: tkipps@ucsd.edu.
References
Author notes
B.C., E.M.G., and L.C. contributed equally to this study.


![Figure 3. ROR1Pos CLL cells have enhanced survival, migration, and proliferation. (A) CLL-cell viability was assessed after 0, 3, 6, 24, or 48 hours and normalized to that of the cells at 0 hour (mean ± standard error of the mean [SEM]; n = 10 for each group). The initial absolute viability of all samples at 0 hour was of 81% ± 3% (mean ± SEM). Significant difference of the percentage live CLL cells between ROR1Pos (n = 10) and ROR1Neg (n = 10) subgroup is indicated by asterisks (Student t test; **P < .01). (B) Percentage of viability after 24 hours (left) and 48 hours (right) incubation. P values were determined by Wilcoxon matched paired nonparametric test. (C-E) After 3 hours of incubation, relative percentage of basal migration (C) and chemotaxis toward CXCL12 (D) or CCL19 (E) of ROR1Neg or ROR1Pos CLL cells, with or without Wnt5a (200 ng/mL), as indicated at the bottom of the graph. P values were determined by Wilcoxon matched paired nonparametric test. (F-H) CD154-induced proliferation of carboxyfluorescein diacetate succinimidyl ester (CFSE)-labeled ROR1Neg (F) or ROR1Pos (G) CLL cells (n = 3 per group) with (right) or without (left) exogenous Wnt5a in the presence of control IgG antibody or UC-961, as indicated in the upper left of each histogram. The results of assays on 1 representative CLL sample are shown with the percent of dividing cells. (H) The bars indicate the mean proportions of ROR1Pos or ROR1Neg CLL cells with diminished CFSE fluorescence from each of 3 different patients for each culture condition indicated at the bottom of the graph. Data are shown as mean ± SEM; **P < .01, *P < .05.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/128/25/10.1182_blood-2016-04-712562/4/m_blood712562f3.jpeg?Expires=1765906094&Signature=NsB51HfAq905nCKfkWTSiUZUiXc1QRwXB4cTSPWt6k9tenBMGfpldFmTsMNcn13SqQpsujShXmIxKe4N6iTxlHTzhcxukry8sqNis0ERvt4yFB2DQdII-VyJpe4YYZj2Fdq35kyLc0LxaUl47sXowbwRx2f5oQIVijX~x9tl07q1QMLyLt2AEoehQd8SgGWPgDCddgAX6LuC3EqcGFrxDfR6T4Ij-ZqMmTcXbIWcdUwyI5WAB8veXv9I0T2Lg2MDHWwLNwy0i-Apu2SNMKAUCA29tLn4UftUMkpQaMrr5TU0WGB-u~AqfhoKsi9FjoMoLqKd5VyqawNrelwPh9YdBQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal